Abstract
The decoding A site of the small ribosomal subunit is an RNA molecular switch, which monitors codon–anticodon interactions to guarantee translation fidelity. We have solved the crystal structure of an RNA fragment containing two Homo sapiens cytoplasmic A sites. Each of the two A sites presents a different conformational state. In one state, adenines A1492 and A1493 are fully bulged-out with C1409 forming a wobble-like pair to A1491. In the second state, adenines A1492 and A1493 form non-Watson–Crick pairs with C1409 and G1408, respectively while A1491 bulges out. The first state of the eukaryotic A site is, thus, basically the same as in the bacterial A site with bulging A1492 and A1493. It is the state used for recognition of the codon/anticodon complex. On the contrary, the second state of the H.sapiens cytoplasmic A site is drastically different from any of those observed for the bacterial A site without bulging A1492 and A1493.
INTRODUCTION
Decoding is the initial process of protein synthesis. It occurs at the aa-tRNA decoding site (A site) on the small ribosomal subunit (30S in bacteria, 40S in eukaryotic cytoplasm and 28S in eukaryotic mitochondria) to recognize the cognate interactions between the aa-tRNA anticodon and the messenger codon (1). Crystal structures of various bacterial 30S ribosomal particles as well as RNA fragments containing the minimal bacterial A site in complex with aminoglycoside antibiotics have revealed the molecular mechanisms of the decoding step at atomic level (2–15). When aa-tRNA is delivered to the A site as a ternary complex with elongation factor EF-Tu and GTP, the A site changes its conformation from an ‘off’ state to an ‘on’ state. In the ‘off’ state, two adenine residues, A1492 and A1493, fold within the internal loop into the shallow/minor groove of the A site (2,7). Recently, other conformations of the ‘off’ state with a single adenine bulging out and the other one paired within the A site were reported (12,14). On the other hand, in the ‘on’ state, the two adenines A1492 and A1493 are fully bulged-out from the A site (3–6,8–15). The bulged-out A1492 and A1493 from helix 44 come together with G530 in the shoulder domain to recognize the first two Watson–Crick base pair of codon–anticodon mini-helix and induce ribosomal transitions from the open to the closed forms (3–6). This conformational change of the A site plays a key role in guaranteeing fidelity of the tRNA selection step during protein synthesis.
Because, in the A site, 11 of the 15 nt of the internal loop are universally conserved except for position 1408, 1410, 1490 and 1491 (Figure 1a) (16), one expects that the decoding mechanism mentioned above is basically conserved in prokaryotes and eukaryotes. In the Homo sapiens cytoplasmic A site, the 1408 residue is a G, while it is an A in all bacterial and mitochondrial A site. In addition, the H.sapiens cytoplasmic A site has a mispair between residues 1409 and 1491 (C1409oA1491) instead of a Watson–Crick C1409 = G1491 base pair. It has been reported that the A1408G mutation in the bacterial 16S ribosomal RNA is sufficient to produce high-level resistance against aminoglycosides (17–21) and that the G1491A mutation is lethal to the growth of Escherichia coli (22). Therefore, the cytoplasmic 18S ribosomal A site may have unique contributions to the decoding mechanism. For the eukaryotic cytoplasmic A site, only two NMR structures of RNA fragment containing the lower eukaryote Tetrahymena thermophila cytoplasmic A site have been reported so far (23,24). In those structures, the secondary structure is identical to that of the A1408G mutant of the bacterial A site. To help decipher the decoding mechanism of higher eukaryote at atomic revel, we have solved a crystal structure of RNA fragment containing two minimal H.sapiens cytoplasmic A site at 2.3 Å resolution.
Figure 1.
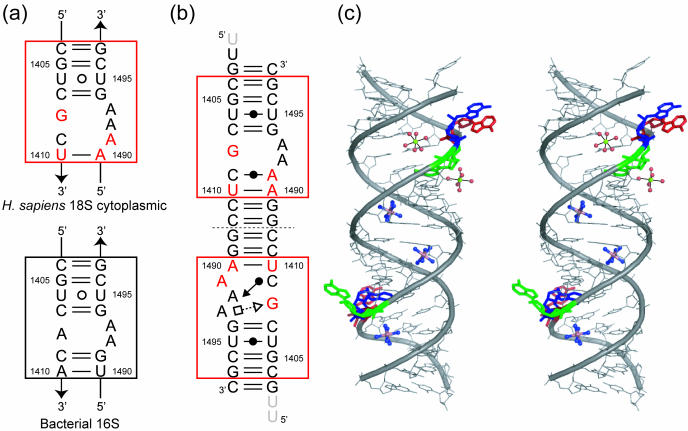
(a) Secondary structures of the H.sapiens 18S cytoplasmic and the bacterial 16S ribosomal decoding A site. The nucleotides different from those in the bacterial A site are colored in red. (b) Secondary structure of the RNA duplex found in the H.sapiens cytoplasmic A site crystal. The unobserved 5′-overhanging uracil residues are shaded. Geometric nomenclature and classification of nucleic acid base pairs are according to the Classification in (47). (c) Stereoview of the RNA duplex. Three adenine residues in the A site, A1491, A1492 and A1493, are colored in green, blue and red, respectively.
In the present structure, one A site forms the ‘on’ state with two fully bulged-out adenines (A1492 and A1493) and a Watson–Crick C1409oA1491 base pair. The second A site forms the ‘off’ state with one fully bulged-out adenine (A1491), one halfway bulged-out adenine (A1493) and a Watson–Crick/Sugar-edge C1409oA1492 base pair. Interestingly, the conformation of the ‘off’ state is almost identical to that observed in the apramycin complex structure (J. Kondo, B. François, A. Urzhumtsev and E. Westhof manuscript submitted). Here, the conformations of the H.sapiens cytoplasmic A site are compared with those of the bacterial 16S and the T.thermophila 18S cytoplasmic A site, and the molecular mechanisms of the decoding process in the H.sapiens 18S rRNA on the 40S subunit are discussed.
MATERIALS AND METHODS
Preparation
Two internal loops of the H.sapiens cytoplasmic A site (Figure 1a) were inserted between Watson–Crick pairs in sequences designed to fold as double helix (Figure 1b). RNA oligomers were synthesized by Dharmacon (Boulder, CO) and purified by high-performance liquid chromatography (HPLC) and reverse phase chromatography. Before crystallization, 1 mM RNA solution containing 50 mM sodium cacodylate (pH 6.5), 12.5 mM sodium chloride and 2.5 mM magnesium sulfate was annealed by heating at 85°C for 2 min followed by slow cooling to room temperature (21–25°C). Crystals of 23mer RNA suitable for X-ray experiments were grown at 20°C by the hanging-drop vapor diffusion method from solutions containing 50 mM sodium cacodylate buffer solution (pH 6.5), 5 mM hexammine cobalt chloride, 200 mM potassium chloride and 1% (v/v) 2-methyl-2,4-pentanediol condition. Fresh crystals were mounted in nylon cryoloops (Hampton Research) with the crystallization solution containing 40% (v/v) MPD as a cryoprotectant and stored in liquid ethane before X-ray experiments.
Data collection
X-ray data of the H.sapiens cytoplasmic A site crystal was collected at 100 K with synchrotron radiation (0.9795 Å) at the BM16 beamline in the European Synchrotron Radiation Facility (ESRF; Grenoble, France). X-Ray diffraction data were recorded on a CCD detector (MarCCD 165) positioned at a distance of 190 mm from the crystal, using 1° oscillations and 60 s exposure per frame, over a total range of 180°. Images were processed to resolution of 2.8 Å with the program MOSFLM (25,26), and the intensity data were scaled and merged using SCALA and TRUNCATE from the CCP4 suite of crystallographic programs (27). The crystal belongs to the space group P21212 with unit cell dimensions of a = 46.1 Å, b = 47.2 Å and c = 56.7 Å. This dataset was used for Molecular Replacement method. The statistics of data collection and the crystal data are summarized in Supplementary Table 1.
For higher resolution data, a second dataset was collected at 100 K with synchrotron radiation (0.9340 Å) at the ID14-1 beamline in the ESRF using a different crystal, with a CCD detector (ADSC Quantum 4R) set 255 mm from the crystal. Oscillations and exposure times were 1° and 10 s. This dataset was processed at a resolution of 2.3 Å in P21212 with unit cell dimensions of a = 46.1 Å, b = 47.4 Å, c = 56.8 Å by using the program Crystalclear (Rigaku/MSC). This dataset was used for structure refinement. The statistics of data collection and the crystal data are summarized in Supplementary Table 1.
Structure determination and refinement
The crystal structure of the RNA duplex containing the bacterial A site complexed with paromomycin (8) was used as a probe for phasing by the Molecular Replacement method. Rotation and translation search with the program AMoRe (28) using the bulk-solvent technique (29,30) gave some candidates with similar R factors and correlation coefficients (CC) but shifts of several base pair distance along the c-axis. We picked up one of these candidates and performed Normal-mode refinement (31) in the resolution range from 10 to 5.0 Å for only the central stem region with eight Watson–Crick base pairs. The model was improved after Normal-mode refinement; initial R and CC values were 40.8 and 20.9% and final Rfree and CCfree values were 30.5 and 32.3%, respectively. Fourier maps with the coefficients 2|Fo| − |Fc| and |Fo| − |Fc| at the resolution from 10 to 3.8 Å were used to trace phosphate-ribose backbone and those calculated with all reflections were used to assign base moieties. After several cycles of model building, two strong densities of hexammine cobalt cations appeared. Since it is known that this cation tends to bind to major groove atoms of guanine–guanine stacks (32,33), the whole RNA model was shifted along the c-axis to make interactions between one hexammine cobalt and the G13–G14 stack as well as between the second ion and the G36–G37 stack. Afterwards, all residues except three of four overhanging uracil (U1, U45 and U46) could be successfully assigned in the electron density map. The atomic parameters of the structure were refined with the program CNS (34) through a combination of rigid-body refinement, simulated-annealing, crystallographic conjugate gradient minimization refinements and B-factor refinements, followed by interpretations of the omit map at every nucleotide residue. At the initial stages of refinement, restraints were applied on all base–base interactions, but in the latter stages, these restraints were removed. In the final refinement step, 3 hexammine cobalt cations, 2 hexa-hydrated magnesium ions and 51 water molecules were included. The final R-factor was 24.7% for 10–2.3 Å resolution data [Rfree = 28.2% for 10% of the observed data (35)]. The root-mean-square deviation (RMSD) between the probe for Molecular Replacement and the present crystal structure optimally superimposed is 2.8 Å for the corresponding atoms of phosphate-ribose backbone. The details of structure determination will be discussed elsewhere. The statistics of structure refinement are summarized in Supplementary Table 2. All figures were drawn using The PyMOL Molecular Graphics system (2002) DeLano Scientific, San Carlos, CA (http://www.pymol.org). The PDB ID entry code for the present structure is 2FQN.
RESULTS
Overall structure
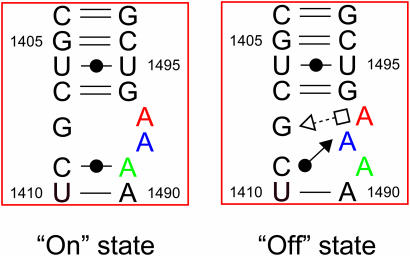
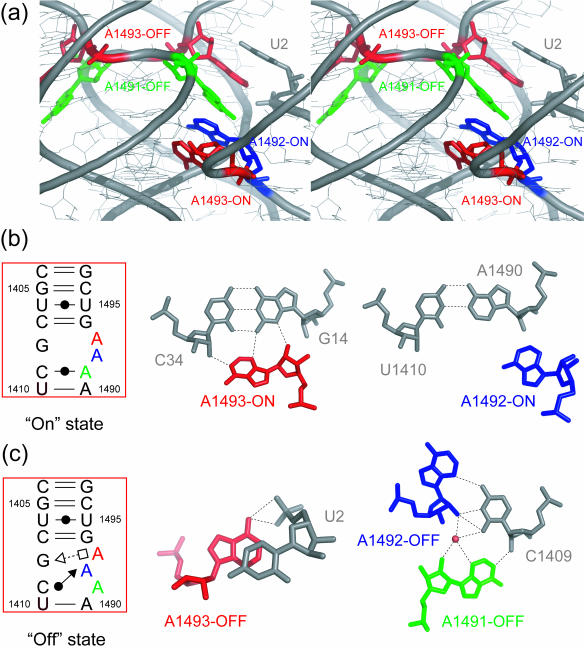
The secondary and overall crystal structures of the RNA fragment containing two H.sapiens cytoplasmic A sites are shown in Figure 1b and c. The two RNA strands form an asymmetrical duplex with each of the two A sites adopting different conformations, one with fully bulged-out A1492 and A1493 and the other one with A1491 bulged-out and A1493 only partly bulged-out. By analogy with the results on the bacterial A site, these two conformations may correspond to the ‘on’ and ‘off’ states of the H.sapiens cytoplasmic A site, respectively. For convenience, we will name them the ‘on’ and ‘off’ states hereafter (Figure 2). Two hexa-hydrated magnesium ions bind to the A site in the ‘on’ state, one in the deep/major groove and the other one in the shallow/minor groove. On the other hand, an hexammine cobalt cation interacts with the A site in the ‘off’ state in the deep/major groove. These two A sites are inserted between regular Watson–Crick stems. At the center of the duplex, four contiguous Watson–Crick G = C base pairs are formed and each G/G stack interacts with an hexammine cobalt cation. Three Watson–Crick G = C base pairs close the stem at both ends. One of the four overhanging uracil residues is involved in crystal packing interaction and the others are disordered in the solvent channel. Hereafter, we will detail the structures of two conformations of the A site using the numbering of H.sapiens cytoplasmic 18S rRNA.
Figure 2.
Secondary structures of the H.sapiens cytoplasmic A site in the ‘on’ and ‘off’ states.
Structure of the cytoplasmic A site in the ‘on’ state
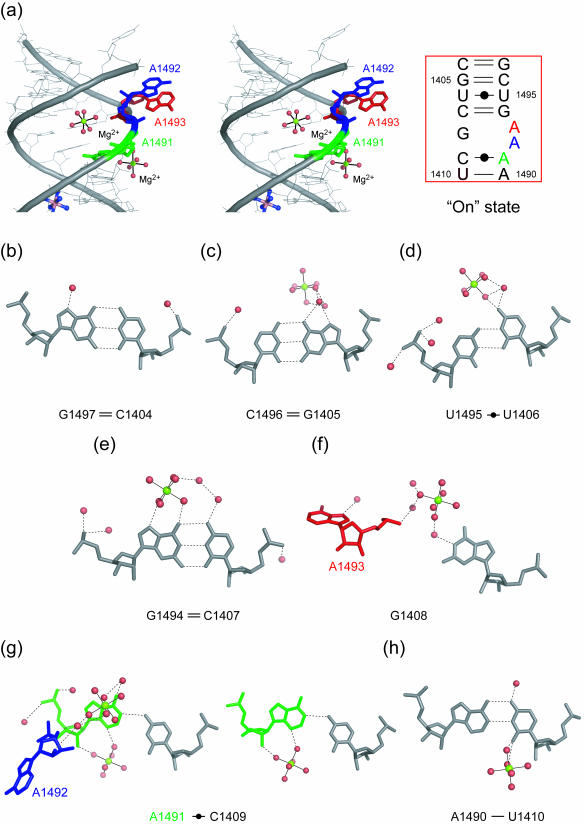
A stereoview with the secondary structure of the H.sapiens cytoplasmic A site in the ‘on’ state is shown in Figure 3a. At both ends of the internal loop of the cytoplasmic A site, four canonical Watson–Crick pairs, C1404 = G1497, G1405 = C1496, C1407 = G1494 and U1410-A1490, are formed (Figure 3b, c, e and h). The U1406 residue forms a non-bifurcated base pair with the U1495 residue by using the Watson–Crick edge (Figure 3d). The G1408 residue, which is universally conserved in all eukaryotic cytoplasmic A site, does not form any base pair. The C1409 residue pairs to the A1491 residue and forms a single H-bond, N4-H···N1 (3.06 Å). The geometry of this Watson–Crick C1409oA1491 base pair corresponds to an open form of the wobble CoA pair without the N1(A)-H···O2(C) H-bond which requires the protonation of the adenine at the N1 position (36). Two adenine residues in the internal loop of the cytoplasmic A site, A1492 and A1493, are fully bulged-out and are involved in crystal packing interaction as described later.
Figure 3.
The H.sapiens cytoplasmic A site in the ‘on’ state. (a) Stereoview and secondary structure of the ‘on’ state. Two adenine residues, A1492 (blue) and A1493 (red), are bulged-out. (b–h) Atomic details of each base pair of the A site. Three adenine residues, A1491, A1492 and A1493, are colored in green, blue and red, respectively. The hydrogen bonds are represented by black dashed lines.
Two hexa-hydrated magnesium ions bind to the ‘on’ state of the cytoplasmic A site. One of them interacts via the magnesium bound water molecules with the Hoogsteen edge of G1494 of the C1407 = G1494 base pair (Figure 3e) and O4 of U1406 of the Watson–Crick U1406oU1495 base pair (Figure 3d). In addition, the hydrated ion bridges the Watson–Crick edge of the G1408 residue and the phosphate backbone of A1492 and A1493 through second shell water-mediated hydrogen bonds (Figure 3f). The other hexa-hydrated magnesium ion directly binds to the shallow/minor groove of A1491 and U1410 of the Watson–Crick C1409oA1491 and U1410-A1490 base pairs (Figure 3g and h).
Structure of the cytoplasmic A site in the ‘off’ state
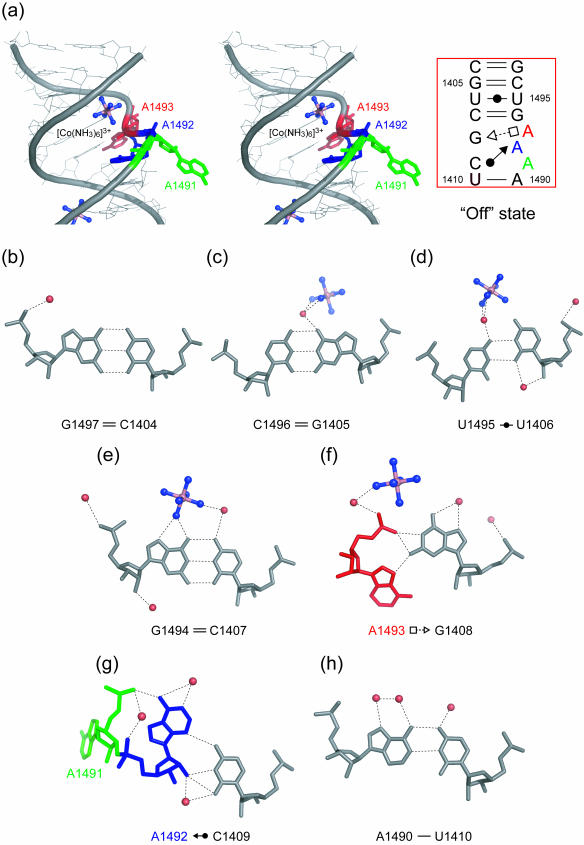
A stereoview with the secondary structure of the H.sapiens cytoplasmic A site in the ‘off’ state is shown in Figure 4a. Base pair geometries at both ends of the internal loop are the same as those in the ‘on’ state of the cytoplasmic A site (Figure 4b–e and h). On the other hand, the conformation of the internal loop of the A site is completely different from that observed for the ‘on’ state. The G1408 residue forms a trans Sugar-edge/Hoogsteen base pair with A1493 with only one H-bond, N2-H···N7 (2.67 Å) (Figure 4f). This base pair is closely related to the trans Sugar-edge/Hoogsteen (sheared) GoA base pair, well known as a motif in the hammerhead ribozyme (37–39). The G1408 residue also makes interactions with the phosphate anionic oxygen atoms of the A1493 residue through N1-H···O2P (2.89 Å) and N2-H···O2P (2.83 Å) H-bonds. In consequence, A1493 is halfway bulging out to the solvent. The A1492 residue, which adopts a bulged-out conformation in the ‘on’ state, is maintained inside the helix by forming a cis Sugar-edge/Watson–Crick base pair with C1409 through three direct H-bonds, O2′-H···O2 (3.32 Å), O2′-H···N3 (2.86 Å) and N3···H-N4 (3.24 Å) (Figure 4g). Residue A1492 also interacts with the phosphate backbone of residue A1491 by using the Hoogsteen edge with one H-bond N6-H···O1P (2.63 Å). Residue A1491 does not have any partner to form a base pair in the internal loop of the A site, and is fully bulging out to the solvent.
Figure 4.
The H.sapiens cytoplasmic A site in the ‘off’ state. (a) Stereoview and secondary structure of the ‘off’ state. Two adenine residues, A1491 (green) and A1493 (red), are bulged-out. (b–h) Atomic details of each base pair of the A site. Three adenine residues, A1491, A1492 and A1493, are colored in green, blue and red, respectively. The hydrogen bonds are represented by black dashed lines.
An hexammine cobalt cation binds to the deep/major groove of the ‘off’ state of the cytoplasmic A site. A cation bound ammine directly interacts with O6 and N7 atoms of G1494 of the Watson–Crick C1407 = G1494 base pair (Figure 4e). In addition, 6 water-mediated H-bonds are observed between the hexammine cobalt cation and RNA atoms.
Crystal packing interactions
As shown in Figure 5a, the crystal packing interface observed in the H.sapiens cytoplasmic A site oligonucleotides is dominated by interactions between duplexes with parallel helical axis. In contrast, in crystals of bacterial A site oligonucleotides, the packing contacts occur between duplexes with helical axis perpendicular to each other (8–12). These two main relative orientations of the duplexes dictate the positions of the bulged-out A1492 and A1493 and the H-bonding contacts made with the neighboring duplexes (Supplementary Figure 1). In the present crystal structure, the two bulged-out adenines from the ‘on’ state of the A site (hereafter A1492-ON and A1493-ON, respectively), those from the ‘off’ state (hereafter A1491-OFF and A1493-OFF, respectively) and the overhanging uracil make an A-A-A-A-U stacked column in the crystal packing interface. In addition, two of these four adenines make contacts with the shallow/minor groove of a neighboring duplex as in A-minor motifs (40). The A1493-ON residue interacts with guanine in an antiparallel fashion [N3···H-N2 (3.22 Å) and O2′-H···N3 (3.00 Å)] and with cytosine in a parallel fashion [N1···H-O2′(2.87 Å)], which correspond to the trans Sugar-edge/Sugar-edge AoG and the trans Watson–Crick/Sugar-edge AoC base pairs, respectively (Figure 5b). The A1492-ON is present at the shallow/minor groove of the A1490 residue of the Watson–Crick A1490-U1410 base pair, but does not make any contacts with A1490 (Figure 5b). The A1491-OFF residue interacts with C1409 in a parallel fashion [N1···H-O2′ (3.22 Å)], which is called the trans Watson–Crick/Sugar-edge AoC base pair (Figure 5c). The A1493-OFF residue makes hydrogen bonds with the O1P and O2P atoms of the overhanging U2 (Figure 5c).
Figure 5.
Crystal packing interactions observed in the H.sapiens cytoplasmic A site crystal. (a) Stereoview of crystal packing interface. Two bulged-out adenines from the ‘on’ state of the A site [A1492-ON (blue) and A1493-ON (red)], those from the ‘off’ state [A1491-OFF (green) and A1493-OFF (red)] and overhanging uracil [U2 (gray)] make an A-A-A-A-U stacked column. (b) Atomic details of each bulged-out adenine from the ‘on’ state. The A1493-ON residues interacts with the Watson–Crick G = C in the central stem region of neighboring duplex. (c) Atomic details of each bulged-out adenine from the ‘off’ state. The A1491-OFF residue interacts with C1409 of the Watson–Crick/Sugar-edge C1409oA1494 base pair of neighboring duplex. The hydrogen bonds are represented by black dashed lines.
DISCUSSION
Structural comparisons between the bacterial and eukaryotic A sites in the ‘on’ state
In the present crystal structure, one of two H.sapiens cytoplasmic A site forms the ‘on’ state with two fully bulged-out adenines (A1492 and A1493) and a Watson–Crick C1409oA1491 base pair (Figure 3 and Supplementary Figure 2a). The conformation of the bacterial A site in the ‘on’ state has been already observed in crystal structures of various bacterial 30S ribosomal particles and RNA fragments in complex with aminoglycoside antibiotics (3–6,8–15) and it is very similar to that observed here for the H.sapiens cytoplasmic A site (Supplementary Figure 2b). The RMSD between the sugar-phosphate backbones of the bacterial (8) and cytoplasmic ‘on’ states of the A site is 2.07 Å (it is 1.81 Å without the bulging As, the positions of which are imposed by the crystal packing as discussed above). The deviations are otherwise due to the U1406oU1495 and C1409oA1491 pairs, which adopt different relative orientations in the bacterial and cytoplasmic A sites. The ‘on’ state of the bacterial A site has the Watson–Crick C1409 = G1491 and A1410-U1490 base pairs instead of the Watson–Crick C1409oA1491 and U1419-A1490 base pairs. The effect of the base change at position 1408 is not apparent on the overall conformation of the A site in the ‘on’ state with fully bulged-out A1492 and A1493. As in the previous structures (8–12), these adenines are involved in crystal packing. In particular, residue A1493 interacts with the shallow/minor groove of the Watson–Crick G = C base pair in the central stem of the neighboring RNA duplex forming an A-minor motif. This packing interaction mimics the potential intermolecular recognition of the Watson–Crick base pair formed in the cognate codon–anticodon stem during the decoding step. It has been shown that aminoglycoside antibiotics specifically bind to the ‘on’ state of the bacterial A site with several conserved contacts (8–15). The binding site of an hexa-hydrated magnesium ion in the present structure occurs in the deep/major groove of the G1494 of the Watson–Crick C1407 = G1494 base pair, a residue which binds specifically ring II of the 2-deoxystreptamine (2-DOS) of aminoglycosides. In this respect, it is worthwhile to note that, in both the ‘on’ and ‘off’ states of the cytoplasmic A site, a cation binds to the Hoogsteen sites of G1494 and G1405 and is nearby G1408.
Structural comparisons between the eukaryotic A sites in the ‘off’ state and complexed with apramycin
One of two H.sapiens cytoplasmic A sites in the present structure forms the ‘off’ state with a single fully bulged-out adenine (A1491), one halfway bulged-out adenine (A1493) and one Watson–Crick/Sugar-edge C1409oA1492 base pair (Figure 4 and Supplementary Figure 2d). We have reported the crystal structure of the H.sapiens cytoplasmic A site in complex with apramycin in another paper (J. Kondo, B. François, A. Urzhumtsev and E. Westhof, manuscript submitted). Apramycin specifically binds to the cytoplasmic A site and stabilizes the ‘off’ conformation (Supplementary Figure 2e). Conformation of the ‘off’ state in RNA–apramycin complex is almost the same as that in the present structure (RMSD = 1.0 Å). The bulged-out A1491 residue and the Watson–Crick/Sugar-edge C1409oA1492 base pair are present in both cases. As discussed (J. Kondo, B. François, A. Urzhumtsev and E. Westhof, manuscript submitted), A1491 might disturb the local conformation of a cytoplasmic ribosomal protein [homologous to S12 in bacteria (41)] thereby maintaining 40S ribosome in an inactive state. The single main difference is that the A1493 residue, which forms the Hoogsteen/Sugar-edge base pair with G1408 in the present structure, is free from any base pair interaction in the RNA–apramycin complex. An hexammine cobalt cation binding to G1494 of the Watson–Crick C1407 = G1494 base pair is replaced with ring II of apramycin. This result suggests that some positively-charged aminoglycosides can specifically bind to the ‘off’ state of the H.sapiens cytoplasmic A site, which may disturb the decoding process by inhibiting translocation of the eukaryotic 40S ribosome (J. Kondo, B. François, A. Urzhumtsev and E. Westhof, manuscript submitted).
Structural comparison with the ‘off’ state of the T.thermophila cytoplasmic A site
Two NMR structures have been solved for the eukaryotic cytoplasmic A site. One is the T.thermophila cytoplasmic A site in the ‘off’ state (23) shown in Supplementary Figure 2f while the other one is in complex with paromomycin (24). The secondary structure of the T.thermophila cytoplasmic A site is different from that of the H.sapiens cytoplasmic A site. Although the T.thermophila cytoplasmic A site has a guanine at position 1408 as the H.sapiens cytoplasmic A site, the A1491 residue and the Watson–Crick U1410-A1490 base pair are replaced by a G1491 residue and a Watson–Crick A1410-U1490 base pair. In other words, the secondary structure of the T.thermophila cytoplasmic A site is identical to that of the A1408G mutant of the bacterial A site. As in the ‘off’ state of the H.sapiens cytoplasmic A site, the A1493 residue of the T.thermophila cytoplasmic A site forms a base pair with G1408 but with a different geometry (distorted trans Hoogsteen/Sugar-edge in the H.sapiens, and distorted cis Watson–Crick in the T.thermophila). Therefore, A1493 in the H.sapiens cytoplasmic A site is halfway bulged-out while that in the T.thermophila cytoplasmic A site stays inside of helix. Residue A1492, which forms the Sugar-edge/Watson–Crick base pair in the H.sapiens cytoplasmic A site, is not involved in any base pair formations in T.thermophila but stays inside the helix by stacking between the A1493 and G1491 residues. Residue G1491 makes a Watson–Crick G = C base pair with C1409 as in the bacterial A site. Many lower eukaryotes, such as T.thermophila and Giardia muris have the Watson–Crick C1409 = G1491 base pair, whereas higher eukaryotes usually have a mispair at those positions (16).
Structural comparison with the ‘off’ states of the bacterial A site
For the bacterial A site, four types of ‘off’ states have been reported. In the ‘off’ state of the Thermus thermophilus 30S subunit (2), the A1492 and A1493 are folded into the shallow/minor groove of the A site (Supplementary Figure 2g). Only A1493 forms a base pair with A1408 in the Watson–Crick geometry with A1492 stacking between A1493 and G1491. In the recent crystal structure of the E.coli 70S ribosome, the two A1492 and A1493 are bulged-in and do not form any hydrogen-bonding contact within the internal loop of the A site, instead there is only stacking interaction between G1491-A1492-A1493 (but see discussion below) (Supplementary Figure 2h) (7). Other examples of the ‘off’ state of the bacterial A site have one bulged-out adenine and one bulged-in adenine (Supplementary Figure 2i and j). In the ‘off’ state with bulged-out A1492 (12), A1493 has a syn conformation around the glycoside bond and forms Hoogsteen/Watson–Crick A1493oA1408 base pair (Supplementary Figure 2i). In the third ‘off’ state with bulged-out A1493 (14), the A1492 residue stays inside of helix and forms a Watson–Crick base pair A1492oA1408 (Supplementary Figure 2j). Another ‘off’ state with two bulged-out As also has been reported, but A1492 and A1493 have poor densities (Supplementary Figure 2k) (5). The C1409 = G1491 base pair is conserved in all four ‘off’ states of the bacterial A site. The important conclusion is that the conformation of the H.sapiens cytoplasmic A site in the ‘off’ state is completely different from any of the four ‘off’ states observed for the bacterial A site (2,5,7,12,14).
CONCLUSIONS
Although A-minor motifs constitute the most frequent occurrences in RNA–RNA contacts (40), several RNA motifs allow for the formation of such contacts. For example, the A-minor contacts formed by the ‘on’ state of the A site are identical to those formed by GNRA tetraloops and helically stacked Watson–Crick base pairs (42). However, the asymmetrical A site internal loop is uncommon in RNA secondary structures and, up to now, there is no other occurrence of an internal loop with a purine residue internal and two bulging As in three-dimensional structures. The present work extends the striking diversity in three-dimensional conformations that such an apparently inconspicuous internal loop can adopt.
In addition, because this work offers two different snapshots of the H.sapiens cytoplasmic A site, plausible insights into the molecular decoding mechanism of the 40S cytoplasmic ribosome can be deduced. Most interestingly, whatever the environment and despite sequence variations, one conformational state is geometrically invariant in all structures of the A site, the state we have called ‘on’, i.e. the one with two bulging As and one internal purine residue. In contrast, the state we have called ‘off’ displays several states both in the prokaryotic and eukaryotic sequences.
The observed conformational similarity between the bacterial and cytoplasmic A sites in the ‘on’ state, i.e. the state used in bacteria for recognition of the complex between 30S and mRNA/tRNA helix, supports the view that a similar recognition mechanism is used in eukaryotes. The present structure is the first illustration of an ‘on’ conformer without bound drug. Interestingly, a bound cation occupies the same place as the positively-charged aminosugar, as was previously proposed (43,44). Until now, such a state could never be observed with an empty bacterial A site, except for the complex between the 30S particle and the initiation factor IF1 (Supplementary Figure 2c) (45), which occupies the same space as the mRNA/tRNA complex and forces the initiator tRNA to bind in the P site.
On the contrary, the ‘off’ state, or the state adopted by a ribosome with an empty A site, adopts a great variety of conformations in crystal structures containing the bacterial or eukaryotic A site (Supplementary Figure 2). As a matter of fact, in structures with empty A sites [30S particles (5) or the recent E.coli ribosomes (7)], the nucleotides belonging to the internal loop have poor densities or are disordered. Thus, it would appear that in the absence of the intermolecular stabilizing interactions made by the bulging adenines, the A site adopts disordered or various conformers. When an aa-tRNA enters the A site and forms cognate codon–anticodon stem with mRNA template, the A site changes its conformation to an identical ‘on’ state. Indeed, the two bulged-out adenines, A1493 and A1492, have to bind to and recognize the first and second Watson–Crick base pair of codon–anticodon, respectively. These intermolecular A-minor-like recognitions contribute to the fidelity of tRNA selection step as observed in the bacterial 30S ribosome (4–6). In the ‘on’ state, the A1491 residue of higher eukaryotes stays inside the helix and forms the Watson–Crick A1491oC1409 base pair. In higher eukaryotic cytoplasm, before decoding, the A site adopts mainly the ‘off’ conformation with bulged-out A1491. The A1491 residue might disturb the active conformation of a cytoplasmic ribosomal protein thereby maintaining 40S ribosome in an inactive state.
It has been long suggested that aminoglycoside binding to the H.sapiens cytoplasmic A site contributes to their toxicity (24,46). Although it is not clear yet how, it is the hope that the present and future crystal structures will allow the design of new aminoglycoside antibiotics with higher selectivity for the bacterial ribosome and less toxicity to mammals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
Dr J. Kondo is supported by Japan Society for the Promotion of Science. The authors are grateful to Dr B. François, Dr B. Masquida, Dr A.C. Vaiana, Dr P. Auffinger, Dr A. Werner, A. Lescoute and Prof. P.S. Ho for helpful discussions and suggestions. The authors thank the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and acknowledge the people of beamline BM16 and ID14-1. Funding to pay the Open Access publication charges for this article was provided by the CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Noller H.F. Ribosomal RNA and translation. Annu. Rev. Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 2.Wimberly B.T., Brodersen D.E., Clemons W.M., Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 3.Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 4.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 5.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 6.Ogle J.M., Carter A.P., Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 7.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 8.Vicens Q., Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A Site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 9.Vicens Q., Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- 10.Vicens Q., Westhof E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 2003;326:1175–1188. doi: 10.1016/s0022-2836(02)01435-3. [DOI] [PubMed] [Google Scholar]
- 11.François B., Szychowski J., Adhikari S.S., Pachamuthu K., Swayze E.E., Griffey R.H., Migawa M.T., Westhof E., Hanessian S. Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site. Angew. Chem. Int. Ed. Engl. 2004;43:6735–6738. doi: 10.1002/anie.200462092. [DOI] [PubMed] [Google Scholar]
- 12.François B., Russell R.J., Murray J.B., Aboul-ela F., Masquida B., Vicens Q., Westhof E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Q., Zhao Q., Fish S., Simonsen K.B., Vourloumis D., Froelich J.M., Wall D., Hermann T. Molecular recognition by glycoside pseudo base pairs and triples in an apramycin–RNA complex. Angew. Chem. Int. Ed. Engl. 2005;44:2694–2700. doi: 10.1002/anie.200500028. [DOI] [PubMed] [Google Scholar]
- 14.Shandrick S., Zhao Q., Han Q., Ayida B.K., Takahashi M., Winters G.C., Simonsen K.B., Vourloumis D., Hermann T. Monitoring molecular recognition of the ribosomal decoding site. Angew. Chem. Int. Ed. Engl. 2004;43:3177–3182. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 15.Zhao F., Zhao Q., Blount K.F., Han Q., Tor Y., Hermann T. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew Chem. Int. Ed. Engl. 2005;44:5329–5334. doi: 10.1002/anie.200500903. [DOI] [PubMed] [Google Scholar]
- 16.Gutell R.R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Stasio E.A., Moazed D., Noller H.F., Dahlberg A.E. Mutations in 16S ribosomal RNA disrupt antibiotic—RNA interactions. EMBO J. 1989;8:1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sander P., Prammananan T., Böttger E.C. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 1996;22:841–848. doi: 10.1046/j.1365-2958.1996.01532.x. [DOI] [PubMed] [Google Scholar]
- 19.Prammananan T., Sander P., Brown B.A., Frischkorn K., Onyi G.O., Zhang Y., Böttger E.C., Wallace R.J., Jr A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 1998;177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 20.Recht M.I., Douthwaite S., Puglisi J.D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böttger E.C., Springer B., Prammananan T., Kidan Y., Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Stasio E.A., Dahlberg A.E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J. Mol. Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- 23.Lynch S.R., Puglisi J.D. Structure of a eukaryotic decoding region A-site RNA. J. Mol. Biol. 2001;306:1023–1035. doi: 10.1006/jmbi.2000.4419. [DOI] [PubMed] [Google Scholar]
- 24.Lynch S.R., Puglisi J.D. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol. 2001;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- 25.Leslie A.G.W. Molecular data processing. In: Moras D., Podjarny A.D., Thierry J.C., editors. Crystallography Computing 5, From Chemistry to Biology. Oxford, UK, Vol. 5: Oxford University Press; 1991. pp. 50–61. [Google Scholar]
- 26.Rossmann M.G., van Beek C.G. Data processing. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 29.Fokine A., Urzhumtsev A. On the use of low-resolution data for translation search in molecular replacement. Acta Crystallogr. A. 2002;58:72–74. doi: 10.1107/s0108767301016476. [DOI] [PubMed] [Google Scholar]
- 30.Fokine A., Capitani G., Grütter M.G., Urzhumtsev A. Bulk-solvent correction for fast translation search in molecular replacement: service programs for AMoRe and CNS. J. Appl. Cryst. 2003;36:352–355. [Google Scholar]
- 31.Delarue M., Dumas P. On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc. Natl Acad. Sci. USA. 2004;101:6957–6962. doi: 10.1073/pnas.0400301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 33.Batey R.T., Gilbert S.D., Montange R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 34.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Brünger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structure. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 36.Leontis N.B., Stombaugh J., Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pley H.W., Flaherty K.M., McKay D.B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 38.Scott W.G., Finch J.T., Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 39.Doudna J.A. Hammerhead ribozyme structure: U-turn for RNA structural biology. Structure. 1995;3:747–750. doi: 10.1016/s0969-2126(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 40.Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cukras A.R., Southworth D.R., Brunelle J.L., Culver G.M., Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 42.Leontis N.B., Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 43.Hermann T., Westhof E. Aminoglycoside binding to the hammerhead ribozyme: a general model for the interaction of cationic antibiotics with RNA. J. Mol. Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]
- 44.Tor Y., Hermann T., Westhof E. Deciphering RNA recognition: aminoglycoside binding to the hammerhead ribozyme. Chem. Biol. 1998;5:R277–R283. doi: 10.1016/s1074-5521(98)90286-1. [DOI] [PubMed] [Google Scholar]
- 45.Carter A.P., Clemons W.M., Jr, Brodersen D.E., Morgan-Warren R.J., Hartsch T., Wimberly B.T., Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 46.Bar-Nun S., Shneyour Y., Beckmann J.S. G-418, an elongation inhibitor of 80 S ribosomes. Biochim. Biophys. Acta. 1983;741:123–127. doi: 10.1016/0167-4781(83)90018-0. [DOI] [PubMed] [Google Scholar]
- 47.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.