Abstract
Enteropathogenic Escherichia coli (EPEC) is an important cause of infantile diarrhea, especially in developing countries. EspB, a key virulence factor of EPEC, is required for the attaching and effacing effect characteristic of EPEC and enterohemorrhagic E. coli and has been posited to play several functions in the process of infection. Attaching and effacing activity is associated with the accumulation of filamentous actin beneath adherent bacteria as measured in the fluorescence actin staining (FAS) test. To determine whether different domains of EspB are responsible for different functions, 42 plasmids carrying mutated espB were introduced into an espB deletion mutant. Two major groups of espB mutants were identified. One group of 17 mutants exhibited positive FAS results and normal levels of hemolytic activity. Another group of 22 mutants exhibited negative FAS results and low levels of hemolytic activity. Three mutants were exceptional. One mutant was FAS positive but had significantly reduced hemolytic activity. Conversely, a second mutant was FAS negative but had full hemolytic activity. A third mutant had a significantly reduced FAS level compared to the wild type but full hemolytic activity. The results of EspF and Tir translocation assays confirmed that FAS-negative insertions disrupt effector translocation and mutants with FAS-positive insertions retain protein translocation activity. These results suggest that EspB has distinct domain functions involved in effector translocation that can be distinguished from its role as a component of the translocation pore.
Enteropathogenic Escherichia coli (EPEC) is a major cause of infantile diarrhea in developing countries. The hallmark of EPEC infection is the ability of the pathogen to attach intimately to the host cell membrane, destroy microvilli, and induce the formation of cup-like pedestals composed of cytoskeletal proteins directly underneath the adherent bacteria. This phenomenon, known as the attaching and effacing (A/E) effect (38), has been observed in vitro in cell culture and in duodenal or rectal biopsy specimens from infants with EPEC infection (45, 58). A 35,624-bp genetic element known as the locus of enterocyte effacement (LEE) is necessary and sufficient for the A/E effect (35, 36). The LEE has 41 open reading frames whose products can be divided into five categories: a type III secretion system (T3SS) composed of Esc and Sep proteins; a filamentous protein translocation apparatus composed of EspA, EspB, and EspD; effector molecules including EspF, EspG, EspH, and Map; regulators known as Ler and GrlA/GrlR; and an outer membrane adhesin known as intimin and the translocated intimin receptor Tir (43).
The LEE is restricted to and conserved among all bacteria capable of A/E, including enterohemorrhagic E. coli (EHEC) and A/E pathogens of animals including the murine pathogen Citrobacter rodentium (11, 14, 44, 47, 64). The functions of the proteins encoded by the LEE have been systematically studied in mice by using C. rodentium as the model pathogen (12).
The A/E effect is dependent on the binding of intimin and Tir. After its translocation into the host cell by the EPEC T3SS, Tir is phosphorylated on two serine residues (61) as well as on tyrosine residue 474 (Y474) (24). The phosphorylation of Y474 is indispensable for actin remodeling in EPEC (23) but is not critical for binding to intimin (21, 24, 51). Phosphorylated Y474 and flanking residues of Tir directly bind the host adaptor protein Nck (3, 19). Nck subsequently recruits the neural Wiskott-Aldrich syndrome protein, which in turn activates the actin-related protein 2 and 3 (Arp2/3) pathway of actin assembly, leading to the formation of a cup-like structure or pedestal directly underneath the bacteria (32). Additional cytoskeletal proteins such as α-actinin, vinculin, cortactin, and talin are recruited to the pedestal and bind the cytoplasmic N terminus of Tir, independent of the C-terminal tyrosine phosphorylation (17). It seems that EPEC uses Tir to subvert fundamental host cell functions to build focal adhesion-like structures that strongly anchor the bacterium to the host cell cytoskeleton. This extracellular connection to the cytoskeleton permits bacteria to move laterally across the epithelial cell surface (46). In addition to Tir, the EPEC T3SS injects several other effector proteins into the host cell, including EspF, EspG, EspH, Map, Cif, and NleA (EspI) (5, 12, 25, 34, 37, 39, 57). These proteins have various effects on the host cell but are not required for the A/E effect.
The ability of the bacteria to deliver effector proteins into host cells is dependent on the presence of a translocation system that comprises filaments that extend from the bacterial surface to the membranes of host cells (28). In EPEC, EscF, EspA, EspB, and EspD are hypothesized to form this protein translocation apparatus. EscF forms a needle-shaped structure projecting approximately 50 nm from the bacterial surface, constituting the major internal structural component of the filament, and is required for protein secretion (49, 62). This needle-like structure is conserved among T3SSs. In the absence of the translocation apparatus proteins EspA, EspB, and EspD, effector molecules can be secreted from the bacteria but not translocated into the host cells. However, in the absence of EscF, effectors can be neither secreted nor translocated. Hence, EscF is part of both the secretion and translocation machinery.
The EspA protein interacts with itself through a coiled-coil domain to form an outer sheath that extends from the EscF needle structure on the bacterial surface by as much as 600 nm (10, 28). The three-dimensional structure of the EspA filament reveals a hollow helical tube with a diameter of 120 Å enclosing a central channel with a diameter of 25 Å through which effector proteins may be transported (9). The authors of a recent bioinformatics study suggest that EspA is related to flagellin (43).
EspB is a 312-amino-acid protein with one putative transmembrane domain and three putative coiled-coil domains. EspB is an essential virulence factor of EPEC in humans and animal models (1, 12, 40, 52). Together with EspA and EspD, EspB forms the translocation apparatus. Without these proteins and without the cytoplasmic protein SepL, effector proteins are secreted but not translocated (41). EspD has two putative transmembrane domains and has been observed in the host cell membrane (59). It is hypothesized that EspD also interacts with itself through a coiled-coil domain to form the main structure of the translocation pore in the host membrane through which the effector molecules pass and that the EspB protein is needed for full pore activity (8, 50, 60). Based on these results and the sequence homology between EspB and EspD and the YopD and YopB proteins, respectively, of Yersinia spp., an interaction between EspB and EspD to form the pore has been proposed but not demonstrated (20). In addition to its role as an essential component of the translocation apparatus, EspB itself is also translocated into the cytosol of host cells (55, 63). Recently, EspB from EHEC was found to bind to the cytoskeleton protein α-catenin in HeLa cells (29). Other authors have found that EPEC EspB can interact with α1-antitrypsin and that this interaction plays a role in pore formation (27). A recent mutagenesis study of yopD demonstrated different functions of different domains (42). Since several functions have been attributed to EspB, we sought to determine through random mutagenesis whether different domains of the molecule are responsible for different aspects of EspB function.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC serotype O127:H6 | 31 |
| UMD864 | E2348/69 ΔespB | 13 |
| UMD870 | E2348/69 espD::aphA3 | 30 |
| UMD872 | E2348/69 espA::aphA3 | 26 |
| Plasmids | ||
| pWSL-1 | Plasmid containing espB cloned into NcoI-EcoRI sites of pACYC184 | This study; 4 |
| pWSL-17 | Minimal plasmid containing espB cloned into SacII-AvaI sites of pACYC184 | This study |
| pTir232-Cya | pBR322-derived plasmid expressing a fusion of the N-terminal 232 amino acids of Tir and adenylate cyclase | 6 |
| pBPM37 | pBluescript-derived plasmid expressing an EspF-adenylate cyclase fusion protein | 37 |
espB mutagenesis.
Two procedures were used to generate random in-frame insertion mutations in the cloned espB gene. Thirty-one codon insertions were made by a previously described method that employs a transposon carried on a bacteriophage and allows screening for in-frame insertions because the insertions result in fusions with beta-galactosidase (33). We amplified a region flanking the espB gene with primers Donne-483 (5′-CCA TGC CAT GGA CTA TTT ACG TTC ATT ACG A-3′) and Donne-484 (5′-CGC GAA TTC GAT CTA CGC GGA TGG ACA A-3′) to create pWSL-1 as a target for this procedure. Plasmid insertion sites of TnlacZ/in were determined by restriction mapping. To convert TnlacZ/in insertion derivatives into 31-codon-insertion mutants, plasmid DNA was cleaved with BamHI, followed by ligation with T4 DNA ligase, transformation of E. coli strain CC118, and plating of transformants onto tryptone-yeast extract supplemented with tetracycline and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). LacZ− transformants were screened for loss of the insertion element on tryptone-yeast extract supplemented with chloramphenicol by patching, and chloramphenicol-sensitive strains were used as a source of plasmid DNA. Plasmid DNA was analyzed by restriction mapping and sequencing to confirm the position of each insert. Five-codon-insertion mutants were generated using the linker scanning mutagenesis GPS-LS kit (New England Biolabs) according to the manufacturer's instructions. To reduce the number of insertions to screen, we created pWSL-17, which has the espB gene with minimal flanking DNA cloned into a derivative of pACYC184 carrying deletions of most of its sequences outside of the origin of replication and the tetracycline resistance gene. For this purpose, we amplified the espB gene of EPEC strain E2348/69 by PCR with Pfu DNA polymerase (sense primer, Donne-534 [5′-ACT CTC GGG TGA TTC TGC GCG AGT GAA TAT-3′]; antisense primer, Donne-533 [5′-CTG ATC CGC GGA TCA ATT ACC CAG CTA AGCG-3′]). The purified PCR product was then cloned into AvaI and SacII sites of pACYC184 and sequenced. The function of the cloned EspB was verified by complementation of espB deletion mutant UMD864 with plasmid pWSL-17 to restore A/E ability (see below). Transposon mutagenesis was performed in vitro according to the manufacturer's instructions by using 80 ng of pWSL-17, and the DNA was used to transform strain DH5α. About 500 transformants were selected on Luria-Bertani (LB) medium containing chloramphenicol and kanamycin, and plasmids were prepared and screened by restriction digestion with BamHI and/or PmeI. Those plasmids with digestion patterns consistent with inserts within the espB gene were religated and sequenced to define the exact insertion sites. Finally, all plasmids with 5- or 31-codon insertions were used to transform UMD864 by electroporation.
Fluorescence actin staining (FAS).
HeLa cell (ATCC CCL2) monolayers were seeded (105 cells/500 μl of Dulbecco's modified Eagle's medium-F12 [DMEM-F12] supplemented with 10% fetal bovine serum [FBS] and 100 μg of penicillin and streptomycin/ml) into chamber slides, incubated at 37°C in an atmosphere of 95% air-5% CO2 for at least 18 h, and grown to 80% confluence. Bacteria from a fresh plate were grown in 1 to 2 ml of LB overnight at 37°C without shaking. The HeLa cells were washed with phosphate-buffered saline (PBS) once and covered with 500 μl of prewarmed DMEM-F12 without additives. After adjustment for the optical density at 600 nm (OD600), approximately 10 μl from each bacterial culture was added to each chamber (multiplicity of infection, ∼100) and slides were incubated for 3 h at 37°C in an atmosphere of 95% air-5% CO2. Slides were washed three times with 500 μl of PBS. Cells were then fixed with 500 μl of 2% formalin for 20 min at room temperature and washed with PBS three times. Cells were then permeabilized by the addition of 0.1% Triton X-100 for 4 min. After washing with PBS three times, cells were incubated with fluorescein isothiocyanate-phalloidin (5 μg/μl) for 20 min. After washing with PBS, the housing was removed and cells were covered with antifade reagent (Molecular Probes) and examined by epifluorescence microscopy without knowledge of strain identity for characteristic bright fluorescence beneath adherent bacteria.
Protein secretion assay.
Bacteria grown in LB overnight were diluted 1:100 into 20 ml of DMEM-F12 without additives and incubated at 37°C and 225 rpm until the OD600 reached 0.8 to 1.0. Bacteria were centrifuged at 3,000 × g and 4°C for 10 min, and the supernatant was sterilized by passage through a 0.4-μm-pore-size filter. Proteins present in the supernatant were precipitated by the addition of trichloroacetic acid (final concentration, 10% [vol/vol]) on ice for 1 h, followed by centrifugation at 10,000 × g at 4°C for 10 min. The pellet was resuspended in 200 μl of sample buffer and heated at 100°C for 10 min. The volume of sample loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis was adjusted according to the OD values of the starting material. Bacterial pellets from the first centrifugation step were washed once with PBS, resuspended in 1 ml of sample buffer, and heated at 100°C for 10 min, and adjusted volumes were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Immunoblotting for EspB was performed using an affinity-purified EspB antiserum (55) with ECL reagents (Amersham).
Hemolytic activity assay.
Sheep red blood cells (RBCs; Rockland Immunochemicals, Gilbertsville, PA) were resuspended in PBS, and 1 ml of a 3% (vol/vol) suspension was added to polylysine-coated 24-well tissue culture plates for 1 h. Nonattached RBCs were removed by careful washing with PBS three times. The resulting RBC monolayer was covered with 500 μl of HEPES-buffered DMEM-F12 without phenol red. Bacterial strains were grown overnight in LB without shaking. An amount of approximately 10 μl of each strain, adjusted according to the OD value, was added in triplicate to each well (multiplicity of infection, ∼30), and the plates were incubated at 37°C for 4.5 h in an atmosphere of 95% air-5% CO2. Following incubation, the culture medium from each sample was transferred into a 1.5-ml tube and centrifuged (12,000 × g, 5 min, 4°C). The amount of hemoglobin released into each sample was calculated by measuring the OD540 (value A). To determine the number of unlysed RBCs in each well, 500 μl of a solution of PBS diluted 30-fold was added and hemoglobin release was measured similarly (value B). Percent hemolysis (P) for each well was calculated using the following formula: P = [A/(A + B)] × 100. In experiments designed to determine whether different EspB proteins resulted in different-sized pores, osmoprotectants polyethylene glycol 2000 (PEG 2000), PEG 3000, and PEG 6000 were included in the assays at a concentration of 30 mM as previously described (22).
Adenylate cyclase protein fusion assay for EspF and Tir translocation.
HeLa cells were trypsinized, seeded into 12-well plates (5 × 105 cells per well), and incubated at 37°C in an atmosphere of 95% air-5% CO2 for at least 18 h. Bacterial strains containing plasmids expressing EspF-adenylate cyclase or Tir-adenylate cyclase fusion proteins were grown overnight at 37°C with aeration at 250 rpm in 2 ml of LB with appropriate antibiotics to retain plasmids. Each culture was diluted 1:100 into DMEM-F12 media including antibiotics and incubated under the same conditions for 4 h. Bacteria were harvested by centrifugation and resuspended in 10 ml of DMEM-F12 without antibiotics to an OD600 of 0.2. HeLa cells were washed twice with PBS and covered with 1.5 ml of prewarmed DMEM-F12 media lacking antibiotics. To each well, 0.5 ml of normalized bacterial culture was added, and the plates were incubated at 37°C in an atmosphere of 95% air-5% CO2 for 1.5 h. After infection, the cells were washed carefully with cold PBS twice to remove nonadherent bacteria. Four hundred microliters of 50 mM HCl was added to each well, and cells were collected into 1.5-ml centrifuge tubes by using disposable cell scrapers and placed in boiling water for 5 min. Tubes were then placed on ice, and 24 μl of 0.5 N NaOH was added to neutralize the acid. Ten-microliter aliquots were reserved for protein determination using the bicinchoninic acid microtiter plate protocol (Pierce). To extract cyclic AMP (cAMP), 800 μl of ethyl alcohol was added to each tube, tube contents were mixed well, and tubes were incubated at room temperature for 5 min. Samples were then centrifuged (10,000 × g, 5 min), and the supernatants were transferred into new 1.5-ml tubes for drying in a SpeedVac for 2 h. Samples were stored at −20°C until assay of cAMP concentrations by using the Biotrak enzyme immunoassay kit (Amersham). Each experiment was performed in triplicate five times. Data were normalized for protein concentration as previously described (37).
Quantification of EPEC adherence and FAS.
HeLa cells were infected according to the standard FAS procedure, washed with PBS, and fixed with 4% formalin for 20 min at room temperature. The slides were blocked with 10% FBS in PBS at 37°C for 1 h. After washing with PBS containing 2% FBS, the slides were incubated with rabbit serum raised against heat-killed EPEC strain E2348/69 (1:250 dilution) for 1 h at 37°C, followed by washing by incubation with rhodamine-conjugated anti-rabbit secondary antibody (1:200; Molecular Probes) for 1 h. HeLa cell nuclei were then stained with 1 μM DAPI (4′-6-diamidino-2-phenylindole) for 1 min, and antifade reagent was added. The slides were then examined without knowledge of strain identity under an epifluorescence microscope (Zeiss), and four representative fields were chosen for acquisition of images representing bacteria (red), actin (green), and nuclei (blue) by using an Axiocam digital camera and AxioVision 3.1 software (Zeiss). Each microcolony was circled, and the fluorescence intensities for actin and bacteria within each circle were measured. For the quantification of A/E, the ratio of the sum of actin intensities to that of bacterial intensities was calculated for each image. For adherence quantification, HeLa cells were counted on the basis of DAPI-stained nuclei and the sum of the bacterial fluorescence intensities on each field was divided by the number of cells. Data are expressed relative to the values obtained for the positive control sample and are representative of results of three independent experiments.
RESULTS
The EspB protein has domains required for overall function and domains highly tolerant to mutagenesis.
We used three different strategies to generate mutations in the cloned espB gene. We subjected pWSL-1 containing the cloned espB gene (Table 1) to an in vivo transposon insertion mutagenesis strategy followed by restriction digestion to generate nine in-frame 93-bp insertions (33). However, we found this procedure to be inefficient and therefore switched to an in vitro mutagenesis strategy. To minimize mutations outside of espB, we cloned the espB gene with minimal flanking DNA into a minimal pACYC184 derivative to create plasmid pWSL-17. Using the in vitro procedure, we generated 29 in-frame 15-bp insertions. This strategy also resulted in two out-of-frame insertions near the 3′ end of the gene, effectively causing deletions. In addition to these random mutations, we used restriction digestion to fuse portions of different plasmids containing in-frame 15-bp insertions, resulting in the in-frame deletion of the intervening fragments. The resulting plasmids were named according to the last amino acid in the EspB protein before the insertion or the amino acids deleted and are listed in Table 2. The locations of the mutations within arbitrarily divided lengths of the espB gene approximate a Poisson distribution and include regions encoding the putative transmembrane domain and coiled-coil regions as well as the N-terminal and C-terminal domains. The mean distance between the insertion sites is 8 amino acids (range, 1 to 36 amino acids; standard deviation, 9 amino acids). Only three stretches of the gene of greater than 45 bp are not represented, those encoding amino acids 102 to 128, 129 to 166, and 204 to 239. We transferred each of these plasmids into the EPEC espB deletion mutant strain UMD864 for analysis. We used UMD864 with plasmids pACYC184 and pWSL-17 as negative and positive controls, respectively, for all assays.
TABLE 2.
Plasmids containing mutated espB alleles and A/E and expression-secretion phenotypes conferred by the mutated alleles generated in this studya
| Plasmid | Type of mutation | FAS resultb | Secretion phenotypec |
|---|---|---|---|
| pEspB4 | 5-Codon insertion | + | + |
| pEspB5/30 | Deletion | − | − |
| pEspB11 | 5-Codon insertion | + | + |
| pEspB13 | 5-Codon insertion | + | + |
| pEspB25 | 5-Codon insertion | + | + |
| pEspB30 | 5-Codon insertion | + | + |
| pEspB36 | 5-Codon insertion | + | + |
| pEspB37 | 31-Codon insertion | + | + |
| pEspB48 | 5-Codon insertion | − | + |
| pEspB60 | 31-Codon insertion | − | + |
| pEspB68 | 5-Codon insertion | − | + |
| pEspB70 | 5-Codon insertion | − | + |
| pEspB75 | 31-Codon insertion | − | + |
| pEspB78 | 31-Codon insertion | − | + |
| pEspB83 | 5-Codon insertion | − | − |
| pEspB98 | 5-Codon insertion | − | + |
| pEspB101 | 5-Codon insertion | − | + |
| pEspB128 | 5-Codon insertion | + | + |
| pEspB166 | 31-Codon insertion | − | + |
| pEspB173 | 5-Codon insertion | − | − |
| pEspB179 | 5-Codon insertion | + | + |
| pEspB180/203 | Deletion | + | + |
| pEspB184 | 5-Codon insertion | + | + |
| pEspB186 | 5-Codon insertion | + | + |
| pEspB188 | 5-Codon insertion | + | + |
| pEspB203 | 5-Codon insertion | + | + |
| pEspB239 | 5-Codon insertion | − | + |
| pEspB241 | 5-Codon insertion | +/− | + |
| pEspB247 | 5-Codon insertion | + | + |
| pEspB252 | 5-Codon insertion | + | + |
| pEspB254 | 5-Codon insertion | − | + |
| pEspB257 | 5-Codon insertion | − | + |
| pEspB260 | 5-Codon insertion | − | + |
| pEspB264 | 31-Codon insertion | − | + |
| pEspB268 | 31-Codon insertion | − | − |
| pEspB282 | 31-Codon insertion | − | + |
| pEspB289 | 5-Codon insertion | − | + |
| pEspB302 | 5-Codon insertion | − | − |
| pEspB304stop | Deletion | − | + |
| pEspB305 | 31-Codon insertion | − | + |
| pEspB316 | 5-Codon insertion | + | + |
| pEspB316stop | Deletion | + | + |
The precise positions of the mutations are indicated by the plasmid names as described in the text.
+, able to restore a positive FAS result to an espB null mutant; −, unable to do so; +/−, able to confer very weak and inconsistent actin staining.
+, detectable levels of protein are expressed and secreted; −, detectable levels of protein are neither expressed nor secreted.
We used the FAS assay to test all 42 espB mutants for their abilities to induce the accumulation of filamentous actin beneath the bacteria in HeLa cells. We consider this test to be a measure of overall EspB function, encompassing all roles that it might potentially play in the formation of actin pedestals as part of the A/E effect. The results are listed in Table 2 and depicted in Fig. 1 in relation to the EspB sequence. These data reveal that two regions within the EspB protein, spanning approximately from resides 40 to 173 and from residue 257 to the carboxyl terminus, are required for overall function. It is noteworthy that these regions include the putative transmembrane domain and the third and strongest predicted coiled-coil domain. While the insertions after residues 98 and 101 are not predicted to destroy the transmembrane domain, the insertions after residues 282 and 289 are predicted to drastically reduce the tendency of this region of the protein to form coiled coils. Similarly, the insertion after residue 203 drastically reduces the tendency of this region to form the second coiled-coil domain. In contrast, two other regions, the amino terminus encompassing the first 37 amino acids and a middle region extending from residue 179 to residue 203, are highly permissive for insertions. To determine whether either of these regions includes any information required for activity, we fused plasmids with insertions at either end of these regions to generate in-frame deletions. Although the region between amino acids 179 and 203 can be removed without affecting activity, the deletion of residues 5 through 30 abolished function. This result is consistent with the notion that the amino terminus of EspB is highly tolerant to mutation but is nevertheless required for EspB function as has been reported for the N termini of other proteins secreted via T3SSs (2). Interestingly, although an insertion of 5 amino acids (MFKHT) at residue 239 totally disrupted function, an insertion at residue 241 (VFKHL) was only partially disruptive. The mutant carrying this insertion revealed an intermediate phenotype with very weak fluorescence under only a small number of adherent bacteria, which was clearly different from both the positive and negative controls. On the other hand, three nearby insertions at residues 247, 252, and 254 did not affect overall activity. We also observed that the extreme carboxyl terminus of EspB is not required for actin accumulation because an insertion at residue 316 and a deletion of the last 6 amino acids of EspB did not disrupt this activity. In contrast, a more extensive truncation after amino acid 303 disrupted function.
FIG. 1.
Sites of amino acid insertions and deletions in espB mutants in relation to FAS activity and to sequence features of EspB. The amino acid sequence of EspB from EPEC strain E2348/69 is shown. Residues conserved among all published EspB sequences are shown in red. Domains predicted to form coiled-coil interactions are shaded in gray. A predicted transmembrane domain is shaded in yellow. The last amino acids before the sites of linker insertion mutations that interfere (red) or do not interfere (green) with overall EspB function are indicated with inverted triangles. Amino acids deleted in certain espB mutants are underlined and color-coded in the same manner as insertion mutations.
We introduced all of the nonfunctional mutant espB alleles into wild-type EPEC strain E2348/69 to determine whether any interfere with activity in the presence of functional EspB. We found that all of the transformants remained FAS positive, indicating that none of the mutations was dominant.
Multiple regions within EspB contribute to its ability to be expressed and secreted.
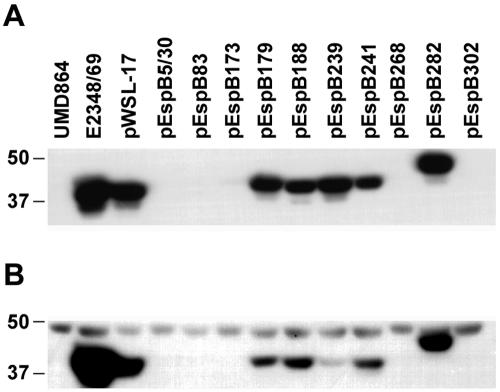
We performed Western blot assays to test the ability of each mutant to express and secrete EspB. For most of the mutants, whether FAS positive or negative, there was no detectable difference from the strain complemented with the wild-type allele in the ability to express and secrete EspB (Table 2). Of note, the levels of EspB in the complemented strain were lower than those in the wild-type strain, probably due to lack of the native promoter on the plasmid. However, a subset of strains with nonfunctional espB mutations had markedly reduced or undetectable levels of secreted EspB. This subset of strains included that with a deletion of the codons for amino acids 5 to 30 and those with insertions after residues 83, 173, 268, and 302 (Fig. 2). As the amount of cellular EspB was also drastically reduced in each of these mutants, it is not clear whether the lack of secretion is a primary or secondary effect. Thus, these mutations could result in a failure of the T3SS to recognize or secrete EspB, with subsequent degradation of cytoplasmic protein (41), or these residues could be important for either efficient EspB production, folding, or stability and the failure of secretion could be secondary to the reduced amounts of protein. All of the strains shown in Fig. 2 expressed and secreted EspA and EspD at levels comparable to those of the espB deletion mutant complemented with the wild-type espB gene (data not shown).
FIG. 2.
Immunoblot of EspB from culture supernatant (A) and bacterial pellets (B) of selected strains. Lane 1, espB mutant UMD864; lane 2, wild-type EPEC strain E2348/69; lanes 3 to 13, UMD864 complemented with plasmids as indicated. The positions of molecular mass markers (in kilodaltons) are shown. In panel B, a nonspecific cross-reactive band migrating slower than EspB serves as a loading control.
The hemolytic activity of EspB correlates imperfectly with overall function.
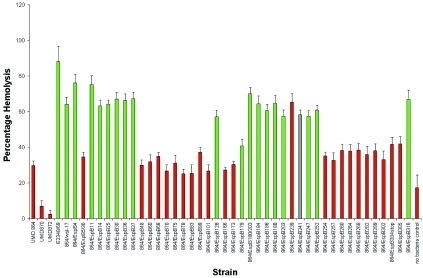
T3SSs of several pathogens including EPEC have been associated with hemolytic activity, a phenotype interpreted as relating to a pore-forming activity of the apparatus. We used a modified hemolytic activity procedure to evaluate the ability of each espB mutant to lyse sheep RBCs. The percentage of total hemoglobin released by each mutant is shown in Fig. 3. In general, mutants that expressed functional EspB, as indicated by positive FAS tests, exhibited levels of hemolytic activity comparable to that of the strain with the plasmid carrying the wild-type espB. Similarly, strains expressing nonfunctional EspB exhibited levels of hemolytic activity comparable to that of the espB deletion mutant strain carrying the control plasmid. These results suggest that the inability of these mutants to induce the accumulation of actin is caused by a deficiency in pore formation in the plasma membranes of host cells. However, two espB insertion mutants, UMD864/pEspB179 and UMD864/pEspB239, exhibited discordant behavior in the FAS and hemolytic assays. UMD864/pEspB179, which is FAS positive, exhibited hemolytic activity comparable to that of the control espB mutant strain UMD864/pACYC184, and UMD864/pEspB239, which is FAS negative, exhibited a level of hemolytic activity comparable to that of the strain complemented with wild-type espB. Similarly, strain UMD864/pEspB241, which has an intermediate FAS phenotype and has an insertion only 2 amino acids downstream from that in strain UMD864/pEspB239, had high hemolytic activity. We next examined the effect of osmoprotectants varying in molecular radii on hemolysis induced by UMD864/pWS-17, UMD864/pEspB179, UMD864/pEspB239, and UMD864/pEspB241. We found that maximum levels of hemolysis by all four strains occurred in the presence of PEG 2000 (molecular radius, 1.2 nm) and that PEG 6000 (molecular radius, 3.2 nm) completely inhibited hemolysis by all four strains (data not shown). PEG 3000 (molecular radius, 1.9 nm) partially inhibited hemolysis by all four strains. Thus, we could detect no drastic differences in the sizes of the pores induced by strains expressing selected mutant EspB proteins. These strains demonstrate that overall EspB function, as determined by FAS activity, can be dissociated from pore-forming activity. Thus, the ability to induce wild-type levels of hemolysis is neither necessary nor sufficient for EspB to support the accumulation of filamentous actin beneath bacteria. Furthermore, the espB deletion mutant UMD864 exhibited significantly higher levels of hemolytic activity (P < 0.05) than either the espA or the espD mutant, which further suggests that EspB does not have a primary role in pore formation and confirms previous observations (50).
FIG. 3.
Hemolytic activity of espB mutants. The percent lysis of sheep erythrocytes after incubation with each mutant and controls is indicated. Strains tested include the wild-type strain E2348/69, the espB mutant UMD864, the espD mutant UMD870, the espA mutant UMD872, and the espB mutant containing plasmids as shown. Green columns represent FAS-negative strains, red columns represent FAS-positive strains, and the gray column represents a mutant with an intermediate FAS phenotype. Data are from five independent experiments. Error bars represent the standard errors of the means.
The translocation activity of EspB correlates well with overall function.
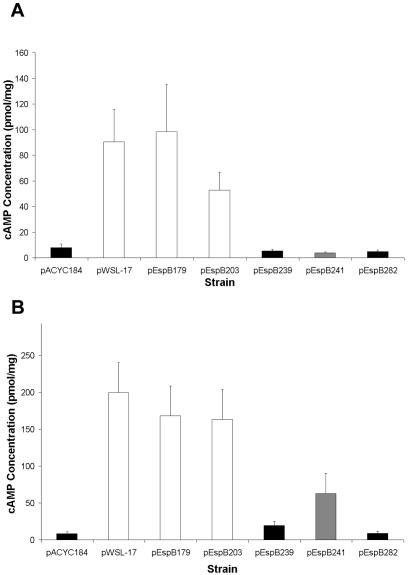
An espB deletion mutant strain is incapable of translocating EspF and Tir into host cells. To determine the effect of espB mutations on protein translocation into epithelial cells, we utilized two fusion genes encoding EspF and Tir fused to adenylate cyclase. We selected espB mutants representative of several classes for this assay. Strain UMD864/pEspB203 represents the FAS-positive and high-hemolytic-activity group, strain UMD863/pEsp282 represents the FAS-negative and low-hemolytic-activity group, strain UMD864/pEspB179 is a FAS-positive mutant with low hemolytic activity, strain UMD864/pEspB239 is a FAS-negative mutant with high hemolytic activity, and strain UMD864/pEspB241 is a mutant with intermediate FAS activity and high hemolytic activity. As expected, strain UMD864/pEspB203 induced high levels of cAMP, both when expressing the EspF fusion and when expressing the Tir fusion (Fig. 4). Conversely, cells infected with strain UMD864/pEspB282 expressing either the EspF or the Tir fusion had low levels of cAMP compared to cells infected with the espB deletion mutant harboring the vector plasmid. These results are consistent with the results of the FAS and hemolytic activity assays. However, cells infected with UMD864/pEspB179, which was FAS positive but had poor hemolytic activity, had cAMP levels comparable to those of cells infected with the strain complemented with wild-type espB. In contrast, cells infected with UMD864/pEspB239, which had a high level of hemolytic activity despite being FAS negative, had low levels of cAMP, indicating lack of translocation of EspF and Tir by this mutant. Another mutant with an insertion at residue 241, which retained pore formation activity, showed partially disrupted protein translocation. This finding demonstrates that pore formation by EPEC during the process of infection is not always coupled with effector protein translocation as previously assumed. Moreover, a domain of EspB including residues 239 and 241 is not required for pore formation but is required for protein translocation, strongly suggesting that EspB plays a role essential for effector protein translocation distinct from its role in pore formation. Thus, EspB has at least two major functions exhibited by different domains of the protein: one is a role in pore formation, indicated by 22 espB mutations that disrupted both pore formation and protein translocation. The other function is a role in protein translocation that is independent of pore formation.
FIG. 4.
Protein translocation activity of selected espB mutants. (A) cAMP levels in HeLa cells infected with espB mutant strain UMD864 transformed with pBPM37 expressing an EspF-adenylate cyclase fusion protein and with control plasmids or plasmids containing selected mutant espB alleles. (B) cAMP levels in HeLa cells similarly transformed with pTir232-Cya expressing a Tir-adenylate cyclase fusion protein and control and espB plasmids. Black columns represent FAS-negative strains, white columns represent FAS-positive strains, and gray columns represent a mutant with an intermediate FAS phenotype. Data are the means of results from five independent experiments performed in triplicate. Error bars show standard errors of the means.
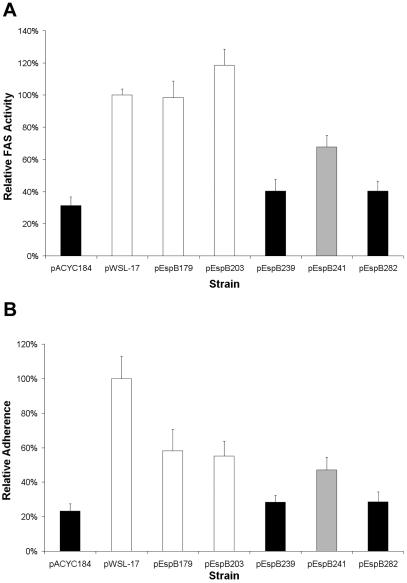
We developed quantitative adherence and FAS assays to better compare selected strains. These assays, performed without knowledge of strain identity, measure fluorescence units after staining of the bacteria with specific antiserum, cell nuclei with DAPI, and actin with phalloidin (see Materials and Methods). Adherence is expressed relative to the number of host cells, and FAS results are expressed as a ratio of actin fluorescence to bacterial fluorescence. The results of the quantitative FAS assay confirmed the impressions made by the standard assay (Fig. 5A). The insertion after residue 239, which abolished translocation without affecting pore formation, reduced the level of FAS to that of the negative control UMD864/pACYC184 and to that of an arbitrarily selected mutant strain that was defective for FAS, hemolysis, and translocation. In contrast, the insertion after residue 179, which reduced pore formation without affecting translocation, had no effect on quantitative FAS activity, as the mutant carrying this insertion was indistinguishable from the positive control strain UMD864/pWSL-17 and an arbitrarily selected mutant strain UMD864/pEspB203 that was positive in all assays. Finally, the quantitative assay confirmed the impression that the mutant UMD864/pEspB241 had an intermediate FAS response, as this mutant, which retained pore-forming activity but had reduced translocation activity, displayed significantly lower FAS activity than the positive control strain and significantly higher FAS activity than the negative control strain. Thus, the quantitative data are consistent with translocation data.
FIG. 5.
Quantitative FAS and adherence activities of selected espB mutants carrying the indicated plasmids. A/E activity was quantified as the ratio of fluorescence intensity of actin staining to that of bacterial staining (A), and adherence was quantified as the mean fluorescence intensity of bacterial staining per HeLa cell as described in Materials and Methods (B). Data are normalized to the values obtained for the espB deletion mutant UMD864 complemented with a plasmid expressing the wild-type EspB protein. Black columns represent FAS-negative strains, white columns represent FAS-positive strains, and gray columns represent a mutant with an intermediate FAS phenotype. Values are the means of results from three independent experiments, each performed in quadruplicate. Error bars show standard errors of the means.
When we quantified the ability of the same selected espB mutant strains to adhere to host cells, we noted a general correlation between adherence and FAS activity (Fig. 5B). Interestingly, compared to the positive control strain UMD864/pWSL-17, each of the selected espB mutants displayed significantly reduced adherence to HeLa cells. However, adherence as low as that of the negative control strain was exhibited only by the arbitrarily selected FAS-negative mutant and by the FAS-negative, pore formation-competent, translocation-defective mutant UMD864/pEspB239. Once again, pore formation-deficient, translocation-proficient strain UMD864/pEspB179 was indistinguishable in adherence from the arbitrarily selected mutant positive in all assays, and UMD864/pEspB241 was intermediate in adherence. The differences in adherence abilities between mutants noted in this quantitative adherence assay do not account for the differences observed in the quantitative FAS assay, as data from the latter assay are expressed as a ratio between actin staining and bacterial staining and thus correct for differences in adherence. Therefore, these data indicate that adherence ability is directly related to translocation and A/E activity and are consistent with the concept that the Tir-intimin interaction makes a strong contribution to overall adherence.
DISCUSSION
EPEC strains share with many gram-negative pathogens a T3SS that secretes effector proteins and translocates them into host cells. In the present study, we sought to learn more about the EspB translocation apparatus protein by testing the hypothesis that different domains of EspB have different functions. We generated 42 espB mutants and determined whether they produced functional or nonfunctional EspB on the basis of their capacities to restore the ability of an espB deletion mutant to induce the accumulation of actin beneath adherent bacteria. We further evaluated these mutants for EspB production and secretion, hemolytic activity, and translocation activity and performed quantitative assays of adherence and FAS activity. In general, there was a strong correlation between overall function, hemolytic activity, and translocation activity. However, 3 of the 42 mutants analyzed displayed unique phenotypes. One had normal levels of hemolytic activity but did not allow for translocation of effector proteins and was therefore nonfunctional; one had normal levels of hemolytic activity, allowed for reduced translocation of effector proteins, and was thus only partially functional; and one allowed for normal translocation despite reduced hemolytic activity and remained functional. Thus, our principle finding is that the role of EspB in forming a pore in the host cell membrane, as determined by hemolytic activity, could be separated from its role in protein translocation.
The concept that bacteria that have T3SSs, such as EPEC and EHEC, form a translocation pore in the cytoplasmic membrane that allows for transfer of effectors into the host cell is not new (16). In the case of EPEC and EHEC, the EspD and EspB proteins have been suggested to make up this pore on the basis of contact hemolytic activity assays that demonstrate a requirement for the T3SS, EspA, EspB, and EspD for activity (22, 50, 59, 60). EspA forms the filament by which the bacteria are tethered to the host cell (28). This filament has a hollow core through which secreted proteins are thought to pass (9). The EspB and EspD proteins are believed to possess the actual pore-forming activity. In the case of atypical, diffusely adhering EPEC strains, contact between the bacteria and erythrocytes is not required for hemolytic activity and the secreted proteins themselves cause hemolysis (22). Osmoprotection assays suggest that the pore created by the conditioned medium containing the secreted proteins is 3 to 5 nm in diameter. Furthermore, using atomic force microscopy, the authors of the osmoprotection study observed structures in the host cell membrane resembling a possible translocation pore with an outer diameter of 80 nm. They also detected an interaction between EspD and EspB by using a gel overlay assay. Our data strongly support the hypothesis that EspB is involved in pore formation because mutagenesis of espB revealed that two major regions (residues 40 to 173 and the C-terminal 55 amino acids) of EspB are required for hemolysis, effector protein translocation, and overall function. Our data are also consistent with the hypothesis that EspB is not the primary component of this pore. In agreement with other authors (50), we found that the residual hemolytic activity of an espB null mutant is higher than that of espD and espA null mutants. Furthermore, we identified espB alleles that support full hemolysis but not protein translocation and vice versa. In particular, the insertion of 5 amino acids (VFKHK) after residue 179 resulted in a protein that had markedly reduced hemolytic activity but retained full translocation activity and function. Thus, we propose that EspB performs a secondary role in pore formation, assisting EspD in forming a fully functional translocation pore.
Results obtained from studying Yersinia enterocolitica and Yersinia pseudotuberculosis also support the concept that EspB plays a secondary role in pore formation. In these organisms, both the YopD protein, which resembles EspB in sequence, and YopB, which resembles EspD, are required for hemolytic activity (20). When Y. pseudotuberculosis is incubated in the presence of liposomes, YopB and YopD are inserted into these structures. When fused with planar lipid bilayers, these liposomes have channel-forming activity (53). This activity is absolutely dependent on YopB, but channels with altered activity are still formed in the absence of YopD. Like EspB, YopD has been detected both in host cell membranes and in the cytoplasm (15, 55, 63). PopB and PopD, the YopB and YopD homologues from Pseudomonas aeruginosa, are also inserted into host membranes (48). When purified PopB and PopD are incubated with liposomes, they interact with each other, as shown by coimmunoprecipitation (18).
In a recent investigation with parallels to the present study, systematic deletions were made in the yopD gene and the resulting alleles were introduced into a null mutant to determine whether different domains of the protein have different functions (42). Interestingly, 4 of the 13 mutants studied were found to retain hemolytic activity but 2 had lost translocation activity. These mutants are therefore similar to the espB mutants we describe with insertions after amino acids 239 and 241. The authors suggested that YopD plays a specific role in translocation of effector proteins beyond its role in pore formation. However, unlike the present study, the previous report described no mutants that lost hemolytic activity but retained translocation activity. Similarly, the finding that overall EspB activity was always associated with protein translocation but not always with hemolytic activity suggests that EspB plays a role in translocation that is distinct from its secondary role in pore formation. Our data suggest that a domain of the protein that includes residues 239 and 241 somehow assists effector proteins in traversing the translocation pore. This hypothesis is consistent with the difference in translocation levels of the EspF-adenylate cyclase and Tir-adenylate cyclase fusion proteins noted for the mutant with the insertion after residue 241, as different effector proteins may have different requirements for translocation. An alternative hypothesis states that a domain including residue 241 plays no specific role in translocation, but the insertion of 5 amino acids in this region obstructs translocation. If the latter hypothesis was correct, one would predict that expression of the mutant allele in the presence of the wild-type allele would interfere with function. However, we observed no such dominant negative phenotype.
One of the strengths of this study was the quantitative data obtained from assays of hemolysis, translocation, adherence, and FAS activity, which were entirely supportive of qualitative observations. Quantitative assessments of adherence and FAS, the latter assay the first of its kind, yielded support for the role of A/E in overall adherence. We found a direct correlation between the ability of each strain to induce pedestal formation and its overall adherence ability. These data are consistent with the notion that the intimin-Tir interaction is paramount to adhesion, as the ability to translocate Tir appears to be required for optimal adherence.
We also tested each mutant for EspB expression and secretion and, as in a prior study, found a strong correlation between secretion and bacterial protein levels (41). This correlation suggests that when EspB is not secreted it is unstable and rapidly degraded. Mutations that interfered with secretion were scattered throughout the sequence, often near the insertion sites of well-expressed and secreted proteins, and included a deletion from residues 5 to 30 and insertions after residues 83, 173, 268, and 303. It would be interesting to determine whether any of these insertions interfered with binding to the EspB chaperone CesAB (7).
In addition to the role of EspB in translocation, it has been speculated that EspB may itself be an effector protein. In agreement with this hypothesis, EspB has been found in the host cytoplasm, where it may have the opportunity to interact with host cell proteins (55). Furthermore, stable expression of EPEC EspB in HeLa cells resulted in a dramatic alteration in cell morphology with loss of actin stress fibers (54). Early speculation that EspB might bind pyridoxal phosphate (13) was not supported by direct evidence (56). However, EHEC EspB was reported to bind to the cytoskeleton protein α-catenin from HeLa cells and to recruit α-catenin to the site of A/E lesions (29). More recently, α1-antitrypsin was found to bind to EspB from atypical EPEC by pull-down and gel overlay assays in vitro (27). Furthermore, addition of α1-antitrypsin reduced EPEC-mediated hemolytic activity in a concentration-dependent manner. Thus, α1-antitrypsin is implicated as a host factor that plays a role in EspB pore-forming activity. However, the data from the present study provide no evidence either for or against a possible function of EspB as an effector protein essential for A/E, as none of the 42 mutants tested lost overall function but retained translocation activity. Thus, for now, the primary role of EspB seems to be to assist proteins to traverse the translocation pore, and much additional work is necessary to determine the precise function of EspB.
Acknowledgments
This work was supported by Public Health Service Award AI32074 from the National Institutes of Health.
We thank Laia Egea Pujol for technical assistance.
Editor: J. B. Bliska
REFERENCES
- 1.Abe, A., U. Heczko, R. G. Hegele, and B. B. Finlay. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188:1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 3.Campellone, K. G., A. Giese, D. J. Tipper, and J. M. Leong. 2002. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 43:1227-1241. [DOI] [PubMed] [Google Scholar]
- 4.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford, J. A., and J. B. Kaper. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855-868. [DOI] [PubMed] [Google Scholar]
- 7.Creasey, E. A., D. Friedberg, R. K. Shaw, T. Umanski, S. Knutton, I. Rosenshine, and G. Frankel. 2003. CesAB is an enteropathogenic Escherichia coli chaperone for the type-III translocator proteins EspA and EspB. Microbiology 149:3639-3647. [DOI] [PubMed] [Google Scholar]
- 8.Daniell, S. J., R. M. Delahay, R. K. Shaw, E. L. Hartland, M. J. Pallen, F. Booy, F. Ebel, S. Knutton, and G. Frankel. 2001. Coiled-coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect. Immun. 69:4055-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniell, S. J., E. Kocsis, E. Morris, S. Knutton, F. P. Booy, and G. Frankel. 2003. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol. Microbiol. 49:301-308. [DOI] [PubMed] [Google Scholar]
- 10.Delahay, R. M., S. Knutton, R. K. Shaw, E. L. Hartland, M. J. Pallen, and G. Frankel. 1999. The coiled-coil domain of EspA is essential for the assembly of the type III secretion translocon on the surface of enteropathogenic Escherichia coli. J. Biol. Chem. 274:35969-35974. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnenberg, M. S., J. Yu, and J. B. Kaper. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175:4670-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. Deng, L.-C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) of enteropathogenic E. coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 15.Francis, M. S., and H. Wolf-Watz. 1998. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol. Microbiol. 29:799-813. [DOI] [PubMed] [Google Scholar]
- 16.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 17.Goosney, D. L., R. DeVinney, R. A. Pfuetzner, E. A. Frey, N. C. Strynadka, and B. B. Finlay. 2000. Enteropathogenic E. coli translocated intimin receptor, Tir, interacts directly with α-actinin. Curr. Biol. 10:735-738. [DOI] [PubMed] [Google Scholar]
- 18.Goure, J., A. Pastor, E. Faudry, J. Chabert, A. Dessen, and I. Attree. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruenheid, S., R. DeVinney, F. Bladt, D. Goosney, S. Gelkop, G. D. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856-859. [DOI] [PubMed] [Google Scholar]
- 20.Håkansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homblé, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 21.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 22.Ide, T., S. Laarmann, L. Greune, H. Schillers, H. Oberleithner, and M. A. Schmidt. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 23.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 24.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 25.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 26.Kenny, B., L.-C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli (EPEC), is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 27.Knappstein, S., T. Ide, M. A. Schmidt, and G. Heusipp. 2004. Alpha 1-antitrypsin binds to and interferes with functionality of EspB from atypical and typical enteropathogenic Escherichia coli strains. Infect. Immun. 72:4344-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kodama, T., Y. Akeda, G. Kono, A. Takahashi, K. Imura, T. Iida, and T. Honda. 2002. The EspB protein of enterohaemorrhagic Escherichia coli interacts directly with alpha-catenin. Cell. Microbiol. 4:213-222. [DOI] [PubMed] [Google Scholar]
- 30.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Sotman, and B. Rowe. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1:1119-1122. [DOI] [PubMed] [Google Scholar]
- 32.Lommel, S., S. Benesch, K. Rottner, T. Franz, J. Wehland, and R. Kuhn. 2001. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2:850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoil, C., and J. Bailey. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J. Mol. Biol. 267:250-263. [DOI] [PubMed] [Google Scholar]
- 34.Matsuzawa, T., A. Kuwae, S. Yoshida, C. Sasakawa, and A. Abe. 2004. Enteropathogenic Escherichia coli activates the RhoA signaling pathway via the stimulation of GEF-H1. EMBO J. 23:3570-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on K-12 E. coli. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 37.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrède, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman, J. V., B. A. Zabel, S. S. Jha, and D. B. Schauer. 1999. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect. Immun. 67:6019-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connell, C. B., E. A. Creasey, S. Knutton, S. Elliott, L. J. Crowther, W. Luo, M. J. Albert, J. B. Kaper, G. Frankel, and M. S. Donnenberg. 2004. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol. Microbiol. 52:1613-1625. [DOI] [PubMed] [Google Scholar]
- 42.Olsson, J., P. J. Edqvist, J. E. Broms, A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2004. The YopD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J. Bacteriol. 186:4110-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallen, M. J., S. A. Beatson, and C. M. Bailey. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perna, N. T., G. F. Mayhew, G. Pósfai, S. J. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothbaum, R. J., J. C. Partin, K. Saalfield, and A. J. McAdams. 1983. An ultrastructural study of enteropathogenic Escherichia coli infection in human infants. Ultrastruct. Pathol. 4:291-304. [DOI] [PubMed] [Google Scholar]
- 46.Sanger, J. M., R. Chang, F. Ashton, J. B. Kaper, and J. W. Sanger. 1996. Novel form of actin-based motility transports bacteria on the surface of infected cells. Cell Motil. Cytoskeleton 34:279-287. [DOI] [PubMed] [Google Scholar]
- 47.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoehn, G., A. M. Di Guilmi, D. Lemaire, I. Attree, W. Weissenhorn, and A. Dessen. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22:4957-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw, R. K., S. Daniell, F. Ebel, G. Frankel, and S. Knutton. 2001. EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol. 3:213-222. [DOI] [PubMed] [Google Scholar]
- 51.Shaw, R. K., S. Daniell, G. Frankel, and S. Knutton. 2002. Enteropathogenic Escherichia coli translocate Tir and form an intimin-Tir intimate attachment to red blood cell membranes. Microbiology 148:1355-1365. [DOI] [PubMed] [Google Scholar]
- 52.Tacket, C. O., M. B. Sztein, G. Losonsky, A. Abe, B. B. Finlay, B. P. McNamara, G. T. Fantry, S. P. James, J. P. Nataro, M. M. Levine, and M. S. Donnenberg. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tardy, F., F. Homblé, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, K. A., P. W. Luther, and M. S. Donnenberg. 1999. Expression of the EspB protein of enteropathogenic Escherichia coli within HeLa cells affects stress fibers and cellular morphology. Infect. Immun. 67:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, K. A., C. B. O'Connell, P. W. Luther, and M. S. Donnenberg. 1998. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect. Immun. 66:5501-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, K. A., C. B. O'Connell, R. Thompson, and M. S. Donnenberg. 2001. The role of pyridoxal phosphate in the function of EspB, a protein secreted by enteropathogenic Escherichia coli. FEBS Lett. 488:55-58. [DOI] [PubMed] [Google Scholar]
- 57.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 58.Ulshen, M. H., and J. L. Rollo. 1980. Pathogenesis of Escherichia coli gastroenteritis in man—another mechanism. N. Engl. J. Med. 302:99-101. [DOI] [PubMed] [Google Scholar]
- 59.Wachter, C., C. Beinke, M. Mattes, and M. A. Schmidt. 1999. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 31:1695-1707. [DOI] [PubMed] [Google Scholar]
- 60.Warawa, J., B. B. Finlay, and B. Kenny. 1999. Type III secretion-dependent hemolytic activity of enteropathogenic Escherichia coli. Infect. Immun. 67:5538-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warawa, J., and B. Kenny. 2001. Phosphoserine modification of the enteropathogenic Escherichia coli Tir molecule is required to trigger conformational changes in Tir and efficient pedestal elongation. Mol. Microbiol. 42:1269-1280. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3:753-762. [DOI] [PubMed] [Google Scholar]
- 63.Wolff, C., I. Nisan, E. Hanski, G. Frankel, and I. Rosenshine. 1998. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 28:143-155. [DOI] [PubMed] [Google Scholar]
- 64.Zhu, C., T. S. Agin, S. J. Elliott, L. A. Johnson, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]