Abstract
The 5′ long terminal repeat of spleen necrosis virus (SNV) facilitates Rev/Rev-responsive element (RRE)-independent expression of intron-containing human immunodeficiency virus type 1 (HIV-1) gag. The SNV RU5 region, which corresponds to the 165-nucleotide 5′ RNA terminus, functions in a position- and orientation-dependent manner to enhance polysome association of intron-containing HIV-1 gag RNA and also nonviral luc RNA. Evidence is mounting that association with nuclear factors during intron removal licenses mRNAs for nuclear export, efficient translation, and nonsense-mediated decay. This project addressed the relationship between the nuclear export pathway of SNV RU5-reporter RNA and translational enhancement. Results of RNA transfection experiments suggest that cytoplasmic proteins are insufficient for SNV RU5 translational enhancement of gag or luc RNA. Reporter gene assays, leptomycin B (LMB) sensitivity experiments, and RNase protection assays indicate that RU5 gag RNA accesses a nuclear export pathway that is distinct from the LMB-inhibited leucine-rich nuclear export sequence-dependent CRM1 pathway, which is used by the HIV-1 RRE. As a unique tool with which to investigate the relationship between different RNA trafficking routes and translational enhancement, SNV RU5 and Rev/RRE were combined on a single gag RNA. We observed a less-than-synergistic effect on cytoplasmic mRNA utilization. Instead, Rev/RRE diverts RU5 gag RNA to the CRM1-dependent, LMB-inhibited pathway and abrogates translational enhancement by SNV RU5. Our study is the first to show that a nuclear factor(s) directs SNV RU5-containing RNAs to a distinct export pathway that is not inhibited by LMB and programs the intron-containing transcript for translational enhancement.
Retroviruses contain structured RNA elements that interact with viral and cellular proteins and modulate nuclear export and efficient translation of intron-containing transcripts (reviewed in references 3, 5, and 11). The long terminal repeat (LTR) of spleen necrosis virus (SNV) facilitates Rev/Rev-responsive element (RRE)-independent expression of unspliced human immunodeficiency virus type 1 (HIV-1) gag reporter RNA (7). Quantitative RNA and protein analysis identified a minor 2-fold increase in cytoplasmic accumulation of gag RNA but a greater-than-100-fold increase in Gag protein production in response to the SNV LTR. The 165-nucleotide 5′ RNA terminus encoded by the RU5 region of the LTR functions in a position- and orientation-dependent manner to direct polysome association and detectable Gag protein synthesis. SNV RU5 functions in a cap-dependent manner (M. Butsch and K. Boris-Lawrie, unpublished data) and is not an internal ribosome entry site (IRES) (35). The SNV U5 region also augments translation of nonviral luciferase (luc) RNA by increasing polysome loading (35). Recently, the R region of human foamy virus (HFV) and RU5 of Mason-Pfizer monkey virus (MPMV) were also shown to program unspliced gag RNA templates for Gag protein synthesis (37; S. Hull and K. Boris-Lawrie, submitted for publication). 5′-terminal translational enhancers appear to be a mechanism shared among divergent retroviruses to achieve efficient expression of viral structural protein. The relationship between nuclear export and productive cytoplasmic utilization directed by these 5′-terminal RNA elements remains an important open question.
Evidence is mounting that nuclear association between nascent RNAs and viral and cellular nuclear-cytoplasmic shuttling proteins choreographs steps of posttranscriptional gene expression (reviewed in references 10, 28, and 38). The HIV-1 Rev protein is an essential activator of nuclear export and cytoplasmic expression of intron-containing viral RNAs (14, 15, 20, 27). Rev interacts with newly synthesized RNA (23) by binding to the RRE, which is within the terminal intron of unspliced and singly spliced viral RNAs (9, 19, 21, 27, 36, 46). Using a leucine-rich nuclear export sequence (NES), Rev acts as an adapter protein that connects RRE-containing RNA to the CRM1/exportin 1 nuclear export receptor (17, 30). CRM1 interacts with FG repeats of nucleoporins and exports RRE-containing RNA by a leptomycin B (LMB)-inhibited pathway that is typically reserved for 5S rRNA and cellular proteins that contain leucine-rich NESs (16, 17). In the cytoplasm, Rev/RRE augments RNA translational efficiency by directing polysome association (1, 7, 12, 26). Results of in situ hybridization assays have shown that Rev transactivation correlates with colocalization of RRE-containing RNAs with the cytoskeleton, which is a supportive framework for interaction of polysomes and mRNA (24, 25). It is not known whether nuclear export by the CRM1-dependent pathway is necessary to program the RRE-containing RNA for subcellular localization with cytoskeletal polysomes and efficient translation.
This project addressed the relationship between the nuclear export pathway and translational enhancement by SNV RU5. Results of RNA transfection and in vitro translation assays indicate that cytoplasmic factors are insufficient for SNV RU5 translational enhancement and suggest that nuclear interactions are necessary. The nuclear export pathway of SNV RU5-containing RNAs is not known. LMB sensitivity assays and RNA analysis show that the nuclear export pathway of SNV RU5 is distinct from the CRM1-dependent pathway accessed by Rev/RRE. Upon combination of SNV RU5 and Rev/RRE on a single gag RNA, we observe that Rev/RRE sequesters the transcript to the leucine-rich CRM1-dependent nuclear export pathway and that this nuclear export pathway abrogates translational enhancement by SNV RU5. Our results are consistent with the model in which 5′-terminal SNV RU5 sequences and nuclear factors direct access to a particular nuclear export pathway that programs intron-containing RNA for efficient translation in the cytoplasm.
MATERIALS AND METHODS
Plasmids.
The previously described plasmids used in this study are pYW99, pYW208, pYW207, pYW205, pYW100, pGem (140-440), pMSBSVT7, pGAPDH (7), pTR103, and pTR105 (35). Plasmids pYW100RRE, pYW205RRE, and pTR155 were constructed from pYW100, pYW205, and pTR147, respectively, by introduction of RRE into a SalI site in the 3′ untranslated region (UTR). Plasmid pTR147 was constructed from pYW100 by exchange of the unique BamHI/ApaI fragment from pYW207 that contains antisense RU5 and HIV 5′ UTR sequences. The RRE was amplified by PCR from HIV-1BRU (coordinates 7217 to 7689). pYW233 contains nine point mutations in RU5 that were introduced into pYW100 by PCR-based site-directed mutagenesis (T. M. Roberts and K. Boris-Lawrie, unpublished data). Plasmids pSVgagpolrre and pRev were kindly provided by David Rekosh, University of Virginia (41).
In vitro transcriptions and RNase protection assays (RPAs).
In the RNA transfection and in vitro translation experiments, DNA templates for in vitro transcription were prepared by PCR with primers that contain the T7 promoter sequence incorporated at the 5′ terminus of the 5′ oligonucleotide. 5′-capped transcripts were synthesized by T7 polymerase with mMessage mMachine (Ambion). The 3′ RNA terminus was polyadenylated in duplicate reactions with poly(A) polymerase and either ATP or [γ-32P]ATP. The concentrations of the in vitro transcripts were determined by spectrophotometry, and the quality was verified by denaturing polyacrylamide gel electrophoresis of the 32P-labeled aliquot and PhosphorImager analysis with ImageQuant version 4.2 software (Molecular Dynamics). RU5 gag and RU5 luc transcripts are 1,970 bases and 1,840 nucleotides in length, respectively.
For RPAs, antisense α-32P-labeled runoff transcripts were synthesized by MAXIscript T7 RNA polymerase (Ambion) in accordance with the manufacturer's instructions. A probe complementary to HIVBRU sequences in the 5′ UTR common to each of the reporter RNAs was in vitro transcribed from template pGEM(140-440) that had been linearized with NotI (7). A probe complementary to cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was transcribed from template pGAPDH, which had been linearized with NcoI. The in vitro-transcribed RNAs were isolated by gel elution, and the RPAs were performed with RPAIII (Ambion) with some modifications. Briefly, 30 μg of cytoplasmic RNA or 15 μg of nuclear RNA was precipitated with 3 × 105 cpm of reporter probe and 2 × 104 cpm of the GAPDH probe. Samples were hybridized at 42°C for 16 h. Samples were digested with an RNase mixture at 37°C for 30 min and then extracted with phenol-chloroform and chloroform and precipitated with ethanol. Pellets were dissolved in loading buffer, heated at 90°C for 3 min, and subjected to denaturing polyacrylamide gel electrophoresis on 5% gels. RNase protection products were visualized by PhosphorImager analysis.
In vitro translations.
RRLs (35 μl; Promega) were programmed with up to 400 ng of capped and polyadenylated transcript in a total volume of 50 μl and incubated at 30°C for 1 h. Translation-competent 293 cell lysates were prepared as described by Carroll and Lucas-Lenard without micrococcal nuclease treatment (8) and were programmed with capped and polyadenylated transcript in a total volume of 25 μl and incubated at 30°C for up to 1 h. Programmed lysates were directly assayed for Gag protein content or Luc activity.
Cells, transfections, and reporter assays.
293 cells are a human embryonic kidney cell line. RNA transfections were performed on 293 cells with dilutions of capped and polyadenylated transcript and 12 μl of Lipofectin (Gibco-BRL) in 1 ml of serum-free Dulbecco modified Eagle medium. The luc and gag RNAs were cotransfected and used reciprocally to normalize transfection efficiency. All transfection assays were performed in triplicate and for at least three replicate experiments. The Lipofectin-RNA mixtures were incubated with 293 cells (8 × 105) in 60-mm-diameter plates for 2 h and then replaced with Dulbecco modified Eagle medium containing 10% fetal calf serum. Cells were harvested in phosphate-buffered saline (PBS) at serial time points between 6 and 36 h posttransfection and resuspended in 55 μl of ice-cold lysis buffer (10 mM HEPES, 10 mM NaCl, 3 mM CaCl2, 7 mM MgCl2, 1 mM EDTA, 0.5% NP-40). Cytoplasmic lysates were harvested by centrifugation at 9,300 × g for 2 min and subjected to a Gag enzyme-linked immunosorbent assay (ELISA) or a luciferase assay. For reporter gene assays and LMB sensitivity experiments, 293 cells (3 × 105) in 60-mm-diameter plates were transfected with 1 μg of a reporter plasmid using a CaPO4 protocol (7). LMB was added 24 h posttransfection, and cells were harvested 24 h later.
HIV-1 Gag levels were quantified by Gag ELISA (Coulter Corp.) and normalized to firefly Luc activity expressed from cotransfected pGL3 (Promega). In DNA transfections that used firefly luc as the reporter gene, pRL-CMV (Promega) was cotransfected and Renilla luciferase activity was used to standardize transfection efficiency. Luc or Renilla luciferase assays were performed with 10 μl of lysate and 100 μl of substrate (Promega), and enzyme activity was quantified in a Lumicount luminometer (Packard).
RNA preparation.
Transfected 293 cells from three 10-cm-diameter plates that were treated with LMB at 0 or 2.5 ng/ml were harvested in PBS and lysed on ice for 15 min in a solution containing 10 mM Tris (pH 8.3), 150 mM NaCl, 1.5 mM MgCl2, and −0.5% NP-40, and nuclei were removed by centrifugation at 10,000 × g for 10 min. The cytoplasmic supernatant was recentrifuged at 10,000 × g for 10 min, and the clarified extract was mixed with Tri-Reagent LS (Molecular Research). The nuclear pellets were washed twice in PBS and mixed with Tri-Reagent (Molecular Research). The RNAs were extracted in accordance with the manufacturer's protocol and treated with DNase I.
RESULTS
Cytoplasmic factors are insufficient for SNV RU5 translational enhancement.
We compared reporter gene activity from 293 cells transfected either with reporter plasmid or liposomes containing in vitro-transcribed capped and polyadenylated reporter RNA. The RNA transfection assay has been used to assess reporter protein synthesis independently of de novo transcription, RNA processing, and nuclear export and provides an in vivo approach by which to determine whether cytoplasmic factors are sufficient for translational enhancement by SNV RU5 (43).
Consistent with our published data, results of DNA transfection assays in 293 cells indicate that SNV RU5 in the sense orientation confers Rev/RRE-independent expression of intron-containing gag RNA (Table 1, pYW100 and pYW208) (7). Furthermore, SNV RU5 augments expression of nonviral luc reporter RNA (compare pTR103 and pTR105) (35). Efficient Gag production is not observed from Gag reporter plasmids that lack SNV RU5 (pYW205, pYW99, and pSVgagpolrre) or contain antisense SNV RU5 (pTR147 and pYW207) or substitution mutant SNV RU5 (pYW233). As expected, cotransfected pRev activates expression from the RRE-containing plasmids (pSVgagpolrre and pTR155).
TABLE 1.
Reporter protein synthesis
| Plasmid | Promoter | RU5 | RRE | Reporter gene | Gag concn (ng/ml) | Luc activity (RLU) |
|---|---|---|---|---|---|---|
| pYW100 | SNV | Sense | Absent | gag | 41.2 ± 1.8 | NAb |
| pYW208 | CMV | Sense | Absent | gag | 18.0 ± 1.1 | NA |
| pYW205 | SNV | Absent | Absent | gag | 5.0 ± 0.9 | NA |
| pYW99 | CMV | Absent | Absent | gag | <MDa | NA |
| pTR147 | SNV | Antisense | Absent | gag | <MD | NA |
| pYW207 | CMV | Antisense | Absent | gag | <MD | NA |
| pYW233 | SNV | Mutant | Absent | gag | <MD | NA |
| pTR155 | SNV | Antisense | Present | gag | <MD | NA |
| pTR155+Rev | SNV | Antisense | Present | gag | 210.0 ± 81.2 | NA |
| pSVgagpolrre | SV40c | Absent | Present | gag | <MD | NA |
| pSVgagpolrre+Rev | SV40 | Absent | Present | gag | 160.5 ± 8.7 | NA |
| pTR103 | SNV | Sense | Absent | luc | NA | 42 ± 2.5 |
| pTR105 | SNV | Absent | Absent | luc | NA | 8 ± 0.1 |
<MD, less than the minimum detectable, 0.02 ng/ml.
NA, not applicable.
SV40, simian virus 40.
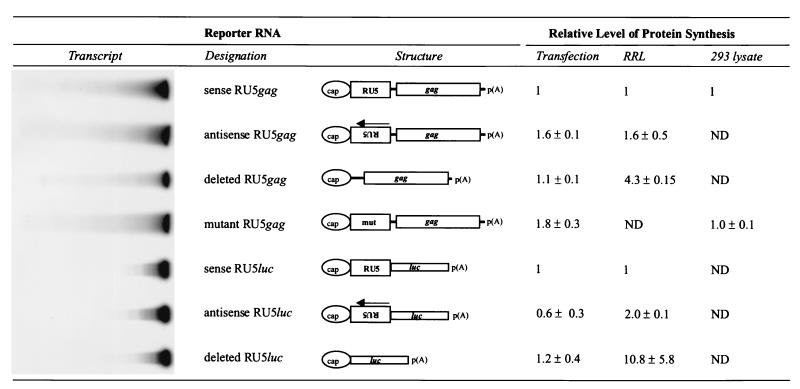
Results obtained with the synthetic capped and polyadenylated gag reporter RNAs indicate that SNV RU5 gag RNA is efficiently utilized as template mRNA for Gag protein synthesis (Fig. 1, 19 ± 0.1 ng/ml, normalized to 1). However, unlike results obtained with the reporter plasmids, similar levels of Gag are synthesized from gag reporter RNAs that contain deleted, antisense, or substitution mutant SNV RU5. Luc activity also remained similar upon transfection with luc RNA that contained sense, antisense, or deleted SNV RU5 (44,000 ± 13,000 relative light units [RLU], normalized to 1). The lack of SNV RU5 stimulation was sustained when dilutions of the reporter RNAs were transfected and when shorter incubation periods were evaluated, indicating that the RNAs were not used at saturating levels and were able to titrate SNV RU5-interactive proteins (data not shown). The results indicate that SNV RU5 does not confer a positive effect on the translation of either gag or luc reporter transcripts that are introduced directly into the cytoplasm. The data imply that interaction between SNV RU5 RNA and a nuclear factor(s) is necessary for the positive effect of SNV RU5. An alternative explanation is that the structural conformation of SNV RU5 is not recapitulated in liposomes. However, Stoneley et al. observed that the human rhinovirus type 2 IRES remains functional in the liposome-mediated RNA transfection assay (43).
FIG. 1.
Protein synthesis from capped and polyadenylated reporter transcripts reveals that cytoplasmic proteins are insufficient for translational enhancement by SNV RU5. Shown are representative transcripts used to program three translation systems and their designations, structures, and relative protein-synthetic activities. RNA transfection of 293 cells was performed with 2.5 μg of transcript and Lipofectamine. RRL and 293 cell lysates were programmed with 400 ng of reporter transcript. In each assay system, luc or gag reporter transcripts were used reciprocally to standardize minor differences in RNA transfection efficiency. Reporter proteins were measured by a Gag ELISA or a Luc enzymatic assay. ND, not determined.
Because a cell-free assay would be a useful tool with which to investigate SNV RU5-interactive proteins, we also investigated the utility of in vitro translation assays. RRL programmed with sense SNV RU5 gag transcripts produced significant Gag protein (Fig. 1, 100 ± 8.7 ng/ml, normalized to 1). Gag production was sustained or increased fourfold in RRL programmed with an antisense SNV RU5 gag or deletion-containing SNV RU5 gag transcript, respectively. Analysis of luc transcripts identified that RRL programmed with sense SNV RU5 luc exhibited Luc activity (8,300 ± 1,700 RLU, normalized to 1), while antisense SNV RU5 luc transcripts increased Luc activity 2-fold and deletion of SNV RU5 increased Luc activity 11-fold. The inhibitory effect of SNV RU5 on protein synthesis in RRL is likely attributable to structural barriers in SNV RU5 RNA. Structural barriers in HIV-1 RU5 and other 5′ UTRs have been previously shown to inhibit protein synthesis in RRL (18, 22, 29). We also performed in vitro translation assays with 293 cell cytoplasmic lysates because 293 cells had been used to characterize SNV RU5 in the DNA transfection assays. The extracts were verified to be translationally competent by [35S]methionine incorporation into nascent polypeptide chains (data not shown) and by programming with reporter transcripts and confirmation of reporter protein synthesis (Fig. 1). Gag production in response to a sense SNV RU5 gag or a mutant SNV RU5 gag transcript was the same (50 ± 7 ng/ml, normalized to 1). The lack of SNV RU5 stimulation in vitro was sustained in the RRL or 293 cell extracts when dilutions of reporter RNA were tested and when shorter incubation periods were evaluated, indicating that the RNAs were not used at saturating levels. The data from these in vitro translation systems are consistent with the RNA transfection results and indicate that the positive effect of SNV RU5 is not recapitulated in these cytoplasmic extracts. The data imply that nuclear interactions are necessary for translational enhancement by SNV RU5. An alternative explanation is that these conventional in vitro translation extracts are deficient in faithful regulation of translation initiation.
LMB does not inhibit cytoplasmic accumulation of SNV RU5 gag RNA.
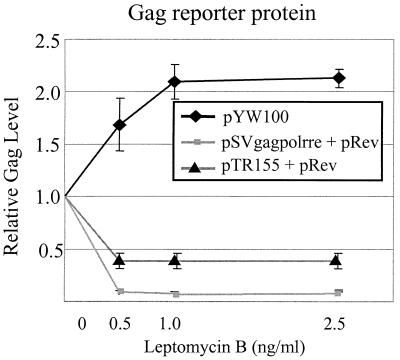
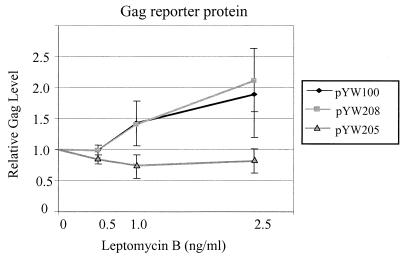
LMB inhibits activation of nuclear export by Rev/RRE by alkylation of CRM1 at Cys-529 and prevention of leucine-rich NES-mediated nuclear export (17, 33, 44). DNA transfection assays were used to investigate whether or not nuclear export of SNV RU5 gag reporter RNA is sensitive to LMB. Triplicate cultures of transfected 293 cells were treated with various concentrations of LMB for 24 h posttransfection, and cell-associated reporter proteins were quantified. Consistent with previous reports, LMB severely reduced Gag production in cells cotransfected with pRev and RRE-containing plasmid pSVgagpolrre (lacks SNV RU5) or pTR155 (contains antisense SNV RU5) (Fig. 2). By contrast, Gag production from pYW100 (contains SNV RU5) was not inhibited by LMB and is reproducibly enhanced in response to LMB. Similar responses were observed when SNV RU5 was positioned downstream of the SNV U3 promoter (pYW100) or the cytomegalovirus (CMV) immediate-early promoter (pYW208) (Fig. 3). Deletion of SNV RU5 (pYW205), which reduces Gag production to low but detectable levels, eliminated the LMB enhancement of Gag production. The data indicate that SNV RU5 confers LMB-enhanced expression in a promoter-independent manner.
FIG. 2.
Effect of LMB on Gag production reveals that SNV RU5 functions independently of CRM1/leucine-rich NES. Results of three to five replicate transfections of 293 cells with the indicated gag reporter plasmids are shown. Reporter proteins were measured 24 h posttreatment with the indicated concentrations of LMB. Gag production is presented relative to that of the mock-treated control.
FIG. 3.
LMB dose-response curves indicate that SNV RU5 confers LMB enhancement in a promoter-independent manner. Results of three replicate transfections of 293 cells with the indicated reporter plasmids are shown. Cell-associated reporter proteins were measured 24 h posttreatment with the indicated concentrations of LMB. Presented is Gag protein production in response to the indicated gag reporter plasmid relative to that of the mock-treated control.
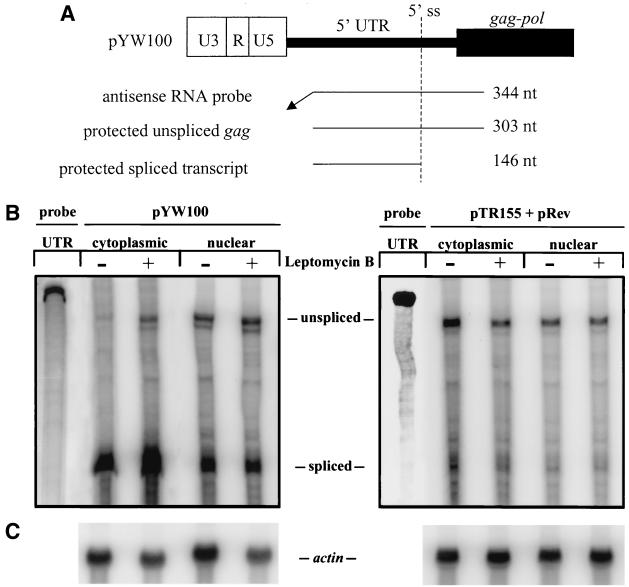
The lack of LMB inhibition of Gag production from SNV RU5 gag reporter RNA suggests that the RNA accesses a nuclear export pathway that is distinct from the leucine-rich NES/CRM1-mediated pathway utilized by Rev/RRE. To evaluate the effect of LMB on cytoplasmic accumulation of SNV RU5 gag RNA, RPAs were performed on nuclear and cytoplasmic RNA from transfected 293 cells treated with LMB at 0 or 2.5 ng/ml. The uniformly labeled HIV antisense RNA probe is complementary to the 5′ UTR and distinguishes unspliced gag mRNA and spliced versions of the primary transcript (Fig. 4A) (7). LMB treatment of cells transfected with pYW100 increased the cytoplasmic accumulation of SNV RU5 gag threefold (Fig. 4B and C). The increase in cytoplasmic SNV RU5 gag RNA is proportional to the fourfold increase in Gag protein observed in response to LMB (Table 2). Cytoplasmic accumulation of spliced SNV RU5 transcripts displayed a similar 2.6-fold increase, indicating that both spliced and intron-containing versions of SNV RU5-containing RNA are affected by LMB. In contrast, cells transfected with the RRE-containing reporter pTR155 and pRev exhibited a reduction in the cytoplasmic accumulation of RRE gag RNA and Gag production by a factor of 0.6. These data indicate that SNV RU5 provides both spliced and intron-containing RNAs access to a nuclear export pathway that is distinct from the NES-CRM1-mediated Rev/RRE pathway. A possible explanation for the LMB enhancement of gag RNA expression conferred by SNV RU5 is that CRM1 inactivation alters the availability of a rate-limiting SNV RU5 nuclear export or RNA stability cofactor(s).
FIG. 4.
RPA of cytoplasmic and nuclear RNAs from transfected 293 cells with or without 24 h of LMB treatment reveals that LMB augments cytoplasmic accumulation of SNV RU5 gag RNA. (A) Relationships among the gag reporter plasmid, an antisense HIV-1 5′ UTR RNA probe, and protected unspliced and spliced transcripts with sizes indicated. ss, splice site; nt, nucleotides. (B) RPA and PhosphorImager analysis of cytoplasmic (30 μg) and nuclear (15 μg) RNAs. Labels indicate the reporter plasmid, LMB treatment, and protected transcript. (C) Northern blot analysis of 10-μg aliquots of the RNA samples hybridized to an actin probe.
TABLE 2.
Effects of LMB on protein and RNA levels
| Plasmid(s) and transcript | Minus LMB
|
Plus LMB
|
Fold increasec | ||||
|---|---|---|---|---|---|---|---|
| Gag concn (ng/ml)a | Nuclear RNAb | Cytoplasmic RNA | Gag concn (ng/ml) | Nuclear RNA | Cytoplasmic RNA | ||
| pYW100 | |||||||
| Unspliced | 363 | 5,028 | 794 | 1,676 | 6,902 | 3,334 | 3.1 |
| Spliced | NAd | 27,774 | 42,012 | NA | 22,897 | 89,606 | 2.6 |
| pTR155 + pRev | |||||||
| Unspliced | 1,442 | 5,409 | 9,691 | 658 | 5,074 | 5,581 | 0.6 |
| Spliced | NA | 3,310 | 6,214 | NA | 3,306 | 4,946 | 0.8 |
Gag protein levels were measured by ELISA.
RNA levels were determined by RNase protection assay and are expressed in PhosphorImager units and normalized to the actin RNA signal.
Ratio of cytoplasmic RNA to nuclear RNA in the presence and absence of LMB.
NA, not applicable.
The combination of SNV RU5 and Rev/RRE augments Gag expression.
We sought to evaluate whether or not the combination of SNV RU5 and Rev/RRE on a single RNA would synergistically augment gag gene expression. The RRE was introduced into pYW100 (contains RU5) and pYW205 (lacks RU5), and Gag production was measured in the presence or absence of Rev. The introduction of RRE had little effect on Gag production in the absence of Rev (Table 3). As expected, cotransfection of Rev transactivated Gag production from pYW100RRE and from pYW205RRE). Notably, the magnitude of the increase in Gag production was smaller in the presence of SNV RU5 than in the absence of SNV RU5 (compare pYW100RRE [34-fold] and pYW205RRE [95-fold]). To address the possibility that the lower fold transactivation is attributable to a maximum threshold of Gag production from pYW100RRE plus Rev, dose-response curves were generated with twofold molar increases of cotransfected pRev. Gag production from pYW100RRE, pYW205RRE, and pTR155 increased progressively in response to increasing pRev and demonstrated that Gag production had not reached a maximum threshold (data not shown). Furthermore, pYW100RRE RNA continued to exhibit a smaller magnitude of Rev transactivation. An alternative explanation for the lower fold Rev transactivation for pYW100RRE is that Rev/RRE disrupts translational enhancement by SNV RU5. Consistent with this possibility, the positive effect of SNV RU5 was reduced in the presence of Rev. The positive effect of SNV RU5 was 7.5-fold in the absence of Rev (compare pYW100RRE [30 ± 1.5 ng/ml] and pYW205RRE [4 ± 0.8 ng/ml]) and was reduced to 2.7-fold in the presence of Rev (compare pYW100RRE plus pRev [1,030 ± 350 ng/ml] and pYW205RRE plus pRev [380 ± 37 ng/ml]).
TABLE 3.
Increase in Gag production due to Rev
| Plasmid | RU5 | Gag concn (ng/ml)
|
Fold increasea | |
|---|---|---|---|---|
| Minus Rev | Plus Rev | |||
| pYW100 | Present | 50 ± 4 (12.5)b | NAc | NA |
| pYW100RRE | Present | 30 ± 1.5 (7.5) | 1,030 ± 95 (260) | 34 |
| pYW205 | Absent | 3 ± 0.7 (0.75) | NA | NA |
| pYW205RRE | Absent | 4 ± 0.8 (1.0) | 380 ± 37 (95) | 95 |
Increase in response to Rev.
In parentheses is the level relative to that obtained with pYW205RRE minus Rev.
NA, not applicable.
NES/CRM1-dependent Rev/RRE nuclear export pathway is dominant and impedes translational enhancement by SNV RU5.
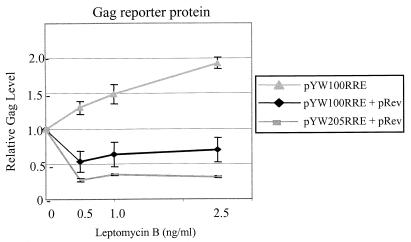
LMB was used to determine whether or not Rev/RRE can divert SNV RU5 gag RNA to the LMB-inhibited CRM1/NES-dependent pathway. In the absence of Rev, pYW100RRE exhibits LMB-enhanced Gag production (Fig. 5), similar to those from pYW100 (Fig. 2A). Gag levels from pYW100RRE increased to 190% ± 8%, indicating that RRE does not disrupt LMB enhancement by SNV RU5. However, in the presence of Rev, Gag production from pYW100RRE was inhibited by LMB (Fig. 5). Gag production was reduced to a level similar to that of pYW205RRE plus pRev (lacks RU5). The shift of pYW100RRE RNA from LMB enhancement in the absence of Rev to LMB inhibition in the presence of Rev indicates that SNV RU5 and Rev/RRE compete for posttranscriptional control of gag RNA. Rev/RRE appears to sequester the SNV RU5 transcripts to the LMB-sensitive NES/CRM1-mediated nuclear export pathway.
FIG. 5.
LMB dose-response curves indicate that LMB enhancement of SNV RU5 gag RNA is mitigated by Rev/RRE. 293 cells were transfected with the indicated gag reporter plasmid, and cell-associated Gag levels are presented relative to that of the mock-treated control.
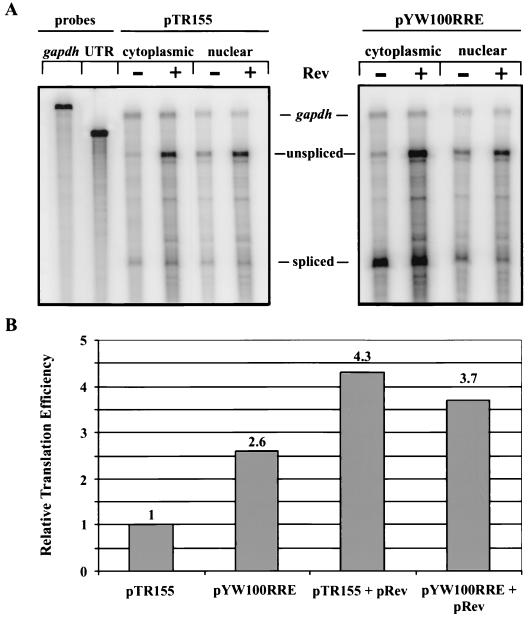
Quantitative RNA and protein analysis was used to determine whether or not sequestration to the Rev/RRE pathway abrogates SNV RU5 translational enhancement. The positive effect of Rev on cytoplasmic accumulation of the gag RNA was similar for pYW100RRE (contains SNV RU5) and pTR155 (antisense SNV RU5) RNAs, 4.5-fold and 2.9-fold, respectively (Fig. 6A; Table 4). Consistent with the results in Table 3, Rev transactivated Gag production from both reporters. Again, the magnitude of the increase was smaller in the presence of SNV RU5 than in the absence of SNV RU5 (14-fold versus 42-fold). The translational efficiency of the gag RNAs remained similar for pYW100RRE and pTR155 in the presence of Rev, 0.26 and 0.30, respectively. However, in the absence of Rev, the translational efficiency of pYW100RRE was greater by 2.6-fold (Fig. 6B). These results indicate that SNV RU5 and Rev/RRE do not function in a synergistic manner to increase the cytoplasmic expression of gag RNA. The augmentation of translational efficiency observed with RU5 alone (compare pTR155 [1.0-fold] and pYW100RRE [2.6-fold]) is abrogated upon Rev/RRE and RU5 combination (compare pTR155 plus Rev [4.3-fold] and pYW100RRE plus Rev [3.7-fold]) (Fig. 6B). Similar results were observed in replicate experiments with pYW205RRE (lacks RU5) and pYW100RRE (data not shown). The elimination of the positive effect of SNV RU5 by Rev/RRE correlates with sequestration to the LMB-inhibited CRM1/NES nuclear export pathway. A possible explanation is that SNV RU5, but not Rev/RRE, recruits a nuclear factor(s) that is necessary to program the RU5 gag RNA for translational enhancement in the cytoplasm.
FIG. 6.
RPA of nuclear and cytoplasmic RNAs from transfected 293 cells to quantify cytoplasmic RNA accumulation in response to Rev/RRE and SNV RU5. Cytoplasmic (30 μg) and nuclear (15 μg) RNAs were protected with a 32P-labeled antisense HIV-1 5′ UTR RNA probe and a GAPDH RNA probe and were visualized by PhosphorImager analysis. Labels indicate the protected transcripts. (A) RNA expressed from pYW100RRE or pTR155 in the absence or presence of Rev. (B) Translational efficiency is presented relative to that of pTR155. The ratio of Gag protein to cytoplasmic RNA is from Table 4.
TABLE 4.
Effects of Rev on protein and RNA levels
| Plasmid and transcript | Minus Rev
|
Plus Rev
|
Fold increasec | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gag concn (ng/ml) | Nuclear RNAa | Cytoplasmic RNA | TEb | Gag concn (ng/ml) | Nuclear RNA | Cytoplasmic RNA | TE | ||
| pYW100RRE | |||||||||
| Unspliced | 219 | 11,599 | 1,194 | 0.18 | 2,985 (14)d | 24,522 | 11,388 | 0.26 | 4.5 |
| Spliced | NAe | 14,236 | 10,675 | NA | NA | 12,405 | 10,417 | NA | 1.1 |
| pTR155 | |||||||||
| Unspliced | 33 | 1,399 | 452 | 0.07 | 1,391 (42) | 5,037 | 4,819 | 0.30 | 2.9 |
| Spliced | NA | 858 | 1,395 | NA | NA | 1,840 | 2,777 | NA | 0.9 |
RNA levels were determined by RNase protection assay and are expressed in PhosphorImager units and normalized to the GAPDH signal.
TE, translational efficiency (ratio of Gag protein production to cytoplasmic RNA level).
Ratio of cytoplasmic RNA to nuclear RNA in the presence and absence of Rev.
Fold increase in response to Rev.
NA, not applicable.
DISCUSSION
Our results uncover a functional linkage between the SNV RU5 nuclear export pathway and translational enhancement of intron-containing retroviral mRNA. Results of RNA transfection assays with synthetic gag and luc RNAs that are capped and polyadenylated indicate that cytoplasmic factors are insufficient for translational enhancement by SNV RU5. We observed that gag RNA introduced into the cytoplasm by RNA transfection is functional template mRNA for protein synthesis. Our data corroborate extensive studies with reporter plasmids and HIV provirus indicating that the Rev/RRE dependence of gag RNA is attributable to derepression of nuclear retention (6, 9a, 14, 26a, 37a). Consistent with previous studies, our in vitro translation data obtained with RRL and 293 cell lysates indicate that neither Rev/RRE nor SNV RU5 is necessary for Gag protein synthesis in cytoplasmic extracts (12). The positive effect of SNV RU5 was not recapitulated in the in vitro translation extracts. This negative result is consistent with a role for nuclear factors or the notion that translational regulation is not faithfully reconstituted in these cell extracts. Recently, a protocol was developed with HeLa cells for the preparation of in vitro translation extracts that faithfully recapitulate the synergistic interplay between the 5′ cap structure and the poly(A) tail during translation initiation (4). Recapitulation of this aspect of translational regulation may be necessary to execute SNV RU5 translational enhancement in vitro.
LMB was used as a tool with which to determine that SNV RU5-containing RNAs achieve nuclear export independently of the leucine-rich NES/CRM1-independent nuclear export pathway utilized by Rev/RRE. Recently, similar CRM1-independent RNA export of intron-containing RNA was reported for the Rous sarcoma virus direct repeat (DR) element, the MPMV constitutive transport element, and the hepatitis B virus posttranscriptional regulatory element (PRE) (33, 34, 45). SNV RU5 is distinct in affecting both intron-containing and intron-lacking reporter RNAs. At high concentrations of LMB (5 to 10 nM), DR element- and PRE-mediated gene expression is increased (33, 34), which is reminiscent of our results obtained with SNV RU5. Although the DR element and the PRE do not share with SNV RU5 a direct effect on translational efficiency, the DR element augments other aspects of cytoplasmic expression, including RNA stability, RNA packaging, and efficient virus assembly (2, 31, 32, 39, 40, 42). SNV RU5 and the DR element may recruit common cellular factors that provide access to a distinct CRM1-independent nucleocytoplasmic export pathway that targets the RNA for cytoplasmic localization at particular subcellular microenvironments, active translation centers in the case of SNV RU5, and virus assembly sites in the case of the DR element.
The combination of SNV RU5 and Rev/RRE on a single RNA provided a unique tool with which to investigate the relationship between utilization of a particular nuclear export pathway and translational enhancement by SNV RU5. First, our data demonstrate that Rev/RRE is significantly more potent than SNV RU5 in its magnitude of posttranscriptional activation. Second, the combination of Rev/RRE and SNV RU5 increases posttranscriptional gene expression in a less-than-additive manner. The small positive effect of the combination of SNV RU5 and Rev/RRE correlates with a small increase in cytoplasmic RNA accumulation of gag RNA and may be attributable to an increase in the stability or export of the RNA. Third, in the presence of Rev/RRE, LMB-inhibited nuclear export of RU5 gag-RRE RNA is observed to function dominantly over the LMB-enhanced pathway. Fourth, sequestration of SNV RU5 gag-RRE RNA to the LMB-enhanced pathway correlated with abrogation of translational enhancement by SNV RU5. The possibility that the translational efficiency of the reporter RNA had reached a maximum level was eliminated by the observation that the gag RNA was competent for increased amounts of Gag protein synthesis when Rev was overexpressed. Our results support the model in which interaction between SNV RU5 and a nuclear factor(s) is necessary for SNV RU5-mediated translational enhancement.
We speculate on two possible roles of nuclear factors in SNV RU5-mediated translational enhancement. First, SNV RU5 may interact with a nuclear factor that is, or recruits, a rate-limiting translation initiation factor. An attractive candidate is eIF4E because eIF4E shuttles between the nucleus and cytoplasm and is the rate-limiting initiation factor for cap-dependent translation (13). In a second model, SNV RU5 interacts with a cellular protein that resembles Rev in the ability to provide nuclear export to a particular RNA export pathway and direct polysome association. This scenario is similar to the recent suggestion that R of HFV directs HFV Gag translation by delivery of the RNA to a translationally active environment in the cytoplasm (37). While Rev functions cotranscriptionally (23) to activate nuclear export by the leucine-rich NES/CRM1-dependent pathway and selectively drives intron-containing gag RNA along a particular cytoplasmic circuit, the hypothetical Rev-like factor would bind SNV RU5 and facilitate export and polysome association along a different cytoplasmic circuit. The dominance of Rev/RRE may be attributable to cotranscriptional interaction of Rev/RRE and host proteins at a point upstream of interaction between SNV RU5 and an RU5-interactive protein(s). We note that SNV RU5 confers LMB enhancement in the context of either the SNV U3 or CMV immediate-early promoter/enhancer, indicating that recruitment of RU5-interactive factors is not strictly dependent on cotranscriptional recruitment by the SNV promoter. SNV, HFV, and MPMV are divergent retroviruses, but they share 5′ RNA elements that may interact with a common nuclear factor(s) that programs the intron-containing RNA for productive cytoplasmic utilization.
Acknowledgments
We thank Minoru Yoshida (University of Tokyo, Tokyo, Japan) for the gift of LMB, Paul Copeland (Cleveland Clinic, Cleveland, Ohio) for expert advice on preparation of 293 cell translation lysates, and Jennifer Frey for plasmid construction. We are grateful to Michael Lairmore for comments on the manuscript and Patrick Green for discussion.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R29AI40851) and the National Cancer Institute (P30CA16058), Bethesda, Md.
REFERENCES
- 1.Arrigo, S. J., and I. S. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 5:808-819. [DOI] [PubMed] [Google Scholar]
- 2.Aschoff, J. M., D. Foster, and J. M. Coffin. 1999. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J. Virol. 73:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. D., K. L. Beemon, and M. L. Linial. 1998. RNA regulatory elements in the genomes of simple retroviruses. Semin. Virol. 8:194-204. [Google Scholar]
- 4.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boris-Lawrie, K., T. M. Roberts, and S. Hull. 2001. Retroviral RNA elements integrate components of post-transcriptional gene expression. Life Sci. 69:2697-2709. [DOI] [PubMed] [Google Scholar]
- 6.Brighty, D. W., and M. Rosenberg. 1994. A cis-acting repressive sequence that overlaps the Rev-responsive element of human immunodeficiency virus type 1 regulates nuclear retention of env mRNAs independently of known splice signals. Proc. Natl. Acad. Sci. USA 91:8314-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butsch, M., S. Hull, Y. Wang, T. M. Roberts, and K. Boris-Lawrie. 1999. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 73:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, R., and J. Lucas-Lenard. 1993. Preparation of a cell-free translation system with minimal loss of initiation factor eIF-2/eIF-2B activity. Anal. Biochem. 212:17-23. [DOI] [PubMed] [Google Scholar]
- 9.Cochrane, A. W., C. H. Chen, and C. A. Rosen. 1990. Specific interaction of the human immunodeficiency virus Rev protein with a structured region in the env mRNA. Proc. Natl. Acad. Sci. USA 87:1198-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cochrane, A. W., K. S. Jones, S. Beidas, P. J. Dillon, A. M. Skalka, and C. A. Rosen. 1991. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65:5305-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 2000. Connections between the processing and nuclear export of mRNA: evidence for an export license? Proc. Natl. Acad. Sci. USA 97:4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Agostino, D. M., B. K. Felber, J. E. Harrison, and G. N. Pavlakis. 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 12:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostie, J., M. Ferraiuolo, A. Pause, S. A. Adam, and N. Sonenberg. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155-1165. [DOI] [PubMed] [Google Scholar]
- 15.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, U., V. W. Pollard, R. Luhrmann, M. Teufel, M. W. Michael, G. Dreyfuss, and M. H. Malim. 1999. Rev-mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle. Nucleic Acids Res. 27:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 18.Geballe, A. P., and M. K. Gray. 1992. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 20:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadzopoulou-Cladaras, M., B. K. Felber, C. Cladaras, A. Athanassopoulos, A. Tse, and G. N. Pavlakis. 1989. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 63:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammarskjöld, M. L., J. Heimer, B. Hammarskjöld, I. Sangwan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heaphy, S., C. Dingwall, I. Ernberg, M. J. Gait, S. M. Green, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 60:685-693. [DOI] [PubMed] [Google Scholar]
- 22.Hemmings-Mieszczak, M., G. Steger, and T. Hohn. 1998. Regulation of CaMV 35S RNA translation is mediated by a stable hairpin in the leader. RNA 4:101-111. [PMC free article] [PubMed] [Google Scholar]
- 23.Iacampo, S., and A. Cochrane. 1996. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, T., I. Hashimoto, M. Nishikawa, and J. I. Fujisawa. 1996. A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal beta-actin and viral protein expression. Biochimie 78:1075-1080. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, T., I. Hashimoto, A. Yamamoto, M. Nishikawa, and J. I. Fujisawa. 2000. Rev-dependent association of the intron-containing HIV-1 gag mRNA with the nuclear actin bundles and the inhibition of its nucleocytoplasmic transport by latrunculin-B. Genes Cells 5:289-307. [DOI] [PubMed] [Google Scholar]
- 26a.Maldarelli, F., M. A. Martin, and K. Strebel. 1991. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J. Virol. 65:5732-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, J. B., A. W. Cochrane, C. V. Johnson, A. Perkins, and C. A. Rosen. 1991. The HIV-1 Rev protein: a model system for coupled RNA transport and translation. New Biol. 3:1220-1232. [PubMed] [Google Scholar]
- 27.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 28.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104:173-176. [DOI] [PubMed] [Google Scholar]
- 29.Miele, G., A. Mouland, G. P. Harrison, E. Cohen, and A. M. Lever. 1996. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 70:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 31.Ogert, R. A., and K. L. Beemon. 1998. Mutational analysis of the Rous sarcoma virus DR posttranscriptional control element. J. Virol. 72:3407-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 70:3834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otero, G. C., M. E. Harris, J. E. Donello, and T. J. Hope. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 72:7593-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paca, R. E., R. A. Ogert, C. S. Hibbert, E. Izaurralde, and K. L. Beemon. 2000. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 74:9507-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, T. M., and K. Boris-Lawrie. 2000. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 74:8111-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen, C. A., E. Terwilliger, A. Dayton, J. G. Sodroski, and W. A. Haseltine. 1988. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 85:2071-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, R. A., Y. Zeng, O. Erlwein, B. R. Cullen, and M. O. McClure. 2001. The R region found in the human foamy virus long terminal repeat is critical for both Gag and Pol protein expression. J. Virol. 75:6817-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 66:150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, S. B., W. Guo, S. C. Winistorfer, R. C. Craven, and C. M. Stoltzfus. 1998. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology 247:86-96. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, S. B., L. Zhang, R. C. Craven, and C. M. Stoltzfus. 1997. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J. Virol. 71:9150-9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, A. J., M. I. Cho, M. L. Hammarskjold, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorge, J., W. Ricci, and S. H. Hughes. 1983. cis-acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J. Virol. 48:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoneley, M., T. Subkhankulova, J. P. Le Quesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES: a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 45.Zang, W. Q., and T. S. Yen. 1999. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology 259:299-304. [DOI] [PubMed] [Google Scholar]
- 46.Zapp, M. L., and M. R. Green. 1989. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature 342:714-716. [DOI] [PubMed] [Google Scholar]