Abstract
The human immunodeficiency virus type 1 (HIV-1) exterior envelope glycoprotein gp120 mediates receptor binding and is the major target for neutralizing antibodies. A broadly neutralizing antibody response is likely to be a critical component of the immune response against HIV-1. Although antibodies against monomeric gp120 are readily elicited in immunized individuals, these antibodies are inefficient in neutralizing primary HIV-1 isolates. As a chronic pathogen, HIV-1 has evolved to avoid an optimal host response by a number of immune escape mechanisms. Monomeric gp120 that has dissociated from the functional trimer presents irrelevant epitopes that are not accessible on functional trimeric envelope glycoproteins. The resulting low level of antigenic cross-reactivity between monomeric gp120 and the functional spike may contribute to the inability of monomeric gp120 to elicit broadly neutralizing antibodies. Attempts to generate native, trimeric envelope glycoproteins as immunogens have been frustrated by both the lability of the gp120-gp41 interaction and the weak association between gp120 subunits. Here, we present solid-phase HIV-1 gp160ΔCT (cytoplasmic tail-deleted) proteoliposomes (PLs) containing native, trimeric envelope glycoproteins in a physiologic membrane setting. We present data that indicate that the gp160ΔCT glycoproteins on PLs are trimers and are recognized by several relevant conformational ligands in a manner similar to that for gp160ΔCT oligomers expressed on the cell surface. The PLs represent a significant advance over present envelope glycoprotein formulations as candidate immunogens for HIV vaccine design and development.
The human immunodeficiency virus type 1 (HIV-1) exterior envelope glycoprotein gp120 and the transmembrane glycoprotein gp41 facilitate virus binding and entry into susceptible target cells (47). The envelope proteins are initially synthesized as highly glycosylated gp160 precursor proteins that oligomerize in the endoplasmic reticulum. After transport to the Golgi apparatus, the cellular protease furin cleaves gp160 into gp120 and gp41 (16). The envelope proteins remain associated through hydrophobic, noncovalent interactions. The mature envelope glycoproteins are transported to the cell surface and from there are incorporated into budding virions (14, 32). Due to the labile gp120-gp41 interaction, a substantial amount of gp120 dissociates from the oligomeric envelope glycoprotein complex (26).
Many lines of evidence suggest that gp120 and gp41 heterodimers form trimers on the viral surface. The HIV-1 ectodomain of gp41 crystallizes as a trimeric coiled coil with interdigitating alpha helices to form a six-helix bundle (8, 38, 44). The trimeric structure of the complete simian immunodeficiency virus (SIV) gp41 ectodomain has been solved by nuclear magnetic resonance (7). The fusion-active or postfusogenic state of HIV-1 and SIV gp41 proteins defined in these studies closely resembles that of the corresponding transmembrane envelope proteins from a number of viruses such as influenza virus (6) and Ebola virus (43). Each of these fusion determinants has been crystallized as helical bundles possessing trimeric coiled-coil motifs. The matrix proteins of HIV and SIVs that interact with gp41 crystallize as trimers (17). The gp160 ectodomain from SIV (gp140) has been shown previously to be trimeric by biophysical analysis (9). Trimerization has also been documented elsewhere for a number of HIV-1 gp120-gp41/gp140 ectodomain constructs (4, 48, 49).
HIV-1 is tropic for cells that express the viral receptor, CD4, and second receptors that belong to the family of the G-protein-coupled, seven-membrane-spanning chemokine receptor proteins (10-12). Binding of gp120 to CD4 induces conformational changes in gp120 that facilitate subsequent binding to the chemokine receptor (41, 46). These events are believed to lead to further conformational rearrangements that expose the gp41 fusion domain, allow for fusion of the viral and cellular membranes, and permit entry into the target cell (47).
In the course of HIV infection, neutralizing antibodies to the envelope glycoproteins are elicited and appear to be an important component of the host immune response. The level of circulating neutralizing antibodies correlated with protection against viral challenge in several animal models (3, 5). Passive immunization with neutralizing antibodies has also been demonstrated previously to protect the host from the establishment of viral infection when administered prior to exposure of the host to HIV-1 (1, 19). While several antibodies effectively neutralize virus that has been adapted to replicate in T-cell lines (TCLA), most clinical, primary isolates are relatively resistant to these antibodies, suggesting that those viruses have been selected in vivo by the presence of neutralizing antibodies. In most infected individuals, two classes of neutralizing antibodies can be distinguished, strain-restricted and broadly neutralizing antibodies. The strain-restricted antibodies are generally directed toward epitopes in the second variable (V2) or third variable (V3) loop of gp120 and appear early during infection (31, 34). These antibodies exhibit only homologous neutralization activity. The broadly neutralizing antibodies appear later, following the establishment of chronic infection (36). These are mostly directed to conformational epitopes, some of which, the CD4 binding site (CD4BS) antibodies, recognize the discontinuous CD4BS and interfere with gp120-CD4 binding (28, 33, 40). Another group of antibodies that neutralize TCLA isolates and a subset of primary isolates (S. H. Xiang, N. Doka, R. K. Choudhary, J. Sodroski, and J. Robinson, unpublished observations) recognize CD4-induced (CD4i) epitopes. These antibodies bind near the gp120 bridging sheet and interfere with gp120-chemokine receptor interaction (18, 39, 46).
Presumably due to its exposed location on the surface of the virus, the gp120 glycoprotein is the major target for neutralizing antibodies. The gp41 glycoprotein also contains one well-defined neutralizing determinant proximal to the membrane-spanning region (22, 29). As a chronic pathogen, HIV-1 has evolved a number of immune escape mechanisms to mitigate the antiviral effects of the host immune response. Monomeric gp120 shed from oligomeric glycoprotein complexes has been suggested previously to be a major immunogen in vivo, eliciting high titers of nonneutralizing antibodies by exposing epitopes that are buried or not formed on the functional trimeric spike (13, 23, 45). The gp120-gp41 interface, for instance, contains immunodominant surfaces on gp120 and gp41 that are not accessible on the functional trimer and therefore not targets for neutralizing antibodies (24, 25, 42). The poor immunogenic potential of gp120 to elicit neutralizing antibodies is also reflected in studies that demonstrate that the relatively rare neutralizing antibodies bind preferentially to oligomeric envelope glycoproteins. No such preference exists for the binding of neutralizing antibodies to monomeric gp120 (15, 30, 35). It is likely that both gp120 and gp41 sample a different range of conformations in the dissociated state from those assumed by the assembled trimer. Recent thermodynamic data suggest that CD4 binding stabilizes the gp120 molecule, whereas free, unbound gp120 is very flexible and is able to sample many conformations (23). As similar results were obtained following deletion of variable regions V1 to V3 and elements of C1 and C5 of gp120, flexibility likely alters the spatial relationship of the gp120 core domains. These core domains consist of an inner domain that interacts with gp41, an outer domain exposed on the trimer, and a bridging sheet. Discontinuous, conserved neutralization epitopes on gp120 related to receptor binding span all three gp120 core domains (47). Thus, the interdomain flexibility could explain in part why potent, broadly neutralizing CD4BS or CD4i antibodies have been difficult to elicit with monomeric gp120. Although the entropic state of gp120 in the context of an assembled trimer is currently unknown, possible differences in the conformations sampled by monomeric and trimeric envelope glycoproteins justify attempts to create immunogens that more accurately mimic the functional envelope glycoprotein spike.
Various strategies have been employed to generate bona fide mimics of oligomeric envelope glycoprotein as found on the viral surface. Soluble gp140 glycoproteins containing the gp41 ectodomain covalently linked to gp120 have been generated by several groups (4, 48). At least two different strategies have been employed to accomplish a stable gp120-gp41 linkage: mutation-deletion of the native gp120-gp41 cleavage site in gp160 and cysteine linkage of gp120 to the gp41 ectodomain while maintaining an intact, functional cleavage site. Additional modifications, such as appending a heterologous trimeric coiled-coil structure (GCN4) to the gp140 cleavage-defective glycoproteins, appeared necessary to maintain trimer stability. In a recent report, soluble trimeric envelope glycoproteins derived from the primary isolate YU2 (gp140-GCN4 YU2 glycoproteins) elicited neutralizing antibodies more effectively than did monomeric YU2 gp120. These molecules elicited antibodies that exhibited limited neutralization of heterologous virus in vitro. Here we describe the creation of solid-phase gp160ΔCT proteoliposomes (PLs) as an approach to generate better mimics of the functional HIV-1 envelope glycoprotein trimer. This approach requires elimination of the gp120-gp41 cleavage site but no other modifications of the envelope glycoprotein ectodomain. An intact gp41 transmembrane region is maintained, allowing the trimeric envelope glycoprotein complex to be embedded in a reconstituted lipid bilayer in a natural context. We present data that strongly suggest that this approach faithfully recapitulates the oligomeric structure found on the surface of HIV-1 envelope glycoprotein-expressing cells.
MATERIALS AND METHODS
Envelope glycoprotein constructs.
The envelope glycoprotein constructs were derived from the primary R5 HIV-1 isolates YU2 and JR-FL and the X4, TCLA isolate HXBc2. The coding sequences for the YU2 envelope glycoprotein were obtained from the pSVIIIenvYU2 expression plasmid. The JR-FL envelope glycoprotein coding sequence, which contains a CD5 heterologous leader sequence in place of the normal JR-FL leader, was obtained from the NIH AIDS Research and Reference Reagent Program and subcloned into the pcDNA3.1(−) (Invitrogen) expression plasmid to include transmembrane region and cytoplasmic tail sequences. The HXBc2 construct was codon optimized for mammalian expression using overlapping primers and PCR and subcloned into pcDNA3.1(+) (Invitrogen). A sequence coding for the heterologous CD5 signal sequence was subcloned to replace the endogenous HXBc2 leader sequence. For all constructs, the cytoplasmic tail truncation was generated by introduction of a stop codon in place of the codon for amino acid 712 and the sequence coding for the C9 tag TETSQVAPA was added according to the QuikChange protocol immediately before the stop codon. To create covalently linked gp120-gp41 glycoproteins, the proteolytic cleavage site between gp120 and gp41 was disrupted by replacing arginines 508 and 511 with serines by QuikChange site-directed mutagenesis. These modifications resulted in constructs encoding cleavage-deficient gp160ΔCT envelope glycoproteins used to generate the PLs. Amino acid residue numbers are designated according to the prototypic HXBc2 sequence. The constructs were sequenced, and introduction of the desired mutations was confirmed by this method.
Envelope glycoprotein expression.
Plasmids expressing the gp160ΔCT glycoproteins (2 μg of DNA per 100-mm-diameter dish of cells) were cotransfected into 293T cells with (YU2) or without (JR-FL and HXBc2) the HIV-1 Tat expressor plasmid pSVTat (0.5 μg), using Effectene reagent (Qiagen) following the manufacturer's protocol. Thirty-six hours after transfection, cells expressing the envelope glycoproteins were harvested with phosphate-buffered saline (PBS) containing 5 mM EDTA.
Coating of M-450 Dynabeads.
Tosyl-activated Dynabeads (Dynal, Inc., Lake Success, N.Y.) were conjugated with the 1D4 antibody (National Cell Culture Center, Minneapolis, Minn.) according to the manufacturer's protocol. The 1D4 murine antibody recognizes the C9 epitope tag and was used to capture the envelope glycoproteins on the Dynal beads.
Preparation of lipid solutions for membrane reconstitution.
Lipids were obtained as chloroform solutions from Avanti Polar Lipids (Alabaster, Ala.). The following lipids were used: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and dimyristoylphosphatidic acid (DMPA) and cholesterol at molar concentrations 45:25:20:10. The lipid mixture was dried in a 2-ml polyethylene tube under a vacuum until all of the solvent was removed. PBS was added to the tube, and a liposomal solution was obtained by ultrasonication for 5 min in an ice bath with an Ultrasonic Processor (Heat Systems, Inc., Farmingdale, N.Y.). Liposomal solutions of the head group-modified synthetic lipids 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-n-(biotinyl) (Biotinyl-DOPE) and dioleoylphosphoethanolamine-lissamine rhodamine B (Rho-DOPE), at a final concentration of 1 mg/ml, were prepared separately using the same protocol.
Formation of PLs.
The formation of PLs is diagrammed in Fig. 1. For the preparation of 4 × 108 PLs, approximately 2 × 107 gp160ΔCT-expressing 293T cells were lysed in 5 ml of solubilization buffer [100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.5), 1% (wt/vol) Cymal-5, and protease inhibitor mixture (one tablet of Complete [Boehringer Mannheim] per 50 ml)] at 4°C for 30 min on a rocking platform. Cell debris was pelleted by centrifugation for 30 min at 13,000 × g. The cleared lysate was incubated with 4 × 108 1D4-conjugated Dynal beads for 16 h at 4°C on a rocking platform. After recovery of the beads, they were extensively washed in solubilization buffer. For the formation of the lipid membrane, beads coated with gp160ΔCT glycoprotein were incubated for 15 min at room temperature with 1 ml of solubilization buffer containing 2 mg of lipid mixture and, if fluorescence labeling or biotinylation was desired, with 1% Rho-DOPE or biotin-phycoerythrin (PE), respectively. The detergent was then slowly removed by dialysis for 24 h at 4°C against PBS, using a 10-kDa-molecular-mass cutoff dialysis membrane (Slide-A-Lyzer 10 K [Pierce, Rockford, Ill.]). The excess of unbound lipid and residual detergent was removed on a magnetic separator in one washing step with PBS. PLs were stored in PBS with 0.1% bovine serum albumin and 0.1% Na2N3 at 4°C for up to 3 months.
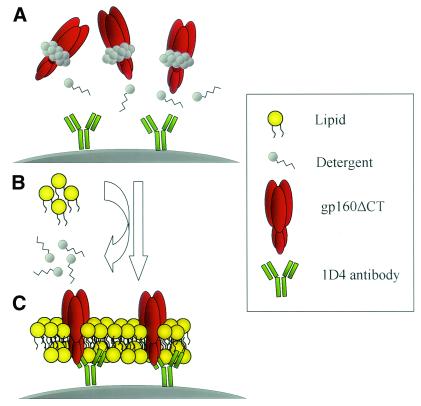
FIG. 1.
Schematic diagram of the formation of PLs. (A) Beads conjugated with the 1D4 antibody recognizing the C9 tag were used to capture the C9-tagged gp160ΔCT glycoproteins from cell lysates. The beads were then washed extensively in detergent-containing buffer to remove non-C9-tagged proteins. (B) The beads were incubated with detergent-solubilized lipid and dialyzed. (C) During dialysis in PBS, the detergent is replaced by a lipid membrane which assembles around the gp160ΔCT glycoproteins.
Analysis of protein composition of the PLs.
Approximately 5 × 107 PLs and control beads were incubated at 100°C in reducing sodium dodecyl sulfate (SDS) sample buffer for 5 min, separated on an SDS-7.5% polyacrylamide gel, and stained with Coomassie blue (Fig. 2A). For the analysis of fractions eluted by molecular exclusion, Western blotting was performed under either nonreducing or reducing conditions. To determine that gp120 was present in the eluted protein peaks, samples from each peak were incubated for 5 min at 100°C in sample buffer containing 2% β-mercaptoethanol (BME) and separated on an SDS-7.5% polyacrylamide gel (Fig. 3B). To confirm that oligomeric forms of envelope glycoproteins could be detected in the high-molecular-weight peaks, the fractions were diluted in sample buffer lacking BME and separated on an SDS-3 to 8% polyacrylamide gel. A sample from the peak consistent with a trimer (fraction 3, Fig. 3C) was also analyzed in the presence of BME. Subsequently, proteins were electrophoretically transferred onto a 0.45-μm-pore-size Hybond-C Extra membrane (Amersham). The gp160ΔCT glycoproteins present in each column fraction were then detected by anti-gp120 rabbit serum and anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (Sigma).
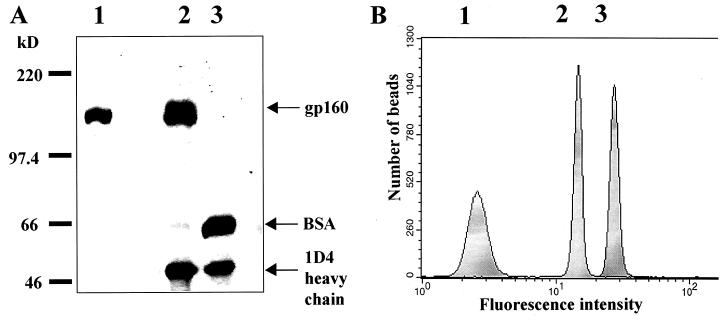
FIG. 2.
Protein composition of PLs. (A) SDS-polyacrylamide gel of gp160ΔCT glycoproteins eluted from 5 × 107 PLs, stained with Coomassie blue. Lane 1, 1 μg of affinity-purified gp160; lane 2, protein eluted from gp160ΔCT PLs; lane 3, protein eluted from beads conjugated with the 1D4 antibody. BSA, bovine serum albumin. (B) FACS analysis of PLs stained with the IgGb12 antibody (peak 2) and HIV-1 patient serum (peak 3), compared to anti-human FITC-conjugated secondary antibody alone (peak 1).
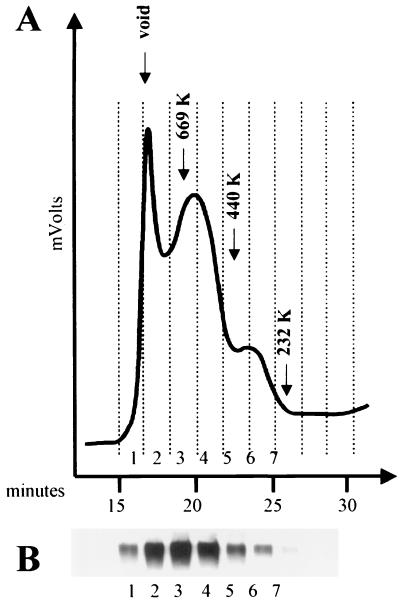
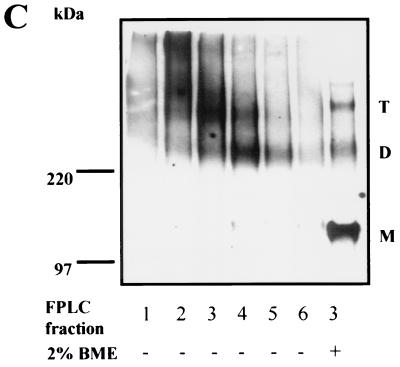
FIG. 3.
Size-exclusion chromatography and Western blotting of JR-FL gp160ΔCT glycoproteins eluted from Dynal beads under native conditions. 293T cells transiently expressing the JR-FL gp160ΔCT C9-tagged glycoproteins were lysed in CHAPS-containing buffer and incubated with Dynal beads conjugated with the 1D4 antibody. Beads were then washed and incubated in buffer containing 0.2 mM C9 peptide and 0.5 M MgCl2 to elute the gp160ΔCT glycoprotein from the beads. (A) Approximately 5 μg of JR-FL gp160ΔCT glycoproteins was analyzed on a Superdex-200 gel filtration column. (B) Eluted fractions were collected, analyzed by SDS-PAGE under reducing conditions (2% BME and 100°C), and analyzed by Western blotting with a polyclonal anti-gp120 rabbit serum. (C) Eluted fractions were analyzed on an SDS-3 to 8% polyacrylamide gradient gel under nonreducing conditions (minus BME and 37°C) and under reducing conditions (2% BME and 100°C), respectively, and detected by Western blotting using a polyclonal anti-gp120 rabbit serum. Protein bands of apparent molecular masses consistent with trimeric gp160ΔCT glycoproteins (T), dimeric glycoproteins (D), and monomeric glycoproteins (M) are marked as indicated. FPLC, fast protein liquid chromatography.
Molecular exclusion chromatography.
The gp160ΔCT glycoproteins captured onto Dynal beads were eluted from the beads for molecular exclusion chromatography under nondenaturing conditions as follows. The beads were incubated in 0.5 M MgCl2-1% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate)-0.2 mM C9 peptide (peptide sequence, TETSQVAPA) at 37°C for 30 min. Approximately 5 μg of eluted gp160ΔCT glycoproteins was loaded onto a Superdex 200 column (Amersham Pharmacia Biotech) in a 200-μl volume. The column was then eluted with PBS containing 1% CHAPS at a rate of 0.5 ml/min for 40 min. The eluted protein was detected by measuring the optical density at 280 nm using a Varian ProStar System (Varian Analytical Instruments). Fractions of the flowthrough were collected at 2-min intervals using a Varian Dynamax fraction collector. The fractions were further analyzed by reducing and nonreducing SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting with a polyclonal anti-gp120 rabbit serum for detection of HIV-1 envelope glycoproteins (Fig. 3B and C). A mixture of high-molecular-weight protein markers (Amersham Pharmacia Biotech) was eluted under identical conditions to calibrate the column.
Flow cytometric analysis of gp160ΔCT on PLs and 293T cells.
For the comparison of antibody binding to either cleavage-defective or cleavage-competent gp160ΔCT glycoproteins, 293T cells were transfected with plasmids expressing the two envelope glycoprotein variants. Approximately 106 cells per sample were harvested with PBS containing 5 mM EDTA and washed once in fluorescence-activated cell sorting (FACS) buffer (PBS, 2% fetal calf serum, 0.02% Na2N3). The cells were incubated for 1 h at room temperature with the indicated amounts of antibodies in a volume of 100 μl. After two washing steps in FACS buffer, the cells were incubated for 30 min with an R-PE-conjugated F(ab′)2 goat anti-human antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.), washed twice, and analyzed with a FACScan flow cytometer and CellQuest software (Becton Dickinson, San Jose, Calif.). For FACS analysis of PLs, staining was performed as described above. Staining for membrane integrity of the PLs was performed with PE-conjugated goat anti-mouse IgG (Boehringer Mannheim, Indianapolis, Ind.) or avidin-fluorescein isothiocyanate (FITC) (Sigma). The following ligands were used for staining of the envelope glycoproteins: soluble CD4 (sCD4), the potently neutralizing CD4BS antibody IgG1b12 (kindly provided by Dennis Burton); the F105 CD4BS antibody (kindly provided by Marshal Posner); the strain-restricted neutralizing V3 loop antibody 39F, the nonneutralizing C1-C4 antibody A32, the nonneutralizing C1-C5 antibody C11, and the CD4-induced 17b antibody (all kindly provided by James Robinson); and the broadly neutralizing gp41 antibody 2F5 (kindly provided by Hermann Katinger).
RESULTS
Creation of gp160ΔCT PLs.
Paramagnetic PLs containing the HIV-1 envelope glycoprotein were created according to methods established for solubilization and membrane reconstitution of the CCR5 chemokine receptor (20). Briefly, cells transiently expressing the HIV-1 YU2, JR-FL, or HXBc2 gp160ΔCT glycoproteins, which contain an alteration of the gp120-gp41 cleavage site and deletion of the gp41 cytoplasmic tail, were lysed in buffer containing Cymal-5 detergent. The gp160ΔCT glycoproteins were then captured onto Dynal beads conjugated to the 1D4 antibody, which recognizes a C9 peptide tag affixed to the gp160ΔCT C terminus. Following addition of membrane lipids and dialysis of the Cymal-5 detergent, an artificial lipid bilayer is formed around the bead surface. Thus, pure, properly oriented HIV-1 envelope glycoproteins, embedded in a natural membrane environment, were incorporated into an easily manipulable solid support.
Analysis of PL protein composition.
The gp160ΔCT PLs were boiled in sample buffer and analyzed by reducing SDS-PAGE to determine their protein composition. As a positive control, cell lysates containing transiently expressed gp160ΔCT glycoproteins were precipitated with the F105 anti-gp120 antibody and protein A-Sepharose and analyzed in parallel. PLs lacking the gp160ΔCT glycoproteins were treated similarly and served as a negative control. As shown in Fig. 2A, a band migrating at a position similar to that of the gp160ΔCT glycoprotein positive control band was observed among the proteins released from the gp160ΔCT PLs. No such band was observed in this molecular weight range in the gp160ΔCT glycoprotein-deficient PL control sample. Apart from a 50-kDa band corresponding to the 1D4 antibody heavy chain, a light chain band not retained on the gel, and the gp160ΔCT band, only minor impurities were detected in the gp160ΔCT PLs. The total amount of gp160ΔCT glycoproteins captured onto 5 × 107 PLs was estimated to be 1 to 2 μg, as determined by the purified recombinant gp160ΔCT glycoprotein control of known concentration.
To examine the exposure of the HIV-1 envelope glycoproteins on the PL surface, the gp160ΔCT PLs were stained with the IgG1b12 anti-gp120 antibody and a mixture of sera from HIV-1-infected individuals and analyzed by FACS. The IgG1b12 antibody recognizes a conformation-dependent gp120 epitope near the CD4BS. The forward scatter versus sideward scatter plot showed mostly single PLs. Doubles and other multiples of PLs were generally observed to be less than 20% of the total events (data not shown). A gate was created to analyze only single PLs and was used for all further FACS analyses. The narrow distribution of the fluorescence intensity associated with each FACS peak following antibody staining suggests that the gp160ΔCT PLs have nearly uniform protein content (Fig. 2B).
Characterization of the size of the gp160ΔCT PL envelope glycoproteins.
The gp160ΔCT glycoproteins were eluted from the reconstituted PLs under nondenaturing conditions by incubation with 0.2 mM C9 peptide in the presence of 1% CHAPS detergent and 0.5 M NaCl. The eluted envelope glycoproteins were analyzed by size-exclusion chromatography on a Superdex 200 column equilibrated in PBS-1% CHAPS buffer. The chromatogram for the HIV-1 JR-FL gp160ΔCT envelope glycoproteins is shown in Fig. 3A. Parallel studies using the YU2 gp160ΔCT glycoproteins yielded a similar profile (data not shown). The fractions of the resolved JR-FL glycoproteins were collected and analyzed by SDS-PAGE and Western blotting (Fig. 3B).
The column was calibrated with molecular mass standards, allowing the apparent molecular size of the major peak to be approximated as 580 kDa. As the gp120 glycoprotein monomer was resolved by size-exclusion chromatography with an apparent molecular mass of 180 kDa (data not shown and reference 48), a mass of 580 kDa is consistent with that of a trimeric gp160ΔCT envelope glycoprotein complex. The fastest-migrating protein peak was detected just after the void volume and apparently consisted of gp160ΔCT glycoprotein aggregates as determined by its migration pattern in the column and by the Western blotting results (Fig. 3). This aggregate peak has previously been observed in molecular exclusion chromatography of soluble GCN4-stabilized gp140 trimers (48). Most of the gp160ΔCT glycoprotein eluted in fractions 3 and 4, with fraction 3 corresponding to the mass of 580 kDa. When all of the gel filtration fractions were subjected to nonreducing SDS-PAGE (with pretreatment in gel loading buffer at 37oC) followed by Western blotting, fractions 3 and 4, corresponding to trimeric envelope glycoproteins, were found to separate into trimers, dimers, and monomers. The greatest number of gel-stable trimeric glycoproteins was detected in fraction 3 (Fig. 3C). Most of the glycoproteins migrating with molecular weights corresponding to trimers in this fraction could be nearly totally reduced to a monomeric gp160ΔCT band by treatment with 2% BME and boiling, although bands migrating in a manner consistent with trimers and dimers could still be observed (Fig. 3C).
Characterization of the PL membrane.
The formation of the lipid membrane was examined by FACS analysis and fluorescence microscopy. According to our model, the murine 1D4 capture antibody would be expected to be partially occluded by a reconstituted lipid membrane (Fig. 1). Therefore, binding of anti-mouse IgG antibody should be impaired on PLs compared to that on beads without a membrane. A more than threefold decrease of anti-mouse-PE signal could be observed on PLs containing a reconstituted lipid membrane compared with that on beads without lipid reconstitution (Fig. 4A, peak 1 compared to peak 2), indicating the presence of at least a partial lipid membrane.
FIG. 4.
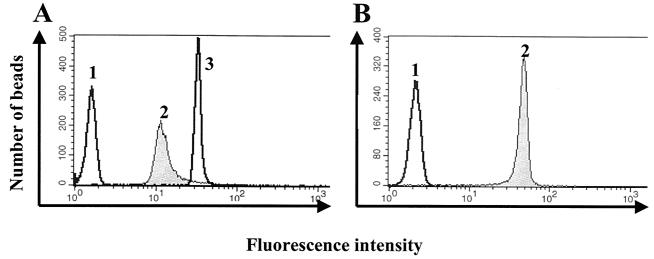
FACS analysis of the reconstituted PL membrane. (A) Occlusion of the 1D4 antibody by lipid membrane reconstitution. gp160ΔCT PLs with (peak 2) and without (peak 3) a reconstituted membrane were probed with anti-mouse Ig-PE antibody. Peak 1 shows staining with the same antibody of nonconjugated beads. (B) PLs with a reconstituted membrane containing 1% biotinylated lipid (peak 2) and beads without a reconstituted membrane (peak 1) were probed with avidin-FITC.
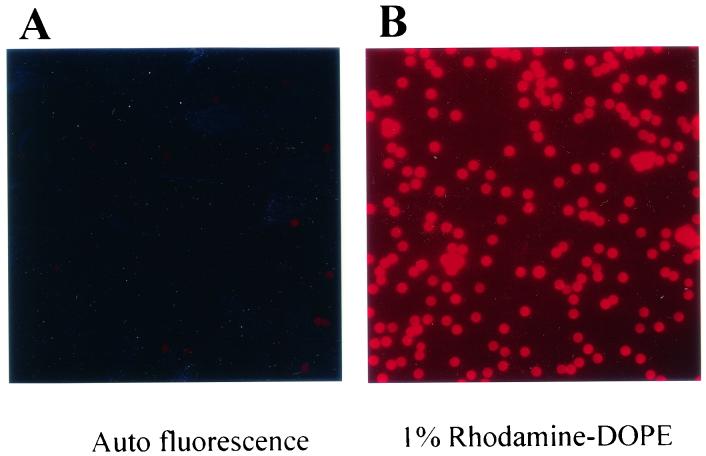
The presence of the lipid membrane was also examined by FACS analysis after reconstitution of the membrane with biotinylated lipid at 1% (wt/wt) total lipids. The PLs were stained with avidin-FITC and showed a ≥20-fold-higher signal on fully reconstituted PLs compared to that on gp160ΔCT glycoprotein-containing beads without lipid reconstitution (Fig. 4B). We then confirmed the presence of a lipid bilayer by visualizing the incorporation of rhodamine-conjugated lipid (rhodamine-DOPE) into the PL membrane by fluorescence microscopy (Fig. 5). Bright fluorescence could be observed in the rhodamine-labeled PLs (Fig. 5B) compared to negligible background fluorescent emission with untreated beads (Fig. 5A).
FIG. 5.
Fluorescence microscopic images of unconjugated beads (A) and PLs reconstituted with a membrane containing 1% DOPE-rhodamine (B).
2F5 antibody binding to gp160ΔCT PLs.
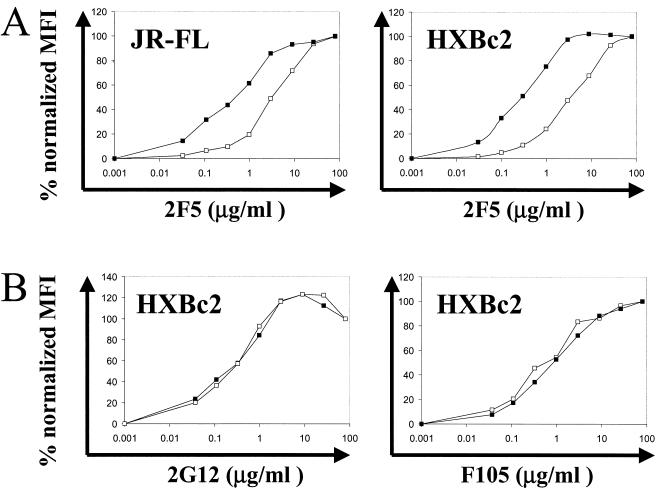
The epitope of the neutralizing antibody 2F5, ELDKWAS, is situated proximal to the viral membrane in the gp41 ectodomain. To determine the influence of the reconstituted membrane on this important neutralizing determinant, we used the 2F5 antibody to probe PLs reconstituted with a membrane and PLs coated with gp160ΔCT glycoprotein but devoid of a membrane. Since sequence variation in the HIV-1 YU2 strain alters 2F5 antibody recognition, PLs with gp160ΔCT glycoproteins derived from the primary isolate JR-FL and the TCLA isolate HXBc2 were examined. The 2F5 antibody bound to HXBc2 gp160ΔCT glycoprotein PLs with a reconstituted membrane with a roughly 10-fold-higher affinity than it did to HXBc2 gp160ΔCT glycoprotein on beads without a membrane (Fig. 6A). The same could be observed with JR-FL PLs, although the effect was less pronounced (Fig. 6A). To show the specificity of the enhanced 2F5 binding observed in the presence of the reconstituted membrane, binding of the gp120-specific antibodies F105 and 2G12 to gp160ΔCT PLs with and without a membrane was examined. No affinity difference could be detected using these antibodies (Fig. 6B).
FIG. 6.
(A) Binding of the gp41 antibody 2F5 to gp160ΔCT from HXBc2 (right) and JR-FL (left) on beads without a membrane (open squares) and fully reconstituted PLs (closed squares). PLs and beads were probed with increasing concentrations of 2F5 antibody and anti-human IgG-PE antibody, respectively, and analyzed by FACS. The mean fluorescence intensity (MFI) was plotted as percent maximal MFI at the given antibody concentration. (B) Binding of the antibodies 2G12 (left) and F105 (right) to the PLs (closed squares) and beads (open squares) was performed as described for 2F5 binding above.
Antigenic characterization of the gp160ΔCT PLs.
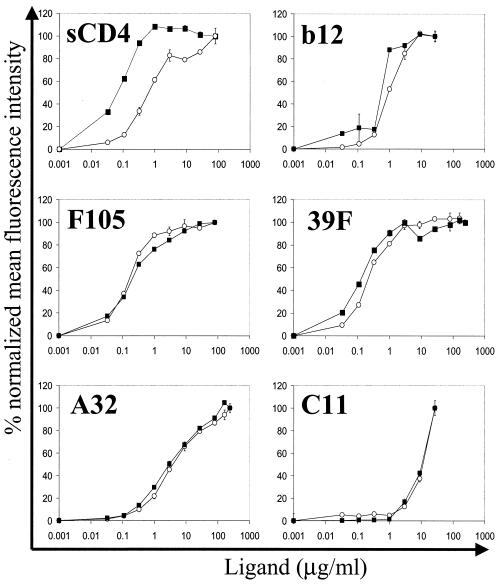
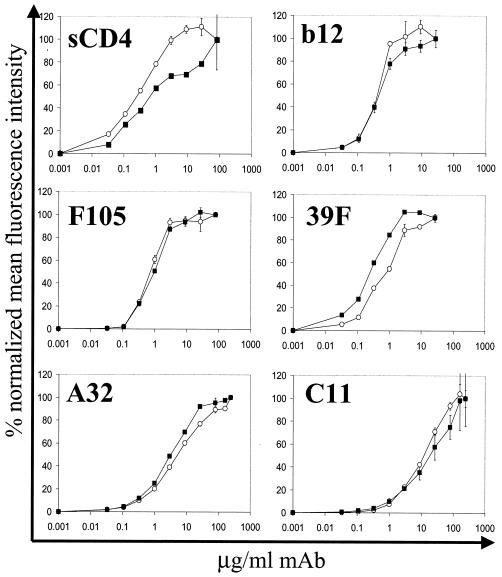
To confirm the conformational integrity of the gp160ΔCT glycoproteins on the surface of the PLs, we compared the binding of a panel of conformationally sensitive ligands to gp160ΔCT glycoproteins on PLs with binding to gp160ΔCT glycoproteins expressed on the surface of transiently expressing cells. The antibodies were selected to probe several faces of the HIV-1 envelope glycoproteins including the neutralization-relevant epitopes overlapping the CD4BS. We used the potently neutralizing CD4BS antibody IgG1b12, the less potent neutralizing CD4BS antibody F105, and sCD4 to confirm the correct native conformation of the CD4BS. In addition, we probed gp160ΔCT PLs with the strain-restricted neutralizing V3-loop antibody 39F and the nonneutralizing antibodies A32 and C11. The binding characteristics of all antibodies to the gp160ΔCT PLs were virtually indistinguishable from those for gp160ΔCT glycoproteins expressed on the cell surface (Fig. 7). The only difference in binding profiles was observed for sCD4. For the PLs, sCD4 displayed an almost-10-fold-higher affinity than it did for gp160ΔCT glycoproteins expressed on cell surfaces. This affinity difference may be a consequence of better exposure of gp160ΔCT glycoproteins on the PLs due to the lack of the cellular glycocalyx and cellular protein components. The observed affinities calculated for the antibodies were in the low nanomolar range, consistent with previously reported values (27, 30). For example, the affinity of the antibody IgGb12, as determined by the concentration necessary to achieve half-maximal binding, was calculated to be 6 nM. The affinity of the nonneutralizing C11 antibody was at least 10-fold lower than that of the neutralizing IgG1b12 antibody. This underestimates the affinity difference between the two antibodies because saturation binding was not achieved with the C11 antibody.
FIG. 7.
Binding of a panel of anti-gp120 antibodies and sCD4 to YU2 gp160ΔCT glycoprotein expressed on 293T cells compared to that to YU2 gp160ΔCT glycoprotein on PLs. Cells (open circles) and PLs (closed squares) were incubated with increasing amounts of the indicated human antibodies, followed by detection with anti-human IgG-PE antibody. For detection of sCD4 binding to the glycoproteins, the rabbit anti-CD4 antibody T45 and anti-rabbit IgG-FITC were used. Following staining, samples were analyzed by FACS. The binding of ligands to gp160ΔCT PLs was plotted as percent normalized mean fluorescence intensity at serially diluted antibody concentrations. The percent normalized mean fluorescence intensity (MFI) values were calculated according to the formula [MFI − MFI (background)] × 100/[MFI (saturation) − MFI (background)]. Error bars indicate the range of values obtained for duplicate samples.
CD4 induction of the 17b epitope.
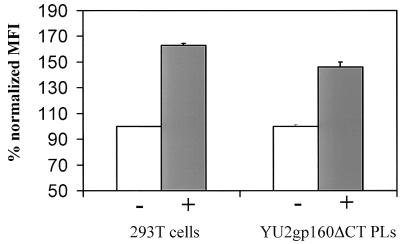
One functional conformational change characteristic of native gp120 is the induction of the 17b antibody epitope by CD4 (37). To test whether the gp160ΔCT glycoproteins on PLs exhibit this property of gp120 expressed on the cell surface, gp160ΔCT glycoprotein PLs and 293T cells expressing cleavage-competent envelope glycoproteins were preincubated with sCD4 prior to binding of the 17b antibody. 17b binding was detected by staining with a PE-conjugated anti-human IgG antibody followed by FACS analysis. The binding of the 17b antibody was increased by 63% after preincubation with sCD4 on cleavage-competent gp160ΔCT glycoprotein expressed on cells compared to a 46% increase on gp160ΔCT PLs (Fig. 8).
FIG. 8.
Induction of the 17b epitope by sCD4. Binding of the 17b antibody to gp160ΔCT glycoprotein on 293T cells and that on PLs were compared. Cells and PLs were incubated with (+) and without (−) sCD4 prior to binding of the 17b antibody and analyzed by FACS. Staining without preincubation with sCD4 was set as 100%, and increase of 17b binding is shown for 293T cells (open bars) and PLs (solid bars). MFI, mean fluorescence intensity.
Characterization of cleavage-deficient gp160ΔCT glycoproteins.
Deletion of the cleavage site for the host protease furin results in the expression of uncleaved gp160ΔCT glycoprotein precursor proteins on the surface of transfected cells. This modification eliminates the dissociation of gp120 from the cell surface and therefore increases the amount of envelope glycoproteins retained on the cell surface or captured on the surface of the PLs. In addition, a cytoplasmic tail deletion was introduced into the gp160 glycoprotein in order to increase cell surface expression (ΔCT). Cell surface expression levels of cytoplasmic tail-deleted envelope glycoproteins increased eightfold over those for full-length constructs containing the intact gp160 cytoplasmic tail (data not shown).
It is possible that the deletion of the cleavage site might distort envelope glycoprotein conformation and result in a conformation not representative of functional, cleaved envelope glycoprotein. To assess the effects of the cleavage site deletion, we analyzed the binding properties of a panel of conformationally sensitive ligands to cleavage-defective and cleavage-competent gp160ΔCT glycoproteins expressed on the cell surface. FACS analysis was performed by staining the gp160ΔCT expressed on 293T cells with increasing concentrations of ligands. No significant affinity difference was observed between cleavage-competent gp160ΔCT glycoproteins and cleavage-defective envelope glycoproteins for any of the ligands tested (Fig. 9). However, one notable difference in the binding profiles was observed. The binding profile of sCD4 on cells expressing cleavage-competent gp160ΔCT glycoproteins had a biphasic shape. As sCD4 is known to induce shedding of gp120 from envelope glycoprotein complexes (21), CD4 binding to the cleavage-competent gp160ΔCT glycoprotein may induce shedding or other conformational changes that result in a biphasic binding profile.
FIG. 9.
Effect of proteolytic cleavage of HIV-1 envelope glycoprotein on ligand binding. Cleavage-competent YU2 gp160ΔCT glycoprotein (closed squares) and cleavage-defective YU2 gp160ΔCT glycoprotein (open circles) were expressed on 293T cells. Cells were incubated with increasing amounts of the indicated ligands followed by detection with anti-human IgG-PE. The anti-CD4 polyclonal rabbit antibody T45 and anti-rabbit IgG-FITC were used for the detection of sCD4 binding. Normalized mean fluorescence intensity was calculated as described for Fig. 7.
To confirm the processing of the cleavage-competent envelope glycoproteins, 293T cells were transfected with cleavage-competent and -defective envelope glycoprotein constructs. Proteins expressed on the cell surface were iodinated by lactoperoxidase. After detergent lysis and precipitation with the F105 antibody, iodinated proteins were analyzed by SDS-PAGE. The ratio of unprocessed gp160ΔCT precursor proteins to cleaved gp120 was determined to be about 1:1 (data not shown).
DISCUSSION
The inability of gp120-based HIV-1 envelope glycoproteins either to elicit broadly neutralizing antibodies or to efficiently select inhibitory ligands from libraries has hampered efforts to develop reagents that intervene in viral envelope glycoprotein-receptor interactions. Recent reports have demonstrated that soluble oligomeric envelope glycoprotein constructs possess characteristics consistent with the trimeric structures found on the surface of virus or infected cells (48). A recent report demonstrated that specific soluble, stable trimeric glycoproteins (YU2 gp140-GCN4 glycoproteins) elicit neutralizing antibodies better than does monomeric gp120 (50). These results support the notion that proteins that more faithfully mimic the viral envelope glycoprotein structure may more efficiently elicit neutralizing antibodies.
There may be several reasons for this better ability of the trimeric gp140-GCN4 glycoproteins than of monomeric gp120 to elicit neutralizing antibodies. The first reason may be that these oligomeric molecules were derived from a primary isolate (YU2), which is highly resistant to most neutralizing antibodies and in particular to strain-restricted V3-loop-directed antibodies. The second possibility is that the YU2 gp140-GCN4 trimeric molecules may reduce the conformational flexibility seen with monomeric gp120 and thereby more efficiently elicit antibodies that recognize the functional trimer. By titration microcalorimetry, we have previously shown that gp120 exhibits a large loss of entropy following interaction with CD4 (23). As this entropy loss is not dependent on the major variable loops V1, V2, and V3, it is likely that the flexibility detected by the thermodynamic analysis involves the core domains of gp120. This flexibility may contribute to the elicitation of many antibodies that do not cross-react with the native trimer. In fact, most antibodies elicited in infected individuals or vaccinated subjects bind with high affinity to monomeric gp120 but not to oligomeric gp120 found on the surface of infected cells (15, 30, 35). Thirdly, the inclusion of gp41 protein sequences might provide additional T-helper epitopes that might quantitatively increase anti-envelope antibody levels. Finally, oligomeric antigens may trigger immune responses more effectively than do monomers.
For these possible advantages, we have sought to construct oligomeric envelope glycoproteins that most closely mimic the trimers found on the surface of the virus or on the surface of envelope glycoprotein-expressing cells. The PLs described here closely fit these criteria, as they possess native gp160ΔCT glycoproteins, as assessed by extensive FACS analysis with conformationally sensitive ligands. The gp160ΔCT glycoproteins captured on the PLs are cleavage defective and thereby possess a covalent linkage between gp120 and gp41 subunits, which enhances spike stability. The envelope glycoproteins exist primarily as trimers on the PL surface as determined by elution and size-exclusion chromatography under nondenaturing conditions. As opposed to our soluble trimeric GCN4-stabilized constructs (48, 49), the native gp41 oligomerization domain in the context of the lipid bilayer is sufficient to maintain the trimeric conformation and no heterologous trimerization motifs need to be appended to the gp160ΔCT glycoprotein. The reconstituted lipid bilayer may provide advantages over the soluble gp140 glycoproteins by both stabilizing known gp41-neutralizing determinants and limiting access by antibody to nonneutralizing envelope glycoprotein determinants.
Other approaches, such as the use of virus-like particles (VLPs), also allow for the presentation of envelope glycoproteins to the immune system as membrane-associated, native entities. However, as VLPs bud from the cell surface, cellular proteins other than envelope glycoproteins are incorporated into VLPs, limiting the purity of these preparations. The well-defined protein content of the PLs described here provides an advance over traditional VLPs, as the affinity capture-purification method renders the PLs devoid of other cellular components. Also, since the bead core of the PLs is optimized for binding of protein at high density, the protein density on the surface of the PLs is likely much greater than that found on VLPs. The PLs have properties that allow for their use in probing libraries of biological or chemical compounds, including phage display libraries. Their paramagnetic properties, although not an absolute requirement for their composition, allow for the fast and complete recovery of the PLs from complex mixtures.
Our results suggest that, antigenically, PLs are virtually indistinguishable from envelope glycoproteins present on the cell surface of gp160ΔCT-expressing cells. For some anti-gp120 ligands, namely, IgGb12 and CD4IgG, the affinity for envelope glycoproteins on PLs is marginally but reproducibly higher than that for envelope glycoproteins expressed on the cell surface. This might be due to better accessibility of the envelope glycoproteins on PLs as a consequence of the lack of other masking proteins and glycocalyx components.
The de novo generation of a lipid bilayer around the individual beads has previously been shown to be an effective method to retain functionality and conformational integrity of the seven-transmembrane proteins CCR5 (20) and CXCR4 (2). When formulated as PLs, the seven-transmembrane CCR5 molecule retained native structure and could withstand changes in salt and pH that denatured detergent-solubilized CCR5 (20). For the gp160ΔCT PLs, reconstitution of the membrane was dependent on the presence of gp160ΔCT glycoprotein on the bead, suggesting that the lipid does not bind to the bead nonspecifically but rather forms in a coordinated fashion. The hydrophobic transmembrane regions of the envelope glycoprotein likely nucleate membrane formation during the reconstitution.
The binding of a secondary antibody to the 1D4 capture antibody was significantly reduced by membrane reconstitution. This indicates that, at least in part, the lipid bilayer was reconstituted around the protein constituents captured on the bead. That the membrane interacts with elements in the gp160ΔCT transmembrane region is supported by the 2F5 antibody binding profiles on PLs with reconstituted membranes compared with those on PLs without membrane. The neutralizing antibody 2F5 recognizes a linear epitope in the gp41 ectodomain (ELDKWAS) directly adjacent to the transmembrane region. The comparison of binding of 2F5 to gp160ΔCT on PLs with binding on beads without a reconstituted membrane shows a roughly 10-fold-higher affinity toward the epitope on the membrane-embedded envelope glycoprotein. Such an affinity difference was not seen with F105 and 2G12, two gp120-directed antibodies (Fig. 6). Although the 2F5 epitope is linear, it might exhibit dependence upon conformation. These requirements of the important but elusive 2F5 epitope seem to be better retained by reconstitution of the lipid environment.
In conclusion, the data presented strongly suggest that the gp160ΔCT PLs provide a native conformation of HIV-1 envelope glycoproteins in a format that will allow testing of whether these unique reagents will effectively elicit neutralizing antibodies or select for novel ligands from phage display peptide or antibody libraries. The gp160ΔCT PLs should also be useful for studies of purified HIV-1 envelope glycoproteins in a natural context.
Acknowledgments
We acknowledge Marshall Posner, Dennis Burton, Hermann Katinger, and James Robinson for generously providing monoclonal antibodies.
This work was supported in part by grants from the National Institutes of Health (5R21AI-44328-02, 1R21AI-44328-02, and AI 31783) and by NIH CFAR grant AI28691. We also acknowledge the support of the G. Harold and Leila Mathers Foundation, the Friends 10, Douglas and Judith Krupp, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, G. J., T. Mirzabekov, W. Wojtowicz, and J. Sodroski. 2001. Ligand-binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 276:38433-38440. [DOI] [PubMed] [Google Scholar]
- 3.Berman, P. W., T. J. Gregory, L. Riddle, G. R. Nakamura, M. A. Champe, J. P. Porter, F. M. Wurm, R. D. Hershberg, E. K. Cobb, and J. W. Eichberg. 1990. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature 345:622-625. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruck, C., C. Thiriart, L. Fabry, M. Francotte, P. Pala, O. Van Opstal, J. Culp, M. Rosenberg, M. De Wilde, P. Heidt, et al. 1994. HIV-1 envelope-elicited neutralizing antibody titres correlate with protection and virus load in chimpanzees. Vaccine 12:1141-1148. [DOI] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chen, B., G. Zhou, M. Kim, Y. Chishti, R. E. Hussey, B. Ely, J. J. Skehel, E. L. Reinherz, S. C. Harrison, and D. C. Wiley. 2000. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J. Biol. Chem. 275:34946-34953. [DOI] [PubMed] [Google Scholar]
- 10.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 11.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 12.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, et al. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 14.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H. D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 17.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 20.Mirzabekov, T., H. Kontos, M. Farzan, W. Marasco, and J. Sodroski. 2000. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat. Biotechnol. 18:649-654. [DOI] [PubMed] [Google Scholar]
- 21.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 22.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oldstone, M. B., A. Tishon, H. Lewicki, H. J. Dyson, V. A. Feher, N. Assa-Munt, and P. E. Wright. 1991. Mapping the anatomy of the immunodominant domain of the human immunodeficiency virus gp41 transmembrane protein: peptide conformation analysis using monoclonal antibodies and proton nuclear magnetic resonance spectroscopy. J. Virol. 65:1727-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palker, T. J., T. J. Matthews, M. E. Clark, G. J. Cianciolo, R. R. Randall, A. J. Langlois, G. C. White, B. Safai, R. Snyderman, D. P. Bolognesi, et al. 1987. A conserved region at the COOH terminus of human immunodeficiency virus gp120 envelope protein contains an immunodominant epitope. Proc. Natl. Acad. Sci. USA 84:2479-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parren, P. W., D. R. Burton, and Q. J. Sattentau. 1997. HIV-1 antibody-debris or virion? Nat. Med. 3:366-367. [DOI] [PubMed] [Google Scholar]
- 27.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7-14. [PubMed] [Google Scholar]
- 28.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 29.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 30.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robey, W. G., L. O. Arthur, T. J. Matthews, A. Langlois, T. D. Copeland, N. W. Lerche, S. Oroszlan, D. P. Bolognesi, R. V. Gilden, and P. J. Fischinger. 1986. Prospect for prevention of human immunodeficiency virus infection: purified 120-kDa envelope glycoprotein induces neutralizing antibody. Proc. Natl. Acad. Sci. USA 83:7023-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robey, W. G., B. Safai, S. Oroszlan, L. O. Arthur, M. A. Gonda, R. C. Gallo, and P. J. Fischinger. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593-595. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, J. E., D. Holton, S. Pacheco-Morell, J. Liu, and H. McMurdo. 1990. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS Res. Hum. Retrovir. 6:567-579. [DOI] [PubMed] [Google Scholar]
- 34.Rusche, J. R., K. Javaherian, C. McDanal, J. Petro, D. L. Lynn, R. Grimaila, A. Langlois, R. C. Gallo, L. O. Arthur, P. J. Fischinger, et al. 1988. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc. Natl. Acad. Sci. USA 85:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to Gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilley, S. A., W. J. Honnen, M. E. Racho, M. Hilgartner, and A. Pinter. 1991. A human monoclonal antibody against the CD4-binding site of HIV1 gp120 exhibits potent, broadly neutralizing activity. Res. Virol. 142:247-259. [DOI] [PubMed] [Google Scholar]
- 41.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J. J., S. Steel, R. Wisniewolski, and C. Y. Wang. 1986. Detection of antibodies to human T-lymphotropic virus type III by using a synthetic peptide of 21 amino acid residues corresponding to a highly antigenic segment of gp41 envelope protein. Proc. Natl. Acad. Sci. USA 83:6159-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 44.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 45.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 47.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 48.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]