Abstract
A mutation that disrupts the interaction between the NS2 protein of minute virus of mice and the nuclear export factor Crm1 results in a block to egress of mutant-generated full virions from the nucleus of infected murine cells. These mutants produce wild-type levels of monomer and dimer replicative DNA forms but are impaired in their ability to generate progeny single-stranded DNA in restrictive murine cells in the first round of infection. The NS2-Crm1 interaction mutant can be distinguished phenotypically from an NS2-null mutant and reveals a role for the Crm1-mediated export pathway at a late step in viral infection.
The autonomous parvovirus minute virus of mice (MVM) produces two nonstructural proteins that play critical roles in the replication of the virus (6-8, 18). The large nonstructural protein, NS1, is a multifunctional protein with site-specific DNA binding, nickase, ATPase, and helicase activities that have been mapped to specific regions of the NS1 protein and are critical for MVM replication (8, 17, 19, 20). The small nonstructural protein, NS2, is also required for MVM replication in a host-cell-specific manner; in murine cells NS2-null mutants (18), as well as a number of other characterized NS2 mutants (5), generate little double-stranded replicative intermediates and little to no progeny single-stranded DNA (ssDNA) is produced. Cells from a variety of other species support MVM replication at near-wild-type levels in the absence of NS2 (18).
The role(s) NS2 plays in MVM replication remains unclear. In the absence of NS2, wild-type levels of the VP1 and VP2 structural proteins are synthesized at early times postinfection; however, they do not efficiently assemble into MVM capsids (5). Although the mechanisms are not well understood, the production of MVM progeny ssDNA is tightly connected to the availability of assembled MVM capsids (8), and so it is likely that, at least to some extent, the lack of progeny ssDNA produced during NS2 mutant infection is a consequence of the lack of proper capsid assembly. The lack of replication of monomer replicative DNA forms (mRF) and dimer replicative DNA forms (dRF) in NS2-null mutant infection is more difficult to explain, as this process is known to proceed efficiently in the absence of capsid production (23).
NS2 was recently shown to interact with two members of the 14-3-3 family of signaling proteins (3) and the nuclear export factor Crm1 (2, 21). Crm1 is a sequence-specific nuclear export receptor that binds leucine-rich sequences within proteins in cooperation with RanGTP and actively transports these proteins from the nucleus to the cytoplasm (9, 24). The NS2 interaction with Crm1 was previously shown to be important in proper localization of NS2 within the cell and was mapped to NS2 amino acids (aa) 81 to 103 by peptide analysis (N. Salome, personal communication) and glutathione S-transferase (GST) pull-down assays (21). The role of the NS2-Crm1 interaction during MVM infection, however, has not been determined.
We have investigated the consequence of the NS2 interaction with Crm1 during viral replication by creating a mutation within NS2 that disrupts its interaction with Crm1. In contrast to MVM NS2-null mutant replication, NS2-Crm1− mutants generate double-stranded replicative-form DNA at near-wild-type levels at early times postinfection; however, there is a striking decrease in the production of progeny ssDNA. We further show that this defect in ssDNA production is associated with a severe defect in the export of full MVM virions from the nucleus to the cytoplasm of the cell. Export of MVM virions from the nucleus is also blocked by the Crm1-specific inhibitor leptomycin B.
MATERIALS AND METHODS
Cells, viruses, and transfections.
Murine A92L, murine ID5, and human NB324K cells were maintained and passaged as described previously (22). Wild-type and mutant the MVM prototype (MVMp) virus was grown from transfection and titers were determined on NB324K cell monolayers. All viral stocks were equilibrated by ssDNA content normalized to a wild-type multiplicity of infection (MOI) as indicated. NS2-null mutant (1989) virus, which contains a single-nucleotide change at the MVM large splice acceptor site at nucleotide (nt) 1989 and makes no NS2 protein, was previously described (18). A92L cells were doubly blocked at the G1/S border where indicated by 44 h of growth in isoleucine-deficient media followed by 12 h of growth in the presence of 12 μg of aphidicolin/ml (Sigma Chemical, St. Louis, Mo.) as described elsewhere (18). Leptomycin B (Sigma Chemical) was added to cells at a final concentration of 10 nM where indicated. Plaque assays were done as previously described (18), using methylene blue to stain fixed cells. Transient-transfection assays were performed by the standard CaPO4 method (5 μg of DNA/60-mm dish) as described previously (23) or with Lipofectamine and Plus reagent (2 μg of DNA/60-mm dish) as recommended by the supplier (Gibco BRL, Grand Island, N.Y.).
Plasmid constructs.
The MVM wild-type and NS2-null (1989) infectious clones were previously described (15, 18). The NS2-Crm1− MVM infectious clone was made by overlap PCR mutagenesis (10), changing MVM nt 1990 to 2008, such that the NS2 amino acids aa 84-FGTLJI-aa 89 were changed to aa 84-VCPVAV-aa 89 without altering the NS1 amino acid sequence. The complete coding region for the mutant protein was sequenced to ensure no additional mutations were introduced. The pCMVNS2 plasmid was previously described (16). The pCMVNS2-Crm1− plasmid was constructed by overlap PCR mutagenesis to incorporate the mutation of aa 84-FGTLJI-aa 89 to aa 84-VCPVAV-aa 89. PGEX-5X-NS2 was previously described (17). pGEX-5X-NS2-Crm1− was made by inserting the EcoRV-XhoI fragment from pCMVNS2-Crm1− into pGEX-5X-NS2. pETCrm1 and pCIHACrm1 were previously described (11) and were gifts of Mark Hannink, University of Missouri—Columbia.
Immunoprecipitation and Western analysis.
A92L cells were transfected as indicated and collected 18 h posttransfection. Immunoprecipitation followed by Western blotting was performed as described previously (16). Antibodies used included rabbit polyclonal anti- (α)-NS2 C terminus, made against a peptide containing NS2 aa 156 to 182, and mouse monoclonal α-HA.11 (Covance, Richmond, Calif.).
Indirect IFAs.
A92L, ID5, or NB324K cells were grown on either coverslips or eight-well slides (Labtech) and infected at the indicated wild-type MOI. Coverslips or slides were processed for immunofluorescence assay (IFA) as described elsewhere (18) at the indicated times postinfection. Images were generated using a SPOT digital camera, and all images from each experiment were obtained at the same exposure time. Primary antibodies were used at a 1:100 dilution and included rabbit polyclonal α-NS2 made against the internal sequences of NS2 (4) and mouse monoclonal α-assembled capsid (12). Secondary antibodies were used at a 1:250 dilution and included fluorescein isothiocyanate (FITC)-conjugated α-rabbit and FITC-conjugated α-mouse antibodies (ICN Biochemicals).
RanGAP protection assay.
pGex-5X-NS2 and pGex-5X-NS2-Crm1− were transformed into and grown in Escherichia coli strain BL21(DE3)pLysS. GST-NS2 and GST-NS2-Crm1− protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and GST-conjugated proteins were purified with glutathione-agarose as described previously (17). pETCrm1 was transformed into and grown in E. coli strain BL21(DE3)pLysS cells. HISCrm1 protein expression was induced with 1 mM IPTG and purified by metal chelate affinity chromatography (Invitrogen) as described elsewhere (11). RanGAP protein was expressed in BLR pREP4 cells and purified by metal chelate affinity chromatography (Invitrogen). RanGAP protection assays were then done as described elsewhere (1, 11). Briefly, Ran protein was incubated with [γ-32P]GTP (10 mCi/ml) for 30 min on ice in buffer containing 20 mM HEPES (pH 7.3), 100 mM potassium acetate, and 5 mM EDTA, followed by gel filtration on a Bio-Spin 6 column (Bio-Rad). Reaction mixtures containing increasing amounts of purified, beaded GST-NS2 or GST-NS2-Crm1− were incubated at 15°C for 30 min with 500 nM purified HisCrm1 and 50 pM [λ-32P]RanGTP in 25 μl of hydrolysis buffer to allow GST-NS2 or GST-NS2-Crm1−-HISCrm1-RanGTP ternary complexes to form. Ten nanomolar RanGAP was then added and the mixtures were incubated at 30°C for 2 min to allow hydrolysis of Ran GTP that was not protected in a ternary complex. The reaction mixtures were filtered through nitrocellulose, the filters were washed, and the filtered lysates were counted in the presence of 3 ml of scintillation fluid. Counts representing percent unbound (unprotected) hydrolyzed RanGTP were plotted against the concentration of NS2 wild-type or mutant protein present.
Southern analysis.
Wild-type and mutant MVM replication forms produced during infection or transfection were visualized by Southern analysis of total cell extracts as previously described (18, 23). To equilibrate virus by ssDNA content, equivalent PFU of wild-type and mutant viral stocks extracted in 50 mM Tris-10 mM EDTA (pH 8.7) were added to micrococcal nuclease buffer and treated with micrococcal nuclease to remove associated MVM double-stranded DNA (dsDNA). Samples were then lysed in 2% sodium dodecyl sulfate (SDS) and treated with proteinase K (0.5 mg/ml) to remove viral capsid proteins. Samples were run on a 1% agarose gel, and Southern blot analysis was performed as described previously (18, 23). Blots were exposed to a Kodak phosphorimaging screen, and the ssDNA was quantitated by using a Bio-Rad Imager FX and Quantity One software where indicated.
Western blot analysis.
Cells collected 18 h posttransfection or viral stocks equilibrated by PFU or ssDNA were lysed in 2% SDS, and Western blot analysis was performed as described elsewhere (16). Antibodies used included rabbit polyclonal α-NS2 made against the NS2 C terminus (aa 156 to 182) and rabbit polyclonal α-capsid antibody (α-allopeptide) that was previously shown to interact with individual MVM capsid proteins VP1 and VP2 and, potentially, assembly intermediates (5).
Hemagglutination assay.
Hemagglutination assays of MVM wild-type and mutant viral stocks equilibrated by ssDNA were performed as described previously (23).
NS1 infectious center assay.
MVM wild-type and mutant viral stocks equilibrated by ssDNA and added at equivalent wild-type MOIs or cell lysates and media isolated at various times postinfection and added at equivalent volumes were used to infect A92L, ID5, or NB324K cells. At 18 h postinfection, the cells were treated for IFA and stained with rabbit polyclonal α-NS1 primary antibody (4) followed by FITC-conjugated goat α-rabbit secondary antibody (ICN Biochemicals). Cells from at least three microscope fields were counted, and the number of NS1-positive cells per 100 total cells was plotted.
Assembled capsid and ssDNA ratio assay.
A92L cells were doubly blocked at the G1/S border by incubation in isoleucine-deficient medium followed by treatment with 12 μg of aphidicolin/ml (18). During the aphidicolin incubation, wild-type and mutant viruses were added at equivalent wild-type MOIs. At 8 and 12 h postrelease from aphidicolin, 35S was added at 100 μCi/ml. At 16 h postrelease from aphidicolin, cells were collected and immunoprecipitated with α-assembled capsid antibody in nondenaturing buffer as previously described (16). After immunoprecipitation samples were split into two, with one half processed for Southern blot analysis to visualize ssDNA while the other half was run on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) to visualize labeled assembled capsid protein. DNA and protein gels were exposed to a phosphorimaging screen and quantitated using a Bio-Rad Imager FX and Quantity One software. Ratios of ssDNA associated with assembled capsid were determined and are shown below. Samples isolated at 8 h postrelease from aphidicolin were immunoprecipitated with the same antibody and processed for Southern blotting as a control for input viral ssDNA.
RESULTS
Mutation of NS2 aa 86 to 91 disrupts the NS2-Crm1 interaction in vivo and functional interaction in vitro and results in a more nuclear-localized NS2.
The region of NS2 involved in interaction with Crm1 has previously been shown to span NS2 aa 81 to 106 by both interfering peptide studies (N. Salome, personal communication) and in vitro GST-NS2 pull-down studies (21). Mutation within the NS2-specific portion of this region, which changed NS2 aa 86 to 91 from FGTLTI to VCPVAV (Fig. 1A), was found to disrupt the NS2-Crm1 interaction. Following cotransfection of wild-type CMVNS2 and a hemagglutinin (HA)-tagged Crm1 expression construct into murine A9 cells, HA-tagged Crm1 could be detected by Western blotting following immunoprecipitation with an NS2-specific antibody (Fig. 1B). When similar levels of mutant NS2 were expressed, however, little to no HA-tagged Crm1 was immunoprecipitated with α-NS2, suggesting that the NS2 interaction with Crm1 was significantly lost in vivo as a result of the 6-aa mutation.
FIG. 1.
Mutation within the NS2 NES disrupts NS2 interaction with Crm1. (A) Graphical representation of the NS2 NES mutant sequence. (B) A92L cells were transfected with HA-Crm1 and either no plasmid, pCMVNS2, or pCMVNS2-Crm1−. At 18 h posttransfection, cells were immunoprecipitated with α-NS2 antibody followed by Western blotting with α-HA antibody (left panel). Parallel samples were subjected to Western blot analysis with either α-NS2 (center panel) or α-HA (right panel) antibody to visualize NS2 and HA-Crm1 protein expression. For reasons not yet determined, the mutant NS2 protein migrates slightly slower than the wild type. The complete coding region for the mutant protein has been sequenced and contains no additional changes. (C) Results of RanGAP protection assay using purified GST-NS2 (•) or GST-NS2-Crm1− (□). Percent GTP hydrolysis is plotted against concentration of wild-type or mutant NS2 protein. (D) Localization of NS2 wild type and NS2-Crm1−. A92L cells were infected with either wild-type MVMp or NS2-Crm1− virus at an MOI of 5. At 24 h postinfection, cells were fixed and stained with α-NS2 rabbit primary antibody and FITC-conjugated α-rabbit secondary antibody. Images were obtained on a SPOT camera at the same exposure time.
Crm1 binds nuclear export signal (NES)-containing proteins cooperatively with RanGTP (9, 14). This interaction protects RanGTP from hydrolysis by RanGAP in vitro, and the level of protection has been used as a quantitative method to measure the functional interaction between Crm1 and NES-containing proteins (1, 11). While 0.063 μM purified wild-type GST-NS2 was required to protect 50% of labeled RanGTP from hydrolysis, it took 13-fold more GST-NS2-Crm1− (0.82 μM) to protect equivalent levels of RanGTP from RanGAP hydrolysis (Fig. 1C). This difference in protection is similar to what has been reported previously for a well-characterized Crm1-NES loss-of-interaction mutant of IκBα (11) and for studies of wild-type versus mutant NS2 NES peptide (1), suggesting that the NS2-Crm1− mutant loses functional interaction with Crm1.
The NS2-Crm1− interaction mutation was then introduced into the infectious clone of MVMp. Similar to other NS2 mutants, the NS2-Crm1− infectious clone was seen to replicate very poorly following transfection of murine A9 cells yet replicated efficiently in NB324K cells, which allowed the generation of mutant viral stocks on permissive cells. A more detailed characterization of the replication of the NS2-Crm1− mutant virus is presented below.
While wild-type NS2 is found in both the nucleus and the cytoplasm of infected cells, it has been previously shown that NS2 is relocalized to the nucleus in the presence of the Crm1 inhibitor leptomycin B (2, 21). We therefore examined the cellular localization of the mutant NS2. Figure 1D shows that there was a significant redistribution of NS2-Crm1− mutant-generated NS2 to the nucleus, providing additional evidence that the 6-aa mutation within the NS2 NES resulted in a loss of in vivo interaction of NS2 with Crm1 during MVM infection.
Characterization of NS2-Crm1− mutant replication following transfection of permissive and nonpermissive cells.
NS2 has been shown to play a critical role in MVM replication in a host-cell-specific manner following either viral infection or transfection of cloned NS2 mutants. There is little production of NS2-mutant dsDNA replication intermediates, and no detectable accumulation of viral progeny ssDNA in murine cells; however, replication is more significant in a variety of permissive cell types of other species (18). In contrast to other described NS2 mutants, the NS2-Crm1− mutant generated dsDNA mRF and dRF at levels nearly equal to those of the wild type at early times following transfection of both permissive NB324K cells and restrictive A9 and ID5 murine cells (Fig. 2). In restrictive murine cells, mutant mRF and dRF did not amplify at the same rate as with the wild type. In contrast, in permissive NB324K cells, the NS2-Crm1− mutant generated mRF and dRF at near-wild-type levels at both early and late times posttransfection. Strikingly, even at early times posttransfection of restrictive murine cells, a dramatic decrease in the accumulation of mutant viral progeny ssDNA was seen, and this decrease became relatively greater than wild-type levels as the transfection proceeded. We consistently detected at least 50-fold less of both cell- or media-associated virus (as assayed by either DNA content or the ability to generate NS1-positive cells) following the transfection into restrictive murine cells of the mutant clone compared to the wild type at times when levels of intracellularly replicating double-stranded replicative forms were similar (data not shown). The decrease in the production of mutant ssDNA was not seen at early times posttransfection of permissive NB324K cells; however, a slight decrease in the production of ssDNA was apparent at later times. These data indicated that the NS2 interaction with Crm1 was not necessary for early steps in MVM dsDNA replication after transfection; however, it was important for the efficient production of progeny ssDNA (required for reinfection) in restrictive cells and to a lesser extent in permissive cell lines.
FIG. 2.
The NS2 interaction with Crm1 is required for production of MVM ssDNA in restrictive cells following transfection. A92L, NB324K, and ID5 cells were transfected by the CaPO4 method with MVMp wild type, NS2-null mutant (A92L cells only), or the NS2-Crm1− mutant infectious clone. Cell samples were isolated at 4, 24, and 48 h posttransfection and processed for Southern blot analysis. Viral monomer (mRF), dimer (dRF), and ssDNA are indicated. Asterisks indicate input plasmid DNA.
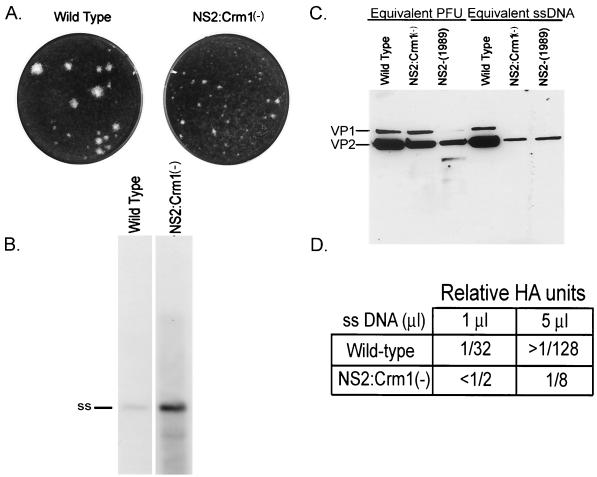
The NS2-Crm1− mutant virus is a small-plaque mutant on NB324K cells and contains approximately 10- to 20-fold less capsid protein per ssDNA than the wild type.
NS2-Crm1− mutant stocks were generated following transfection of NB324K cells. These stocks formed very small plaques on NB324K cells compared to wild-type virus (Fig. 3A). Surprisingly, however, the NS2-Crm1− mutant virus contained approximately 10-fold greater amounts of ssDNA per PFU than did the wild type (Fig. 3B), suggesting that 10-fold more NS2-Crm1− mutant-encapsidated full virions, compared to that with the wild type, were required to form a plaque. When virus was normalized by ssDNA and total capsid protein was visualized by Western blotting, the mutant contained 10- to 20-fold less capsid protein per ssDNA molecule than the wild type (Fig. 3C). This suggested that mutant virus stocks had a low capsid protein/ssDNA ratio compared to the wild type. Hemagglutination assays were done to measure the assembled capsid associated with equivalent wild-type and mutant ssDNA. The wild-type virus had approximately 15 times more hemagglutination activity per ssDNA unit than the mutant (Fig. 3D), suggesting that the mutant virus stocks contain relatively fewer empty capsids than wild-type stocks do.
FIG. 3.
NS2-Crm1− mutant virus characterization. (A) The NS2-Crm1− mutant is a small plaque mutant. Seven days postinfection with wild-type or NS2-Crm1− virus, NB324K cells were fixed and stained to visualize plaques. (B) The NS2-Crm1− mutant contains approximately 10 times more ssDNA per PFU than the wild type. Equivalent wild-type and NS2-Crm1− PFU were processed for Southern blot analysis. Micrococcal nuclease-resistant, encapsidated ssDNA is indicated. (C) NS2-Crm1− mutant viral stocks contain approximately 10 times less total capsid protein than the wild type does. Equivalent PFU or equivalent ssDNA units of wild-type and mutant viral stocks were run on SDS-PAGE gels, and Western blot analysis was performed using rabbit α-allotropic primary antibody and horseradish-conjugated α-rabbit secondary antibody. (D) The NS2-Crm1− mutant contains approximately 15-fold less hemagglutination activity than the wild type.
When equilibrated by ssDNA, the NS2-Crm1− mutant initiates infection at wild-type levels.
Mutant and wild-type viral stocks were adjusted to equal levels of encapsidated ssDNA (Fig. 4A), and their abilities to initiate infection, as monitored by NS1 production, were compared following infection of either murine A92L or ID5 cell lines or permissive NB324K cells. Although there was a slight decrease in the number of NS1 infectious centers formed on A92L cells, the NS2-Crm1− mutant virus initiated infections at levels similar to wild-type levels on both permissive (NB324K) and restrictive (A9, ID5) cell lines (Fig. 4B). These data suggested that mutant virus was not defective in steps required prior to the appearance of NS1 (e.g., cellular entry, delivery of DNA to the nucleus, or conversion of genomic ssDNA to mRF) but at a later step during MVM infection.
FIG. 4.
Equivalent NS2-Crm1− mutant ssDNA initiates infection at wild-type levels but is defective in ssDNA production following infection of murine cells. (A) Equivalent wild-type and NS2-Crm1− mutant ssDNA titers determined in previous experiments were validated by Southern blot analysis. (B) Equivalent titers of wild-type, 1989 (the NS2-null mutant; not shown in panel A), and NS2-Crm1− virus from stocks shown in panel A were used to infect A92L, NB324K, and ID5 cells at a wild-type MOI of 5. At 16 h postinfection, cells were fixed and processed for immunofluorescence using a rabbit α-NS1 primary antibody and FITC-conjugated α-rabbit secondary antibody. The number of NS1-positive cells per total cells in each field was counted and plotted on the graph. At least three separate fields in two separate experiments were counted and averaged. (C) A92L, ID5, and NB324K cells were infected with wild-type, NS2-null (1989), and NS2-Crm1− mutant virus at equivalent ssDNA titers at a wild-type MOI of 5. At 12, 20, and 48 h postinfection cells were collected and processed for Southern blot analysis. MVM mRF, dRF, and ssDNA (ss) are indicated. At 12 h postinfection, 0.05 U of neuraminidase/ml (+N) was added to one set of 48-h samples to prevent reinfection of surrounding cells in these samples.
Characterization of NS2-Crm1− mutant virus replication on permissive and restrictive cells.
When assayed for replication following infection using viral stocks adjusted to contain an equal ssDNA content, the NS2-Crm1− mutant generated viral mRF to near-wild-type levels in the first round of infection of both restrictive (A9, ID5) and permissive (NB324K) cell lines (Fig. 4C). This result is in contrast to results obtained with other NS2 mutants, which generate only very low levels of double-stranded replicative forms in the first round of infection in restrictive murine cells. This observation distinguishes the NS2-Crm1− mutation from NS2-null mutants and suggests that this mutation abrogates only a subset of NS2 functions. Again, however, as seen following transfection of the mutant infection clones, there was little to no detectable mutant ssDNA produced late in infection in restrictive murine A9 or ID5 cells. By 48 h postinfection it is clear that wild-type virus has exited and has been rebound to the surface of infected cells (compare 48-h lanes with and without neuraminidase). This finding suggested that our assay had likely detected the majority of virus produced and further demonstrated that disrupting the NS2 interaction with Crm1 resulted in a late defect causing decreased accumulation of cell-associated progeny virus. Similar to what was seen following transfection, the mutant generated wild-type levels of mRF and dRF in permissive NB324K cells at both early and late time points following infection, suggesting that the mutant was generating near-wild-type levels of progeny ssDNA (Fig. 4C); however, for reasons not yet determined, we have observed that it is difficult to recover progeny ssDNA produced during infection of NB324K cells, and it is therefore difficult to measure ssDNA production in these cells.
NS2-Crm1− mutant virus produces assembled MVM capsids that remain strikingly nuclear localized throughout infection.
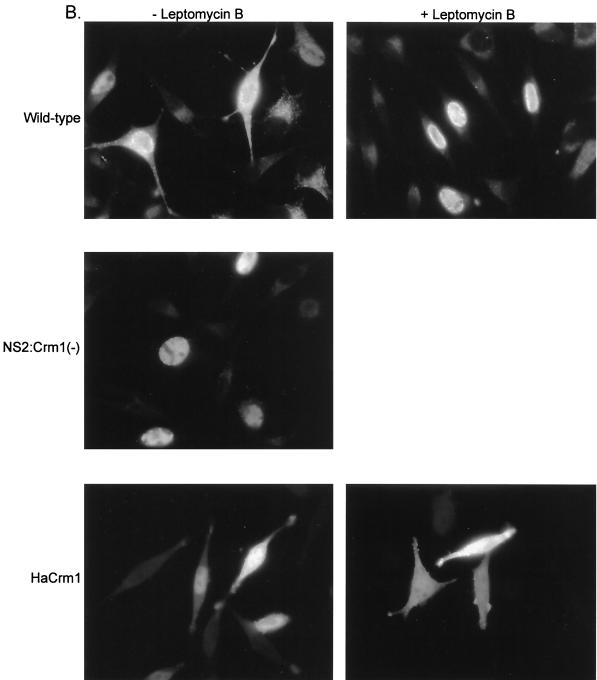
Examination of MVM assembled capsids at 16 to 20 h postinfection by IFA using an antibody shown to react exclusively with assembled capsids (13) showed that the mutant produced assembled capsids in amounts that were qualitatively similar to wild-type amounts in restrictive A92L cells at early times postinfection. However, whereas wild-type assembled virions were localized in both the nucleus and the cytoplasm, NS2-Crm1− mutant assembled virions were found predominantly in the nucleus of the infected cells (Fig. 5A, top panel). At later times during infection (48 h), the NS2-Crm1− mutant assembled capsids were still predominately nuclear, while wild-type assembled capsids were found primarily in the cytoplasm. Cytoplasm-localized wild-type virus seen at this time point are presumed to be full assembled capsids that have moved into a second round of infection of surrounding cells (Fig. 5A, middle panel); they are not seen when neuraminidase is added to prevent reinfection (data not shown). In permissive cells at both early and late times, there was also an increase in mutant nuclear-associated assembled capsids compared to that in the wild type. Some of the mutant virus, however, had clearly escaped the nucleus, as the overall distribution of assembled capsids in the permissive NB324K cells more closely resembled that of wild-type virus infection (Fig. 5A, bottom panel). Our results suggested that the NS2-Crm1− mutant is defective in export of assembled capsids out of the nucleus.
FIG. 5.
NS2 interaction with Crm1 is important for nuclear export of MVM virions during infection of murine cells. (A) A92L (top and middle panels) and NB324K (bottom panel) cells were infected with equivalent ssDNA titers of wild-type and NS2-Crm1− virus at a wild-type MOI of 5. (B) A92L cells were infected with wild-type (top panel) or mutant (middle panel) virus (MOI = 5) or transfected with pCIHACrm1 (bottom panel). At 12 h following infection or transfection, cells were treated with 10 nM leptomycin B where indicated and further incubated for 10 h. Cells in both panels A and B were fixed and processed for immunofluorescence using a mouse monoclonal α-assembled capsid primary antibody and a FITC-conjugated α-mouse secondary antibody.
Treatment of wild-type infected cells with the Crm1-specific inhibitor leptomycin B 12 to 20 h postinfection also resulted in retention of assembled capsids in the nucleus of treated cells (Fig. 5B, top panel). This effect was specific, because similar treatment did not affect the typical whole-cell distribution of expressed Crm1 protein (Fig. 5B, bottom panel). These data corroborate a role for the Crm1-mediated export pathway in the nuclear export of MVM virions.
Nuclear-localized assembled capsids produced during NS2-Crm1− infection of restrictive cells contain ssDNA and remain fully infectious.
Although at early times postinfection the NS2-Crm1− mutant capsids assembled at near-wild-type levels as measured by IFA, we could detect little progeny ssDNA on restrictive cells as measured by whole-cell Southern blotting. This suggested that either the mutant assembled capsids did not contain ssDNA or that the amount of ssDNA produced during mutant infection was below the limit of detection of our assay. Figure 6A shows that although there was an overall decrease in ssDNA-containing full virions produced in infection of A9 cells with the mutant compared to that with the wild type, there was a significant amount of full virus present, suggesting that our inability to detect ssDNA following transfection and infection of murine cells was likely to be because the amount of ssDNA produced in the mutant was below our limit of detection. However, there was also a concomitant decrease in the amount of assembled capsid protein produced on infection with the mutant; the relative ratio of capsid protein to ssDNA was approximately the same as that with the wild type. This suggested that at early times mutant virus encapsidation proceeded normally; however, when egress of capsids from the nucleus did not occur, continued production of assembled capsids and additional ssDNA was blocked.
FIG. 6.
The NS2-Crm1− mutant assembled capsids contain ssDNA and are fully infectious. (A) Highly synchronous A92L cells were infected with equivalent ssDNA titers of wild-type and NS2-Crm1− mutant virus at a wild-type MOI of 5. At 8 h postrelease, 100 μCi of Tran35S-label/ml was added. At 8 h after label was added, cells were collected and lysed in Tris-EDTA (pH 8.7) to release all of the virus from the cells. The pH was adjusted to 7.5 and immunoprecipitation was performed using a mouse monoclonal α-assembled capsid antibody. Following immunoprecipitation, the sample was split into two, half of which was processed for Southern blot analysis (left panel) and half of which was run on an SDS-PAGE to visualize labeled MVM capsid proteins (right panel). Protein and ssDNA counts were quantitated on a phosphorimaging screen; data from a representative experiment are shown. (B) A92L cells were infected at equivalent ssDNA titers with wild-type or NS2-Crm1− virus at a wild-type MOI of 5. At 16 h postinfection, cells and media were collected, the cells were lysed in Tris-EDTA (pH 8.7) to release all associated virus, and cell-associated virus and media collected from each sample were used at equivalent volumes to infect NB324K cells. At 20 h postinfection, cells were fixed and processed for immunofluorescence using a rabbit polyclonal α-NS1 primary antibody and a FITC-conjugated α-rabbit secondary antibody. The number of NS1-positive cells per total cells in at least three different fields was counted and plotted on the graph.
The full virus made in infection of restrictive A9 cells with the mutant was also shown to be fully infectious. Cell lysates and media taken early (16 to 20 h postinfection) from mutant or wild-type infection of A9 cells were used at equivalent volumes to infect NB324K cells, and resulting NS1 infectious centers were measured by IFA (Fig. 6B). The NS2-Crm1− mutant generated levels of cell-associated infectious virus that approached 80% of wild-type levels, further demonstrating that assembled capsids visible in nuclei by IFA in NS2-Crm1− infection contain progeny ssDNA. Based on the experiments shown in Fig. 4C, cell-associated virus included any virus that had exited and rebound to cells. In addition, the culture media from the mutant at early times postinfection was shown to contain 10-fold less infectious virus than the culture media collected from the wild-type MVM-infected cells (Fig. 6B). At late times postinfection (48 h), as predicted from the large differences in ssDNA production, there were 50- to 100-fold less mutant cell- and media-associated infectious virions than that with the wild type (data not shown). This further suggested that viable mutant progeny are produced in A9 infection; however, the full virions primarily remain cell associated.
DISCUSSION
In this study, we showed that the interaction between NS2 and the nuclear export protein Crm1 is important for viral infection. Mutants deficient in this interaction are impaired in their ability to generate wild-type levels of progeny ssDNA, and assembled, full, mutant-generated capsids are retained in the nucleus.
The Crm1 interaction mutant that we have generated can be distinguished from the NS2-null mutation. In contrast to the NS2-null mutant (18) and a number of other characterized NS2 mutants (5) which are blocked in an early step required for the production of mRF and dRF, the Crm1 interaction mutant generates mRF and dRF to wild-type levels early in infection of restrictive murine A9 and ID5 cells, but it is blocked at a later step in replication. This observation confirms the suspicion that NS2 has multiple functions and is required for both early and late steps during replication, and the Crm1− mutation abrogates only a subset of these. Since the NS2-null mutant does not generate assembled capsids in murine cells, capsid localization in restrictive cells in the absence of NS2 cannot be assessed.
The primary defect in the Crm1 interaction mutant is most likely to be capsid egress from the nucleus. The capsids produced in infection of restrictive murine cells with the mutant initially assemble normally, encapsidate DNA, and are infectious. The full virions that are produced are not transported out of the nucleus, however, and this results in a late block in the first round of infection, causing a significant reduction in the accumulation of ssDNA. The defect in ssDNA accumulation is seen within 16 h of primary infection of restrictive cells and is therefore not merely due to the absence of reinfection, which does account for the large decrease in mutant mRF, dRF, and progeny ssDNA accumulation seen later (48 h) in infection. How a failure to export assembled capsids leads to the inhibition of further ssDNA production in restrictive cells is not clear, but this observation suggests that there may be a feedback mechanism between the exit of full virions from the nucleus and either continued genome production or capsid assembly. It is well known that capsid formation and ssDNA production are intimately linked (8, 23).
It is not yet known how the NS2 interaction with Crm1 is involved in egress of assembled capsids from the nucleus. Perhaps NS2 interacts directly with full virions to facilitate their exit through a Crm1-mediated pathway. However, no direct interactions between NS2 and assembled capsids have yet been demonstrated. A more likely possibility is that the NS2-Crm1 interaction is important for the regulation of a cellular protein that is required for nuclear export of virus. It has been shown that the phosphorylation of the amino terminus of VP2 is critical for viral exit (B. Maroto and J. M. Almendral, VIIIth Parvovirus Workshop, Quebec, Canada, p. 32, 2000). Perhaps the NS2-Crm1 interaction influences the nuclear location of a kinase or phosphatase important for this modification. As expected for a Crm1-dependent process, export of assembled MVM capsids from the nucleus was blocked in the presence of leptomycin B.
It is important to note that the Crm1 interaction mutant-generated capsids also concentrate in the nucleus of permissive NB324K cells. In this case, perhaps enough capsids exit the human cell nucleus to allow the continuation of single-strand production or new capsid assembly. Alternatively, the feedback mechanism which halts single-strand production in murine cells may not be active in human NB324K cells. It is relevant to note here that NS2-null mutant-generated capsids, which are produced in NB324K cells, do not localize predominantly in the nucleus. This may suggest that the NS2-Crm1− mutation is acting in a dominant-negative fashion in this cell type.
It seems that at least at early times in infection parvoviruses can exit from cells prior to cell lysis. Little is known about how nonenveloped viruses exit from the nucleus prior to cell lysis, and parvoviruses seem to provide a valuable system to investigate this process.
Acknowledgments
We thank Nathalie Salome (DKFZ-Heidelberg) for sharing information prior to publication. We also thank Mark Hannink and Sang-hyun Lee (University of Missouri—Columbia) for helpful discussion, reagents, and help with RanGAP functional assays, Greg Tullis for helpful discussion, and Lisa Burger for excellent technical assistance. This work was supported by Public Health Service grants AI21302 and AI46458 from the National Institute of Allergy and Infectious Diseases to D.J.P.
REFERENCES
- 1.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salome. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salome. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemens, K. E., D. R. Cerutis, L. R. Burger, C. Q. Yang, and D. J. Pintel. 1990. Cloning of MVM cDNAs and preliminary analysis of individual viral proteins expressed in murine cells. J. Virol. 64:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotmore, S. F., A. J. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., L. Sturzenbecker, and P. Tattersall. 1983. The autonomous parvovirus encodes two nonstructural proteins in addition to its capsid polypeptides. Virology 129:333-343. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177:477-487. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6:271-281. [Google Scholar]
- 9.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 10.Haut, D. D., and D. J. Pintel. 1998. Intron definition is required for excision of the minute virus of mice small intron and definition of the upstream exon. J. Virol. 72:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. H., and M. Hannink. 2001. The N-terminal nuclear export sequence of IκBα is required for RanGTP-dependent binding to CRM1. J. Biol. Chem. 276:23599-23606. [DOI] [PubMed] [Google Scholar]
- 12.Lombardo, E., J. Ramirez, A. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virology 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroto, B., J. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melchior, F., and L. Gerace. 1998. Two-way trafficking with Ran. Trends Cell Biol. 8:175-179. [DOI] [PubMed] [Google Scholar]
- 15.Merchlinsky, M. J., P. J. Tattersall, J. J. Leary, S. F. Cotmore, E. M. Gardiner, and D. C. Ward. 1983. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J. Virol. 47:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 17.Mouw, M., and D. Pintel. 1998. Amino acids 16-275 of minute virus of mice NS1 include a domain that specifically binds (ACCA)2-3-containing DNA. Virology 251:123-131. [DOI] [PubMed] [Google Scholar]
- 18.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of MVM is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 20.Nuesch, J. P., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. [DOI] [PubMed] [Google Scholar]
- 21.Ohshima, T., T. Nakajima, T. Oishi, N. Imamoto, Y. Yoneda, A. Fukamizu, and K. Yagami. 1999. CRM1 mediates nuclear export of nonstructural protein 2 from parvovirus minute virus of mice. Biochem. Biophys. Res. Commun. 264:144-150. [DOI] [PubMed] [Google Scholar]
- 22.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus MVM is not required for encapsidation of progeny single-strand DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullman, K., M. Powers, and D. Forbes. 1997. Nuclear export receptors: from importin to exportin. Cell 90:967-970. [DOI] [PubMed] [Google Scholar]