Abstract
Infection with the human gastric pathogen Helicobacter pylori can give rise to chronic gastritis, peptic ulcer, and gastric cancer. All H. pylori strains express the surface-localized protein HpaA, a promising candidate for a vaccine against H. pylori infection. To study the physiological importance of HpaA, a mutation of the hpaA gene was introduced into a mouse-adapted H. pylori strain. To justify that the interruption of the hpaA gene did not cause any polar effects of downstream genes or was associated with a second site mutation, the protein expression patterns of the mutant and wild-type strains were characterized by two different proteomic approaches. Two-dimensional differential in-gel electrophoresis analysis of whole-cell extracts and subcellular fractionation combined with nano-liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometry for outer membrane protein profiling revealed only minor differences in the protein profile between the mutant and the wild-type strains. Therefore, the mutant strain was tested for its colonizing ability in a well-established mouse model. While inoculation with the wild-type strain resulted in heavily H. pylori-infected mice, the HpaA mutant strain was not able to establish colonization. Thus, by combining proteomic analysis and in vivo studies, we conclude that HpaA is essential for the colonization of H. pylori in mice.
Helicobacter pylori is a gram-negative bacterium that colonizes the human gastric and duodenal mucosa of more than half of the world's population, with the majority of infections occurring in the developing world (26). Infection with the bacterium can give rise to chronic gastritis, peptic ulcer, and gastric cancer in a subpopulation of individuals (6). Both bacterial and host genetic determinants may contribute to these various disease outcomes. Due to the important role of membrane proteins in adhesion and survival in the host, membrane proteins are known to play an essential role in the pathogenicity of the bacteria and are potential targets in vaccine development.
In order to establish and maintain infection, H. pylori expresses a variety of different types of colonization and virulence factors. This includes the urease enzyme that neutralizes gastric acidity in order to create a favorable environment for the bacterium and the flagella, which provide motility. Animal studies have shown that urease and flagella are essential for colonization, because strains lacking any of these proteins cannot colonize mice (13, 33). Additionally, H. pylori produces a number of factors harmful to the host, e.g., the CagA pathogenicity island, H. pylori neutrophil-activating protein, and the VacA cytotoxin. However, infection studies in animal models with H. pylori strains lacking either cagA or vacA have shown that they are not important for colonization (31, 38).
Many H. pylori proteins have been implicated to have a role in the attachment of the bacteria to epithelial cells, for example, the blood group antigen binding adhesin (BabA) and the sialic acid binding adhesin (SabA), which bind to Lewis b (Leb) and sialyl Lex receptors, respectively, on epithelial cells in the gastric mucosa (10, 16, 18)
H. pylori adhesin A (HpaA) is a surface-located (7, 14, 20) lipoprotein (25) that was initially described as a sialic acid binding adhesin, but supportive evidence is still lacking. It is recognized by antibodies from H. pylori-infected individuals (23, 39), and the expression of the HpaA protein has previously been found to be highly conserved among H. pylori isolates (9, 39). Furthermore, genomic studies (2, 32) show no significant sequence homologies of HpaA with other known proteins. Taken together, this makes HpaA a putative candidate as a vaccine antigen against H. pylori infection.
In this study, we have constructed an HpaA mutant in the mouse-adapted H. pylori Sydney strain 1 (SS1) to examine the role of HpaA in colonization. Because of cotranscription, constructed gene mutations have the potential to cause polar effects, i.e., inhibiting expression of downstream genes in an operon. In addition, it has been shown that knocking out one gene can affect other genes in an unpredicted manner (30). Thus, when studying a mutant, proteomic analysis offers a convenient method to monitor changes in protein expression without prior knowledge of what those changes might be.
Studies of protein expression profiles are usually obtained by two-dimensional (2-D) gel analysis of cell extracts from the cells or tissue of interest. However, despite a wealth of experimental progress, critical limitations, such as discrimination against low-abundance and highly hydrophobic proteins, are still a problem in standard 2-DE gels.
The introduction of differential in-gel electrophoresis (DIGE) (34), which allows separation of two sets of protein mixtures from different sources (e.g., wild type and mutant), has minimized previous difficulties with reproducibility associated with 2-D gels. Moreover, recent studies that focused on subproteome analysis combining subcellular fractionation with 2-D gel analysis or non-gel-based proteomic techniques, such as liquid chromatography (LC) coupled with mass spectrometry (MS), have shown impressive results regarding improved dynamic range and identification of hydrophobic proteins (3, 8, 40). Several previous studies have focused on the identification of outer membrane proteins (OMPs) from H. pylori (5, 12, 15, 24).
The first aim of this study was to examine the overall protein profile, including the protein expression of the genes located downstream of hpaA, of the mouse-adapted SS1 strain and its isogenic HpaA mutant. This was achieved by a proteomic approach where whole-cell extracts of the bacteria were compared by DIGE analysis. We also combined subcellular fractionation and one-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis with nano-LC Fourier transform (FT) ion cyclotron resonance (ICR) (FT-ICR) MS and tandem MS (MS/MS) analyses in order to compare the OMP profiles of the SS1 wild-type and mutant strains. To determine whether HpaA is essential for survival in the host, mice were infected with either H. pylori SS1 or the HpaA mutant strain, and the colonization levels and immune responses were assessed.
MATERIALS AND METHODS
Construction of SS1 hpaA-negative/deficient mutant SS1(ΔhpaA).
The hpaA mutant was originally constructed in H. pylori strain CCUG 17874 by a two-step amplification resulting in a 450-bp deletion of the hpaA gene (kindly provided by P. Doig et al., Astrazeneca Research Centre, Boston, MA) and insertion of a 1.4-kb kanamycin cassette (25). The mutation was transferred from H. pylori CCUG 17874 to the mouse-adapted SS1 strain by natural transformation. Five kanamycin-resistant transformants were analyzed by PCR with two HpaA-specific primers (forward primer, 5′-GGCGTAGAAATGGAAGCG-3′; reverse primer, 5′-CCCAAGCTTCATCAGCCCTTAAATACACG-3′) (21) to confirm that the kanamycin cassette was inserted in the hpaA gene, resulting in a larger PCR product than of that of the wild-type SS1 strain. One of the transformants with the correct insertion was further characterized by SDS-PAGE and immunoblotting with the monoclonal antibody HP30-1:1:6, specific for HpaA (9). This strain, SS1(ΔhpaA), was negative in the immunoblot.
Strains and culture conditions.
The mouse-adapted H. pylori strains SS1 (CagA+ VacA+ Ley) (19) and SS1(ΔhpaA) were used in all experiments and stored at −70°C as stock cultures. For preparation of antigens from SS1 and SS1(ΔhpaA), the bacteria were grown on Colombia-Iso agar plates to confluence for 3 days under microaerophilic conditions (10% CO2, 6% O2, and 84% N2). SS1(ΔhpaA) was cultured in the same way as SS1 throughout the experiment, with the exception of the cultures being supplemented with 25 μg/ml kanamycin.
Growth curves.
SS1 and SS1(ΔhpaA) were first grown on Colombia-Iso plates to confluence for 2 to 3 days and then resuspended in 2 ml Brucella broth (Difco Laboratories) to an optical density at 600 nm (OD600) of 0.3 (1.5 × 109 bacteria/ml). The bacteria were then transferred to a 250-ml flask containing 25 ml Brucella broth supplemented with 10 μg/ml vancomycin and 5 μg/ml trimethoprim for 7 days. The liquid culture was exposed to a gas mixture consisting of 10% CO2, 6% O2, and 84% N2 and incubated in a rotary shaker at 150 rpm. Samples were taken at several time points over 7 days, and the OD600 was determined as a measure of the growth of bacteria. The viable count of the bacteria was also performed by making serial dilutions from the liquid culture, and 100 μl of the diluted suspensions was spread on horse blood agar plates and incubated under microaerophilic conditions. The bacterial colonies were then counted.
Sample preparation for 2-D DIGE.
Bacterial cells from culture dishes (Colombia-Iso plates) were dissolved in sample buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 10 mM Tris-HCl, pH 8.5}, lysed by sonication (five 15-s pulses on ice), and centrifuged at 12,000 × g for 10 min. Protein extracts from three unique preparations for each strain were pooled, and the protein concentrations of the samples were determined by the Coomassie Plus protein assay as recommended by the manufacturer (Pierce, Rockford, IL). Stock cyanine fluorescent dyes (Cy2, Cy3, or Cy5) (34) (1 nmol/μl; GE Healthcare, Uppsala, Sweden) were reconstituted in anhydrous N,N-dimethylformamide (Fluka, Buchs, Switzerland) to 400 pmol/μl, and 8 pmol dye was added per μg protein in the cell lysate. The sample was vortexed, centrifuged briefly, and left on ice in the dark for 30 min. The reaction mixture was quenched by the addition of 10 mM lysine followed by incubation on ice for a further 10 min.
According to the instructions from the manufacturer (GE Healthcare, Uppsala, Sweden), two out of four samples from the wild-type strain were labeled with Cy3, and the other two samples were labeled with Cy5; the same was done for the mutant. An internal standard composed of equivalent amounts of proteins from the wild-type strain and the mutant was labeled with Cy2, mixed with the labeled wild-type and mutant lysates, and run in one gel. One additional preparative gel that contained 500 μg of the pooled samples was also run, so a total of five gels were run within the same set.
2-D electrophoresis.
Prior to isoelectric focusing (IEF), the sample was mixed with rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 1% IPG buffer [pH 3 to 10], 13 mM dithiothreitol) before being applied to 24-cm DryStrips (pH 3 to 10 [nonlinear]; Amersham Bioscience, Uppsala, Sweden) in a strip holder for active rehydration. The first-dimension separation was carried out with an isoelectric focusing system (IPGphor; Amersham Bioscience, Uppsala, Sweden). The total focusing time was 60 kVh. After equilibration, second-dimension electrophoresis was performed in an ETTAN Dalt (Amersham Bioscience) at 15 mA/gel for 15 h on 12% polyacrylamide gels cast in 20- by 24-cm glass plates. After 2-DE, gels were scanned with the 2920 Master Imager (Amersham Bioscience) using excitation/emission wavelengths specific for the different CyDyes.
Image analysis.
2-D images were created and analyzed by DeCyder (GE Healthcare) software. The differential in-gel analysis module was used to define spot boundaries and to filter out spots resulting from nonprotein sources (e.g., dust particles). The gels were normalized against the internal standard and grouped. The biological variation analysis mode of DeCyder was then used to match all images for comparative cross-gel statistical analysis of the two groups (wild type and mutant) using a Student's t test. Comparison of test spot volumes with the corresponding standard spot volumes gave a standardized abundance for each matched spot; these values were averaged across the four replicates and compared between the SS1 strain and its isogenic hpaA mutant. Spots displaying P values of less than 0.05 were selected for identification by MS.
OMP preparation.
The sarcosine-insoluble outer membrane fraction of H. pylori was prepared as described previously (5), with minor modifications. H. pylori cells were harvested by centrifugation (5,000 × g, 10 min) and washed with 20 mM Tris-HCl (pH 7.5). The cells were suspended in 20 mM Tris-HCl (pH 7.5) and then disrupted with an ultrasonicator. DNase (Benzonase; Sigma-Aldrich, Steinheim, Germany) and protease inhibitors (Complete EDTA-free; Roche Diagnostics, Mannheim, Germany) were added to the cell suspension, and the mixture was incubated at room temperature for 30 min. Unbroken cells were removed by centrifugation (12,000 × g, 20 min, 4°C). Total membrane proteins were collected by centrifugation (45,000 × g, 30 min, 4°C), resuspended in 20 mM Tris-HCl (pH 7.5) containing 2.0% (wt/vol) sodium lauryl sarcosine (USB, Cleveland, OH), and incubated at room temperature for 30 min. OMPs were collected by centrifugation (45,000 × g, 30 min, 4°C).
The OMP pellet was dissolved in SDS sample buffer and separated by SDS-PAGE (10% Bis-Tris gel; Novex, San Diego, CA). After staining of the gels with either Sypro ruby (Molecular Probes, Eugene, OR) or Bio-Safe Coomassie (Bio-Rad), all visual protein bands were excised for further analysis.
The method described previously by Shevchenko et al. (29) was applied, with some minor modifications. Briefly, the gel pieces were destained by washing three times with 25 mM NH4HCO3 in 50% CH3CN. Gel pieces were dried in a vacuum centrifuge and incubated with digestion buffer (50 mM NH4HCO3, 10 ng/μl trypsin) at 37°C overnight. Peptides were extracted in 50% CH3CN-1% CH3COOH, and the supernatant was evaporated to dryness in a vacuum centrifuge.
Nano-LC FT-ICR MS and MS/MS analyses.
Protein digests were reconstituted in 0.1% HCOOH (Merck, Darmstadt, Germany) and separated by chromatography in a 17-cm fused silica column (50-μm inner diameter) packed in-house with reverse-phase C18-AQ 3-μm porous particles with a 50-min gradient from 0 to 50% CH3CN (Merck)-0.1% HCOOH at a flow rate of approximately 100 nl/min. A tapered fused silica (20-μm inside diameter) was used as an emitter.
Mass analyses were performed with a hybrid linear ion trap FT-ICR mass spectrometer equipped with a 7-T ICR magnet (LTQ-FT; Thermo Electron, Bremen, Germany). The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey MS spectra (from m/z 400 to 1,600) were acquired in the FT-ICR, and the three most intense doubly or triply charged ions in each FT scan were fragmented and analyzed in the linear ion trap. Proteins were identified by automated database searching (Matrix Science, London, United Kingdom) of all tandem mass spectra using the NCBI database. The search parameters were set as follows: MS accuracy, 15 ppm; MS/MS accuracy, 0.5 Da; one missed cleavage allowed; fixed propionamide modification of cysteine; and variable modification of oxidized methionine.
Infection of mice with H. pylori. (i) Animals.
Six- to eight-week-old C57BL/6 female mice were purchased from B&K Universal (Sollentuna, Stockholm, Sweden) and housed in microisolators at the Laboratory for Experimental Biomedicine, Göteborg University, Göteborg, Sweden, during the study. All experiments were approved by the Ethical Committee for Laboratory Animals in Göteborg (92/03).
(ii) Bacteria and culture conditions for infection.
H. pylori strain SS1 (19) was prepared for infection of mice as previously described (28).
(iii) Infection.
Sixteen female C57BL/6 mice were orally infected with approximately 109 CFU of H. pylori SS1 or SS1(ΔhpaA) in Brucella broth under anesthesia (Isoflurane; Abbott Scandinavia Ab, Solna, Sweden) as previously described (27).
Detection of H. pylori SS1 (wild type) and SS1(ΔhpaA) in infected mice. (i) Quantitative culture.
The kinetics of SS1 in the colonization of mice have been well characterized, showing stable colonization between 2 and 8 weeks of infection (27). To determine the kinetics of colonization by SS1(ΔhpaA) in mice, animals were killed at various time points after infection (3 days, 3 weeks, and 8 weeks). The stomachs were removed and washed with phosphate-buffered saline to remove food residues. One half of the stomach was used for quantitative culture as previously described (27), and the other half was used for detection of H. pylori-specific genes by PCR. The stomach homogenates from the SS1(ΔhpaA)-infected mice were cultured on blood Skirrow plates both with and without kanamycin to examine if they had lost their antibiotic resistance during the gastric infection.
(ii) H. pylori-specific PCR.
For detection of H. pylori-specific DNA from the infected mice, the mucus layer was removed from the other half of the stomach by using a glass slide and put into an extraction buffer (20 mM Tris, pH 8.0, 0.5% Tween 20, 0.5 mg/ml proteinase K). The tubes were incubated for 1 h at 55°C and thereafter for 10 min at 98°C before they were centrifuged for 5 min at maximum speed. The supernatants were then transferred to a new tube. The sample was stored at −20°C before analysis by an H. pylori ureA-specific PCR performed as previously described (11).
Detection of Omp18 (HP0796) mRNA by RT-PCR.
RNA was isolated from SS1 and SS1(ΔhpaA) isolates cultured in Brucella broth for 24 h by using an RNeasy kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. Reverse transcription (RT) was performed on DNase-treated total RNA using a Sensiscript RT kit (QIAGEN) with a reverse primer specific for the omp18 (HP0796) gene (5′-ACC CAA TTC TAT CGC CCA ATT C-3′). The cDNA was thereafter amplified with the primer used in the RT step and a forward primer (5′-ACC AGC GGT TTT TAG CCA AGA-3′). An RT reaction without RNA and the same sample not subjected to RT were used as negative controls in PCR. PCR was done with an annealing temperature of 55°C for 2 min and a 2-min elongation step for 30 cycles. Samples were analyzed on a 2% agarose gel.
RESULTS
Comparison of the major proteome components in H. pylori strains SS1 and SS1(ΔhpaA).
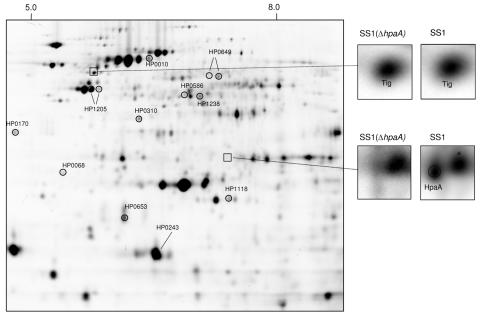
To identify that no specific protein expression change had followed the construction of the HpaA mutant, we analyzed the proteome of H. pylori strain SS1 and its isogenic mutant by the 2-DE-based DIGE system. By use of cell lysis buffer compatible with the DIGE technology and isoelectric focusing at a pH interval of 3 to 10, over 800 distinct protein spots from each sample in the four replicates were detected by the DeCyder software and subsequent manual correction. The analysis of the expression profiles in strain SS1 and the SS1(ΔhpaA) mutant resulted in the identification of a minor number of spots (13) with a significantly changed level (P < 0.05). Of these spots, eight were found to be down-regulated and five spots were found to be upregulated in the SS1(ΔhpaA) mutant (Fig. 1). For identification of proteins, one preparative gel was stained with Sypro ruby, and spots were digested in gel and analyzed by nano-LC FT-ICR MS and MS/MS. We successfully identified the proteins shown in Table 1. Notably, the trigger factor encoded by the tig gene located downstream of hpaA showed similar levels of expression in both strains (Fig. 1 and 2). However, Omp18 (HP0796) was detected in neither the wild-type strain nor the mutant. Thus, to ascertain that the disruption of the hpaA gene had not affected the transcription of its downstream gene, omp18, an omp18-specific RT-PCR was performed on SS1 and the SS1(ΔhpaA) mutant strain, which showed that Omp18 was transcribed in both strains (data not shown).
FIG. 1.
DIGE analysis. Protein spots with changes in expression levels between wild-type and mutant strains are marked and identified by mass spectrometry (Table 1). The upper inserts show the presence of the trigger factor (tig) in both the wild-type and mutant strains. The insert below illustrates the presence of the HpaA protein in the gel from the wild type and the lack of HpaA in the corresponding area in the gel from the mutant.
TABLE 1.
Protein spots showing altered expression levels by use of DIGE technology
| Gene locus | Protein name | Function | Molecular mass (kDa) | pI | Avg ratioa |
|---|---|---|---|---|---|
| HP1118 | Gamma-glutamyltransferase | Transpeptidation reaction | 61.1 | 9.27 | ↓ 0.65 |
| HP0649 | Aspartate ammonia lyase | Amino acid biosynthesis | 52.5 | 6.37 | ↓ 0.68 |
| HP0649 | Aspartate ammonia lyase | Amino acid biosynthesis | 52.5 | 6.37 | ↓ 0.60 |
| HP0586 | 2-Oxoglutarate oxidoreductase | Oxidoreductase activity | 41.7 | 5.98 | ↓ 0.32 |
| HP1238 | Aliphatic amidase | Hydrolase activity | 38.5 | 6.20 | ↓ 0.36 |
| HP0310 | Hypothetical protein | Hydrolase activity | 33.6 | 5.42 | ↓ 0.42 |
| HP0653 | Nonheme ferritin protein | Iron storage protein | 19.3 | 5.40 | ↓ 0.42 |
| HP0243 | Neutrophil-activating protein | Neutrophil activation | 16.8 | 5.69 | ↓ 0.52 |
| HP0010 | Chaperone and heat shock protein | Chaperone and heat shock protein | 58.3 | 5.55 | ↑ 1.43 |
| HP1205 | Elongation factor EF-TU | Elongation factor | 43.7 | 5.17 | ↑ 2.09 |
| HP1205 | Elongation factor EF-TU | Elongation factor | 43.7 | 5.17 | ↑ 2.56 |
| HP0170 | Hypothetical protein | Unknown | 28.7 | 4.63 | ↑ 1.47 |
| HP0068 | UreG | Incorporating Ni to Urease | 22.1 | 5.03 | ↑ 2.27 |
Protein expression level in the SS1(ΔhpaA) mutant compared to that of the wild-type strain. Unchanged protein expression is set to 1.
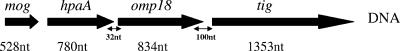
FIG. 2.
Gene location of hpaA (HP0797). The gene order and sizes of genes have been obtained from the H. pylori TIGR genome database (http://www.tigr.org). nt, nucleotide.
Analysis of surface-exposed proteins.
It is well known that 2-DE gel analysis is not the method of choice for studies of hydrophobic and low-abundance proteins (i.e., OMPs). Therefore, we combined subcellular fractionation of the sarcosine-insoluble outer membrane and one-dimensional SDS-PAGE analysis with nano-LC FT-ICR MS and MS/MS analyses in order to compare the expression of outer membrane proteins in H. pylori strain SS1 and its isogenic mutant.
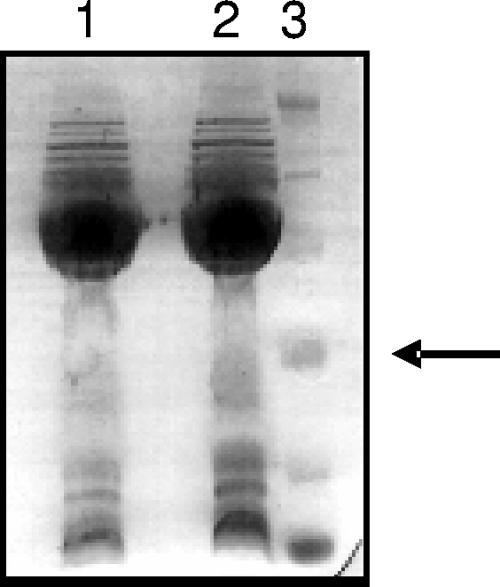
The separation of the OMP fraction by SDS-PAGE revealed an almost-identical protein pattern between the two strains (Fig. 3). We identified over 50 proteins with considerable overlap from each of the strains. Of these proteins, 25 are known to be outer membrane proteins or putative outer membrane proteins, 15 are hypothetical proteins with unknown function and localization, 7 are flagellar proteins, and 1 additional protein is involved in flagellar motility (Table 2). In addition, several cytoplasmic proteins known to be localized at the bacterial surface, including urease, catalase, Hsp60, Nap, and VacA, were identified in both strains. The HpaA protein was identified in the OMP fraction from the SS1 strain.
FIG. 3.
SDS-PAGE analysis of the outer membrane fractions. Lane 1, SS1; lane 2, SS1(ΔhpaA); lane 3, molecular mass marker. The arrow indicates 31 kDa.
TABLE 2.
Selected subset of identified proteins from the enriched outer membrane fractions of H. pylori strain SS1 and its isogenic mutant, SS1(ΔhpaA)
| Protein name | Gene locus | Protein identitya
|
|
|---|---|---|---|
| SS1 | SS1(ΔhpaA) | ||
| Adhesin-thiol peroxidase (TagD) | HP0390 | • | • |
| Catalase | HP0875 | • | • |
| Flagellar basal-body rod protein (FlgG) | HP0547 | • | • |
| Flagellar hook FlgE | HP0870 | • | • |
| Flagellar hook-associated protein 1 (HAP1) | HP1119 | • | • |
| Flagellar hook-associated protein 2 (HAP2) | HP0752 | • | • |
| Flagellar hook-associated protein 3 (HAP3) | HP0295 | • | • |
| Flagellin A | HP0601 | • | • |
| Flagellin B | HP0115 | • | • |
| H. pylori adhesin A (HpaA) | HP0797 | • | |
| HofC | HP0486 | • | • |
| Iron-regulated outer membrane protein | HP1512 | • | • |
| Membrane-associated lipoprotein lpp20 | HP1456 | • | • |
| Neutrophil-activating protein (Nap) | HP0243 | • | • |
| Omp1 (HopZ) | HP0009 | • | • |
| Omp2 | HP0025 | • | • |
| Omp5/Omp29 | HP0227/1342 | • | • |
| Omp6 | HP0229 | • | • |
| Omp10 | HP0324 | • | • |
| Omp11 | HP0472 | • | • |
| Omp12/Omp22 | HP0477/0923 | • | • |
| Omp14 | HP0671 | • | • |
| Omp15 (HopE) | HP0706 | • | • |
| Omp16 | HP0722 | • | • |
| Omp21 (AlpB) | HP0913 | • | • |
| Omp23 | HP1107 | • | • |
| Omp25 | HP1156 | • | • |
| Omp27 (HopQ) | HP1177 | • | • |
| Omp28 (BabA) | HP1243 | • | • |
| Omp30 | HP1395 | • | • |
| Omp31 | HP1469 | • | • |
| Secreted protein involved in flagellar motility | HP1462 | • | • |
| Urease α | HP0073 | • | • |
| Urease β | HP0072 | • | • |
| Urease accessory protein (UreG) | HP0068 | • | • |
A • indicates protein identity significant at the 99% level.
Furthermore, a Western blot performed on SS1 and SS1(ΔhpaA) OMP fractions using an HpaA monoclonal antibody (9) clearly confirmed the presence of HpaA in the preparation from SS1 but not in the preparation made from SS1(ΔhpaA) (data not shown).
Growth of bacteria in vitro.
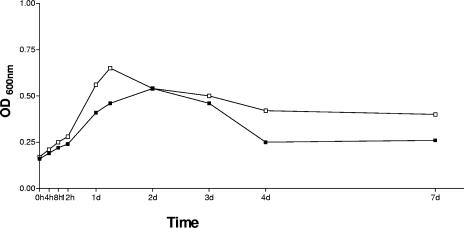
To ensure that the mutant strain had not significantly altered its growth properties compared to SS1, both strains were cultured in liquid shaking culture for a week. Samples were taken at various time points, and the OD and viability of the bacteria were measured. As shown in Fig. 4, no obvious differences in growth properties in vitro were seen when the mutant strain was compared with wild-type SS1. The viabilities of the two strains were also comparable (data not shown).
FIG. 4.
Growth curves of SS1 (▪) and SS1(ΔhpaA) (□). Bacteria were grown in liquid shaking culture, and the OD600 was measured at 0, 4, 8, and 12 h and at days (d) 1, 2, 3, 4, and 7. Data are shown as a representative experiment of two independent experiments.
Detection of bacteria in infected mice.
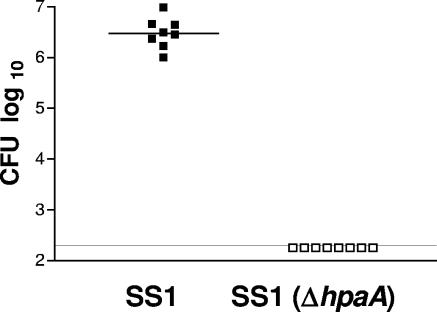
Colonization of H. pylori was detected both by quantitative culture and by H. pylori-specific PCR. To evaluate the colonization pattern for SS1(ΔhpaA), mice were infected with either SS1(ΔhpaA) or SS1 as a reference and then killed at various time points ranging from 3 days to 2 months. Mice infected with SS1 showed a massive colonization at all time points studied, but bacteria could not be detected in the stomachs of mice infected with SS1(ΔhpaA) either by culture (Fig. 5) or by H. pylori-specific PCR at any time point (data not shown). To ascertain that SS1(ΔhpaA) had not lost its kanamycin resistance during the colonization in the stomach, the bacteria were grown on plates with and without kanamycin. However, no bacteria could be detected after culture on plates without kanamycin either (data not shown).
FIG. 5.
Colonization of SS1 (▪) and SS1(ΔhpaA) (□) 3 weeks postinfection. The horizontal lines indicate the median for each group. No SS1(ΔhpaA) could be detected by quantitative culture. The line designates the detection limit of 200 bacteria/stomach. Data shown are pooled from two independent experiments.
DISCUSSION
Many colonization and virulence factors have been evaluated as protective antigens in immunization studies in animal models (17, 22). For a bacterial protein to be considered as a candidate vaccine antigen, it should preferably be conserved (i.e., present in all strains), secreted or surface localized, and immunogenic (i.e., capable of stimulating the immune system). HpaA fulfills all these criteria; the gene encoding HpaA is present in and expressed by all H. pylori isolates (9, 39), indicating that it is valuable for the bacterium. Furthermore, H. pylori-infected subjects mount serum antibody responses against HpaA, which decline after eradication of the bacterium (23, 37), and HpaA induces maturation and antigen presentation of dendritic cells, showing its immunogenicity (36). In addition, it has been shown that HpaA is expressed both intracellularly and on the bacterial surface (20, 25).
To investigate the importance of HpaA in H. pylori infection, a previously described mutation of HpaA (25) was introduced into the mouse-adapted strain SS1, and the mutant strain was tested for its colonization ability and immunogenicity in a well-established animal model.
In order to verify that the mutation had not caused any damage on downstream genes or second-site mutations, we performed 2-D DIGE analysis to examine the overall protein expression pattern of H. pylori strain SS1. All the detected protein spots in the wild-type strain, with the exception of HpaA, were found in the mutant strain. However, 13 spots corresponding to 11 unique proteins showed small changes in expression levels in the mutant compared to the wild-type strain; of these, seven proteins were found to be down-regulated and four proteins were up-regulated. These identified proteins do not seem to be related on either the genetic or the functional level. In addition, it has been shown that minor changes in the protein expression level normally occur within a bacterial strain (35) (E. Carlsohn et al., unpublished data). The most important finding in the DIGE analysis of the wild type and its isogenic mutant was that the trigger factor encoded by the tig gene located downstream of hpaA showed similar levels of expression in both strains.
It is well known that OMPs tend to be discriminated in standard 2-DE displaying total cell extract. This is due both to poor solubility and low expression levels of the proteins of interest, and it is therefore important to design an appropriate isolation procedure for this protein species. We performed subcellular fractionation of OMPs in combination with one-dimensional PAGE analysis and nano-LC FT-ICR MS and MS/MS analyses of tryptic peptides. By use of this novel approach, we identified over 20 outer membrane proteins and 8 flagella-associated proteins in both investigated strains. All OMPs present in the wild-type strain, with the exception of HpaA, were also expressed in the mutant strain. The cotranscription of hpaA and the downstream gene omp18 has previously been described (20). It was therefore of interest to study the expression of the omp18 gene product in the constructed HpaA mutant to investigate possible polar effects on surrounding genes in the mutant. Unfortunately, the Omp18 protein was not detected in any of the strains. However, RT-PCR analysis of omp18 mRNA from the wild-type and mutant strains clearly showed that omp18 was transcribed in both strains, indicating that disruption of hpaA did not have any polar effects on its downstream genes (data not shown). In addition, to the best of our knowledge, the Omp18 protein has never been detected, suggesting that it might not be translated but that it might only be present on the mRNA level. Because no major differences between the two strains could be detected, we proceeded to an animal model for evaluation of the physiological importance of HpaA.
In vivo studies showed that while mice infected with the wild-type SS1 strain were heavily colonized, its isogenic mutant failed to colonize the mice at all time points examined. Thus, the fact that the mutant did not show significant differences in growth under laboratory conditions suggests that the observed phenotype is strictly in vivo dependent.
HpaA was originally pointed out as a putative N-acetylneuraminyllactose-binding hemagglutinin, and several studies have tried to elucidate the function of HpaA in in vitro adhesion studies, but the results are not conclusive. For example, bacterial binding to gastric cell lines in vitro was not affected by an inactivated hpaA gene (25). However, epithelial cell lines have been demonstrated to respond quite differently to bacterial stimulations compared to freshly isolated epithelial cells (4). Furthermore, deletion of the hpaA gene did not influence the glycosphingolipid recognition pattern of the bacteria, as evaluated by binding of the bacteria to previously identified H. pylori-binding glycosphingolipids on thin-layer chromatograms (1). Thus, both the parent SS1 strain and the HpaA knockout mutant bound to lactosylceramide, gangliotetraosylceramide, lactotetraosylceramide, and Leb-terminated glycosphingolipids (S. Teneberg et al., unpublished data). One may therefore speculate whether HpaA itself directly mediates receptor binding or whether it is involved in facilitating the adhesin transport and folding, or if it exerts regulatory functions. The role of HpaA needs to be elucidated in further investigations.
In conclusion, we have shown that the disruption of the HpaA-encoding gene did not induce any major differences in the protein expression pattern in the mutant compared with the wild-type strain. We have also demonstrated that HpaA is essential for bacterial colonization in the gastric mucosa of mice, establishing for the first time a physiological role of HpaA in vivo.
Acknowledgments
We thank Hasse Karlsson for the nano-LC MS setup and preparations of the nano columns and Thomas Larsson for help with data evaluation. Susann Teneberg is gratefully acknowledged for the adhesion analysis.
Financial support was provided by the Swedish Research Council (in medicine; projects 06X-09084, 03X-14183, and 03BI-14930), Swedish Society for Medical Research. The purchase of the FT-ICR mass spectrometer was made possible through a grant from the Knut and Alice Wallenberg Foundation.
Editor: J. B. Bliska
REFERENCES
- 1.Achtman, M., and S. Suerbaum. 2001. Helicobacter pylori: molecular and cellular biology, p. 185-206. Horizon Scientific Press, Norfolk, England.
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Backert, S., T. Kwok, M. Schmid, M. Selbach, S. Moese, R. M. Peek, Jr., W. Konig, T. F. Meyer, and P. R. Jungblut. 2005. Subproteomes of soluble and structure-bound Helicobacter pylori proteins analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proteomics 5:1331-1345. [DOI] [PubMed] [Google Scholar]
- 4.Backhed, F., B. Rokbi, E. Torstensson, Y. Zhao, C. Nilsson, D. Seguin, S. Normark, A. M. Buchan, and A. Richter-Dahlfors. 2003. Gastric mucosal recognition of Helicobacter pylori is independent of Toll-like receptor 4. J. Infect. Dis. 187:829-836. [DOI] [PubMed] [Google Scholar]
- 5.Baik, S.-C., K.-M. Kim, S.-M. Song, D.-S. Kim, J.-S. Jun, S.-G. Lee, J.-Y. Song, J.-U. Park, H.-L. Kang, W.-K. Lee, M.-J. Cho, H.-S. Youn, G.-H. Ko, and K.-H. Rhee. 2004. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J. Bacteriol. 186:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161:626-633. [DOI] [PubMed] [Google Scholar]
- 7.Blom, K., B. S. Lundin, I. Bolin, and A. Svennerholm. 2001. Flow cytometric analysis of the localization of Helicobacter pylori antigens during different growth phases. FEMS Immunol. Med. Microbiol. 30:173-179. [DOI] [PubMed] [Google Scholar]
- 8.Blonder, J., M. B. Goshe, W. Xiao, D. G. Camp II, M. Wingerd, R. W. Davis, and R. D. Smith. 2004. Global analysis of the membrane subproteome of Pseudomonas aeruginosa using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 3:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Bolin, I., H. Lonroth, and A. M. Svennerholm. 1995. Identification of Helicobacter pylori by immunological dot blot method based on reaction of a species-specific monoclonal antibody with a surface-exposed protein. J. Clin. Microbiol. 33:381-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 11.Clayton, C. L., H. Kleanthous, P. J. Coates, D. D. Morgan, and S. Tabaqchali. 1992. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J. Clin. Microbiol. 30:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doig, P., and T. J. Trust. 1994. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect. Immun. 62:4526-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, D. G., D. J. Evans, Jr., J. J. Moulds, and D. Y. Graham. 1988. N-Acetylneuraminyllactose-binding fibrillar hemagglutinin of Campylobacter pylori: a putative colonization factor antigen. Infect. Immun. 56:2896-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exner, M. M., P. Doig, T. J. Trust, and R. E. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falk, P., K. A. Roth, T. Boren, T. U. Westblom, J. I. Gordon, and S. Normark. 1993. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proc. Natl. Acad. Sci. USA 90:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy, B., C. Hessler, S. Fourage, J. Haensler, E. Vialon-Lafay, B. Rokbi, and M. J. Millet. 1998. Systemic immunization with urease protects mice against Helicobacter pylori infection. Vaccine 16:850-856. [DOI] [PubMed] [Google Scholar]
- 18.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 19.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 20.Lundstrom, A. M., K. Blom, V. Sundaeus, and I. Bolin. 2001. HpaA shows variable surface localization but the gene expression is similar in different Helicobacter pylori strains. Microb. Pathog. 31:243-253. [DOI] [PubMed] [Google Scholar]
- 21.Lundstrom, A. M., I. Bolin, M. Bystrom, and S. Nystrom. 2003. Recombinant HpaA purified from Escherichia coli has biological properties similar to those of native Helicobacter pylori HpaA. APMIS 111:389-397. [DOI] [PubMed] [Google Scholar]
- 22.Marchetti, M., M. Rossi, V. Giannelli, M. M. Giuliani, M. Pizza, S. Censini, A. Covacci, P. Massari, C. Pagliaccia, R. Manetti, J. L. Telford, G. Douce, G. Dougan, R. Rappuoli, and P. Ghiara. 1998. Protection against Helicobacter pylori infection in mice by intragastric vaccination with H. pylori antigens is achieved using a non-toxic mutant of E. coli heat-labile enterotoxin (LT) as adjuvant. Vaccine 16:33-37. [DOI] [PubMed] [Google Scholar]
- 23.Mattsson, A., A. Tinnert, A. Hamlet, H. Lonroth, I. Bolin, and A. M. Svennerholm. 1998. Specific antibodies in sera and gastric aspirates of symptomatic and asymptomatic Helicobacter pylori-infected subjects. Clin. Diagn. Lab. Immunol. 5:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson, C. L., T. Larsson, E. Gustafsson, K. A. Karlsson, and P. Davidsson. 2000. Identification of protein vaccine candidates from Helicobacter pylori using a preparative two-dimensional electrophoretic procedure and mass spectrometry. Anal. Chem. 72:2148-2153. [DOI] [PubMed] [Google Scholar]
- 25.O'Toole, P. W., L. Janzon, P. Doig, J. Huang, M. Kostrzynska, and T. J. Trust. 1995. The putative neuraminyllactose-binding hemagglutinin HpaA of Helicobacter pylori CCUG 17874 is a lipoprotein. J. Bacteriol. 177:6049-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pounder, R. E., and D. Ng. 1995. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol. Ther. 9(Suppl. 2):33-39. [PubMed] [Google Scholar]
- 27.Raghavan, S., M. Hjulstrom, J. Holmgren, and A. M. Svennerholm. 2002. Protection against experimental Helicobacter pylori infection after immunization with inactivated H. pylori whole-cell vaccines. Infect. Immun. 70:6383-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghavan, S., A. M. Svennerholm, and J. Holmgren. 2002. Effects of oral vaccination and immunomodulation by cholera toxin on experimental Helicobacter pylori infection, reinfection, and gastritis. Infect. Immun. 70:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 30.Somigliana, E., P. Vigano, P. Filardo, M. Candiani, M. Vignali, and P. Panina-Bordignon. 2001. Use of knockout transgenic mice in the study of endometriosis: insights from mice lacking beta(2)-microglobulin and interleukin-12p40. Fertil. Steril. 75:203-206. [DOI] [PubMed] [Google Scholar]
- 31.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.Tsuda, M., M. Karita, T. Mizote, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. Essential role of Helicobacter pylori urease in gastric colonization: definite proof using a urease-negative mutant constructed by gene replacement. Eur. J. Gastroenterol. Hepatol. 6(Suppl. 1):S49-S52. [PubMed] [Google Scholar]
- 34.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071-2077. [DOI] [PubMed] [Google Scholar]
- 35.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voland, P., N. Hafsi, M. Zeitner, S. Laforsch, H. Wagner, and C. Prinz. 2003. Antigenic properties of HpaA and Omp18, two outer membrane proteins of Helicobacter pylori. Infect. Immun. 71:3837-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voland, P., D. L. Weeks, D. Vaira, C. Prinz, and G. Sachs. 2002. Specific identification of three low molecular weight membrane-associated antigens of Helicobacter pylori. Aliment Pharmacol. Ther. 16:533-544. [DOI] [PubMed] [Google Scholar]
- 38.Wirth, H.-P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, J., Y. F. Mao, and Z. X. Shao. 2005. Frequencies of the expression of main protein antigens from Helicobacter pylori isolates and production of specific serum antibodies in infected patients. World J. Gastroenterol. 43:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, N., N. Li, and L. Li. 2004. Liquid chromatography MALDI MS/MS for membrane proteome analysis. J. Proteome Res. 3:719-727. [DOI] [PubMed] [Google Scholar]