Abstract
Prolonged expression of a ras oncogene in primary cells accelerates the natural process of senescence. This ras-induced permanent growth arrest is bypassed in cells expressing the simian virus 40 large T antigen. Previously we showed that two regions of T antigen, a region consisting of the N-terminal 147 amino acids and a region consisting of amino acids 251 to 708 (T251-708), independently overcome ras-induced senescence. Coexpression of either T-antigen fragment and Ras results in the appearance of dense foci of transformed cells. Using a series of mutants that produce shorter T-antigen fragments, we show that the C-terminal limit of the N-terminal T-antigen fragment that cooperates with Ras lies between amino acids 83 and 121. The N-terminal limit of the C-terminal T-antigen fragment lies between amino acids 252 and 271. In addition, we present evidence that cooperation between the N-terminal fragment and Ras depends upon an intact T-antigen J domain and the ability of the T antigen to bind and inactivate the growth-suppressive effect of the tumor suppressor Rb. Introduction of specific amino acid substitutions surrounding residue 400 into T251-708 prevented the fragment from cooperating with Ras. T251-708 proteins with these same substitutions inhibited the transcriptional transactivating potential of p53 as effectively as did the wild-type protein. Thus, at least one activity contained within T251-708, other than inactivating p53 as a transcriptional transactivator, is likely to be required to bypass Ras-induced senescence.
Primary cells in cultures naturally lose replicative potential with successive subculturing until the end point of division, senescence, is reached (reviewed in references 5 and 68). The expression of simian virus 40 (SV40) T antigen allows cells to bypass natural senescence and thereby become capable of an indefinite number of cell divisions (immortalized). The expression of a ras oncogene alters the process of senescence. Transient expression of ras is mitogenic. ras expression increases the level of cyclin D in cells (13, 41). Cyclin D in concert with cyclin E and their associated kinases is responsible for phosphorylating the tumor suppressor protein Rb (reviewed in reference 36). In its hypophosphorylated form, Rb binds to E2F transcription factors and represses the E2F-dependent transcription of S-phase genes (reviewed in reference 22). Rb phosphorylation releases E2F from complexes with Rb, promoting entry into S phase. However, prolonged expression of ras results in transactivation of the E2F-responsive p16 and p19ARF genes (48, 50) and acceleration of the senescence process (29, 50). Both p16 and p19ARF proteins contribute to senescence. p16 is one of a class of cyclin-dependent kinase (CDK) inhibitors which act specifically on cyclin D-dependent kinases (reviewed in reference 49). High levels of p16 prevent Rb phosphorylation and the consequent release of E2F. The p19ARF protein is an inhibitor of the oncoprotein MDM2. MDM2 forms complexes with p53 and targets p53 for degradation (18, 27). The formation of complexes between p19ARF and MDM2, however, promotes the rapid degradation of MDM2 instead (73). As a result, p53 accumulates and is available to transactivate the p21WAF1,CIP1 gene. p21 is one of a class of CDK inhibitors with broader activity and inhibits several CDKs, including cyclin D- and E-dependent kinases (69). This protein further limits Rb phosphorylation and promotes the accumulation of the hypophosphorylated, growth-suppressing form of the protein. While both p53 and p16 are essential for ras-induced senescence, genetic interruption of either pathway is sufficient to prevent the permanent G1 arrest characteristic of senescence (29, 50) in rat cells. The requirement for p53 and p16 in ras-induced senescence is consistent with the demonstrated involvement of p53-inducible p21 and p16 in the early and late stages of natural senescence, respectively (59).
The expression of the SV40 large T antigen allows primary rodent cells to overcome ras-induced senescence (reviewed in references 3, 33, and 53). Two regions of the large T antigen, amino acids 1 to 147 (T1-147) and amino acids 251 to 708 (T251-708), were previously shown to independently cooperate with a ras oncogene to produce dense foci on monolayers of rat embryo fibroblasts (REF) (6). Each of these regions contains activities that could be involved in bypassing ras-induced senescence. N-terminal segments of T antigen that retain the capacity to bind and inactivate hypophosphorylated Rb (21, 29, 30) would be expected to abrogate ras-induced senescence. Similarly, segments of T antigen that can bind p53 should diminish the ability of the tumor suppressor to transactivate p53-responsive genes such as p21.
Hypophosphorylated (active) Rb (31, 32) and related family members p107 and p130 (8) (hereafter, the designation Rb refers to the three family members, unless stated otherwise) bind to a region located between amino acids 106 and 114 of T antigen (reviewed in reference 22). Disruption of the LXCXE motif between amino acids 103 and 107 prevents binding. As a result of binding to T antigen, the active forms of the negative growth regulators are prevented from binding members of the E2F family of transcription factors and repressing genes required for maintenance of the S phase of the cell cycle (reviewed in reference 38).
Certain T-antigen functions depend on Rb binding. These include the ability to override p53-mediated growth arrest (16, 46) and the ability to bind the glial cell transcription factor Tst-1/Oct6/SCIP (55). Other T-antigen activities, such as alteration of the phosphorylation state of p107 and p130 (61), degradation of p130 (60), the ability to override Rb, p107, and p130 growth arrest (70), disruption of p130-E2F complexes and consequent relief of Rb- and p130-dependent E2F repression (70), enhancement of Ts-1/Oct6/SCIP (55), and growth in low-serum medium (61, 65), depend upon not only the Rb-binding motif but also one or more functions within amino acids 1 to 82 of T antigen. The first 82 amino acids of T antigen are similar to the J domains of the DnaJ family of molecular chaperones (4, 43). Within this region, the hexapeptide motif (HPDKGG) is highly conserved among papovaviruses. The J domain of T antigen interacts with the ATPase domain of DnaK protein Hsc70 through the DNA J-HDP loop and stimulates Hsc70 ATPase activity (4, 43). Hydrolysis of ATP can alter the conformation of bound substrates so as to change the folding status of the substrate or disrupt multiprotein complexes.
It is not known whether the ability of an N-terminal T-antigen segment to cooperate with Ras in transforming primary rodent cells depends on Rb binding, a J-domain activity, or both. Similarly, it is not known whether inhibiting p53 as a transcriptional transactivator is sufficient for a C-terminal T-antigen segment to cooperate with Ras.
In the investigation reported here, we further defined the limits of the T-antigen regions involved in Ras cooperation and examined the contributions of individual functions within each region to this transformation process. The results showed that Ras cooperation by the N-terminal fragment depended upon binding and functional inactivation of the growth-inhibiting properties of the tumor suppressor Rb in a J-domain-dependent fashion. Within the C-terminal T251-708 segment, specific amino acid substitutions surrounding residue 400 abrogated Ras cooperation. In spite of these mutations, the C-terminal fragments maintained the ability to reduce the transactivating potential of p53. It appears, therefore, that at least one activity contained within T251-708, other than inactivating the transcriptional transactivating potential of p53, is needed to bypass ras-induced senescence.
MATERIALS AND METHODS
Plasmids.
Plasmid pPVU0 (23) produces both full-length large T antigen (T1-708) and small t antigen. Plasmid dl536 (56) produces small t antigen only. Plasmid dl2005 (54) produces T1-708 but no detectable small t antigen. Plasmid dl1135 (44) encodes T antigen missing amino acids 17 to 27 (T1-708 dl17-27) and small t antigen with the same deletion. Plasmid dl2441 (74) produces T antigen missing amino acids 105 to 108 (T1-708 dl105-108) and small t antigen. Plasmid pSelectESV contains the SV40 enhancer-promoter and early-region sequences from the KpnI to BamHI sites cloned into the corresponding sites of the vector pSelect (Promega). Plasmid pSelectESV E107K was generated from pSelectESV by oligonucleotide-directed mutagenesis with an Altered Site System (Promega). Plasmid dl400 (25) produces T antigen in which amino acid 400 is replaced with three amino acids (Arg Ile Arg) encoded by an EcoRI linker (T1-708 dl400). pBluescript SK(+) plasmids encoding T antigens with the amino acid substitutions A382P, M388L, G390A, A392G, L394V, H395N, C396S, L397V, L398V, K400R, D402E, D402H, S403A, V404L, D407E, F408Y, K410R, and C411R were kindly provided by D. Simmons. The constructs encoding these T antigens contain the SV40 genome at the BamHI site of pBluescript II SK(+) and were described previously (30).
Plasmid pCAV251-708 (6) encodes a fusion protein that originates in β-galactosidase and contains T251-708. Each of the amino acid substitutions between residues 388 and 411, indicated above, was introduced into T251-708 in the following manner. Prior to cloning of the mutations into pCAV251-708, the orientation of the viral genomes in pBluescript II SK(+) were reversed by releasing the inserts and religating the DNA fragments to produce plasmids in which the SV40 early region was adjacent to the SalI site of the vector. From each of these constructs, a 1,320-bp fragment containing the mutation was released by digestion with SalI and NdeI, purified, and cloned into the corresponding sites of modified pBluescript SK(+)T251-708 (6), containing an MluI site at nucleotide 977, a BglII site at nucleotide 330, and the T251-708 sequence. Then, the fragment was released by MluI and BamHI inserted into plasmid pCAV251-708 as described previously (6).
Additional plasmids encoding T antigens with amino acid substitutions E107K, D402N, M401T, K400E, P399L, L397W, C3965 and L397W (C396S/L397W), and C396S were generated by oligonucleotide-directed mutagenesis. The presence of the expected nucleotide changes was confirmed by DNA sequence analysis. Then, the 1,287-bp segment released by digestion with NdeI and SalI was cloned into pCAV251-708.
Plasmid pT1-82 (7) produces a polypeptide containing only the first 82 amino acids of large T antigen and small t antigen. Plasmid pVBt2tk5 (47) was subjected to DNA sequence analysis and found to encode a large T antigen containing amino acids 1 to 110 (T1-110) and small t antigen. Plasmid pT147NS (57) encodes T1-147; the construct does not encode small t antigen. Plasmid dl2420 (67) encodes T antigen containing amino acids 1 to 138 (T1-138) and small t antigen. Plasmid dl1137 (44) encodes T antigen containing amino acids 1 to 121 (T1-121) and small t antigen. Plasmid dl1137 t− (42, 57) encodes T1-121 only. Plasmid pT1-127 (64) encodes T antigen containing amino acids 1 to 127 (T1-127) and small t antigen. Plasmid pCAV83-708 (7) encodes T antigen containing amino acids 83 to 708 (T83-708); small t antigen is not produced. Plasmid pCAV251-708 (6) encodes T251-708; small t antigen is not produced. A derivative of pT1-127 that cannot produce small t antigen was generated by fragment exchange between pSelect dl2005, which contains the early region of mutant dl2005 extending from the KpnI site (nt 294) to the BamHI site (nt 2533), and pT1-127. Specifically, the PflMI (nt 4558)-BamHI (nt 2533) segment in pSelect dl2005 was replaced with the corresponding region from pT1-127. The resulting plasmid, pT1-127 t−, encodes T1-127 only.
The E107K substitution was introduced into T1-121 and T1-127 following PCR amplification of nucleotides 4947 to 4476 from pSelectESV E107K in such a way as to generate an XhoI site between nucleotides 4488 and 4483 without altering the corresponding amino acid sequence. Then, the region between nucleotides 4496 and 3621 was PCR amplified from pT1-127 in such a way as to generate an XhoI site between nucleotides 4489 and 4482. The resulting fragments were cleaved with BstXI and XhoI or XhoI and HpaI to generate appropriate ends for insertion of the two fragments between the BstXI and HpaI sites of plasmid pPVU0. The same strategy was used to introduce the E107K mutation into plasmid dl1137.
Plasmids pwt2 P43L/K45N (45) and pwt2 D44E/G47R, which express small t antigens with the amino acid substitutions indicated, and the small-t-antigen cDNA expression plasmid wt2 were kindly provided by K. Rundell. These amino acid substitutions were introduced into T1-708, T1-127, and T1-121 by replacing the region between the BstXI and BamHI sites of the inserts in pwt2 P43L/K45N and pwt2 D44E/G47R with the corresponding region from pSelectESV (full-length T antigen expressed in the presence of small t antigen [T1-708,t]), plasmid dl1137, plasmid dl1137 t−, pT1-127, or pT1-127 t−.
Plasmid Sp72-ras (6) contains the oncogene H-ras (17) at the BamHI site of the vector Sp72. Plasmid pPUR, purchased from Clontech, contains the puromycin N-acetyltransferase gene and confers puromycin resistance to mammalian cells. Plasmid LTRp53cG-ala (11) contains a wild-type murine p53 cDNA and was provided by A. Levine. Plasmid pCMV-Rb, kindly provided by L.-S. Chang, contains the mouse Rb cDNA under the control of the cytomegalovirus immediate-early enhancer-promoter.
The reporter p50-2luc (71) contains the p53-responsive elements from the murine muscle-specific creatine kinase gene cloned 5′ of the luciferase gene and was kindly provided by G. Zambetti. The reporter pAluc expresses luciferase under the control of a 7-kb cyclin A promoter region (19).
Cells and cell lines.
Primary Fischer 344 REF were generated as described previously (6). SAOS-2 is a cell line devoid of functional p53 and Rb (9). All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 0.03% glutamine, 0.075% NaHCO3, 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, and 100 U of penicillin per ml (D10×1) unless stated otherwise.
Ras cooperation assay.
Primary REF at passage 1 were recovered from liquid nitrogen storage, seeded into a T75 flask containing 20 ml of D10×1, and incubated overnight at 37°C. On the following day, the medium was replaced with fresh D10×1, and incubation was continued. At 1 or 2 days later, the culture was passaged 1:2 and incubated overnight at 37°C. Then, for each DNA to be tested, each of two T75 flasks was seeded with 5 × 105 cells in 20 ml of D10×1, and the flasks were incubated overnight. Cells were transfected by using the calcium phosphate precipitation method with 1 μg of T-antigen expression plasmid, 1 μg of plasmid Sp72-ras, and 10 μg of salmon sperm DNA exactly as described previously (6). On the following day, the medium was removed from the transfected cultures and replaced with fresh D10×1. Subsequently, the medium was changed every 3 to 4 days. As needed, the amount of NaHCO3 in the medium was increased to control acidification of the medium. Within 2 weeks, dense colonies appeared in flasks that received plasmids expressing T1-708 and Ras. In experiments involving T251-708 and mutant derivatives of T251-708, foci appeared by 3 weeks. Colonies either were removed by using cloning pipettes and expanded into cell lines or were stained with crystal violet staining solution and counted.
Transactivation of a p53-responsive reporter.
Confluent cultures of SAOS-2 cells were passaged 1:2 and incubated overnight at 37°C. For each DNA combination tested, four 60-mm culture dishes were seeded with 3 × 105 cells in 5 ml of D10×1, and the cultures were incubated overnight. Each culture was transfected with 5 μg of salmon sperm DNA, 2.5 μg of Bcl2 expression plasmid, 1 μg of wild-type mouse p53 expression plasmid, 2.5 μg of the p53-responsive reporter p50-2luc, and various amounts of T-antigen expression plasmids by the calcium phosphate method. Specifically, 0.5 ml of calcium phosphate DNA precipitate was added directly to the culture medium, and the cultures were incubated at 37°C for 6 h. After the incubation period, the culture medium was removed. The cell monolayers were washed twice with DMEM supplemented with 0.03% glutamine, 0.075% NaHCO3, 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, and 100 U of penicillin per ml (DMEM0×1), and fresh D10×1 was added to the cultures. Incubation was continued for an additional 24 h, after which time the culture medium was replaced with fresh D10×1. Samples were harvested 48 h after transfection and assayed for luciferase activity by using a Promega luciferase assay system according to the manufacturer's instructions.
Transactivation of the cyclin A promoter.
TC7 cells were seeded in 60-mm dishes containing 5 ml of Dulbecco's medium with 10% fetal bovine serum, 0.15% NaHCO3, and 25 mm HEPES at a density of 6 × 105 cells per dish and incubated at 37°C overnight. The cells were transfected by using the DEAED-chloroquine method exactly as described previously (6), except that each dish received 2.5 μg of reporter (pAluc) and 5 μg of T-antigen expression plasmid or calf thymus DNA. Forty-eight hours later, the cultures were harvested and processed for the luciferase assay as indicated above.
Rb-induced growth inhibition assay.
The flat cell assay (20) was used to test the ability of T antigens to overcome the growth-inhibiting effects of overexpressed Rb. Briefly, for each DNA tested, three 35-mm culture dishes each were seeded with 3 × 105 SAOS-2 cells that had been passaged 1:2 on the previous day. After overnight incubation at 37°C, each dish was transfected by adding 0.3 ml of calcium phosphate DNA precipitate containing 0.75 μg of pCMV-Rb, 0.6 μg of pPUR, and 3.6 μg of T-antigen expression plasmid directly to the medium on the plate. After 6 h of incubation at 37°C, the medium containing the precipitate was removed, and the cell monolayers were washed twice with 5 ml of DMEM0×1. Then, 5 ml of D10×1 was added, and the cultures were incubated overnight at 37°C. The medium was replaced with fresh D10×1, and incubation was continued. Forty-eight hours later and daily thereafter, the medium was replaced with D10×1 containing 0.25 μg of puromycin per ml. Nine days after transfection, the plates were stained with crystal violet. For each dish, the number of large cells in a total of 1,000 cells was determined.
Immunoprecipitation and immunoblot analysis.
For detection of SV40 T antigens, 6 × 106 SAOS-2 cells were transfected with various amounts of T-antigen-encoding plasmids, 10 μg of salmon sperm DNA, 5 μg of Bcl2 expression plasmid, 5 μg of reporter p50-2luc, and 2 μg of the p53 expression plasmid LTRp53cG-ala according to the procedure indicated above for the transfection of SAOS-2 cells. Forty-eight hours after transfection, protein extracts were prepared as described previously (25). T antigens were immunoprecipitated from the extracts as previously described (18) by using monoclonal antibody PAb901, which recognizes an epitope in the C terminus of T antigen, or PAb416, which recognizes an epitope in the N terminus of T antigen adjacent to but not overlapping the J domain. The immunoprecipitated proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and probed for T antigens by using PAb901 or PAb416 as described previously (25).
RESULTS
Role of the J domain and an Rb-binding motif in Ras cooperation.
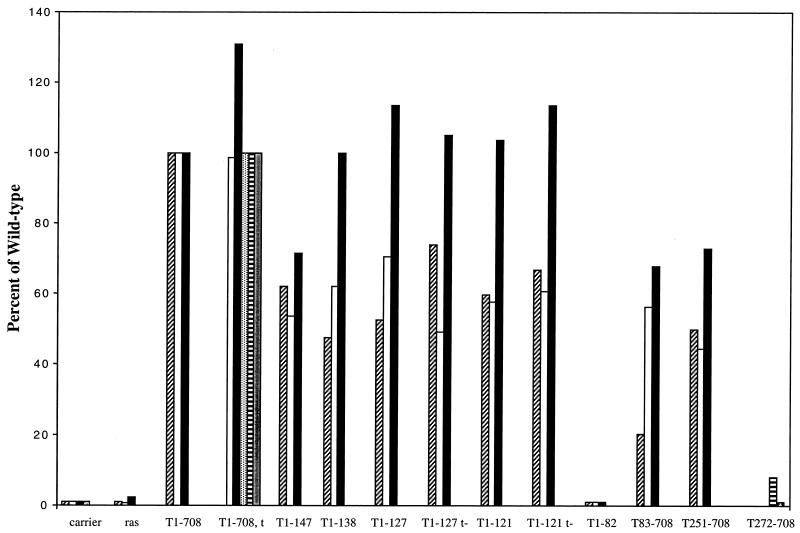
Initially, N-terminal fragments of T antigen were examined for the ability to cooperate with Ras. The results of multiple independent experiments are shown in Fig. 1. As shown previously, full-length T antigen (T1-708) cooperated efficiently with Ras. N-terminal fragments T1-147, T1-138, T1-127, and T1-121 also cooperated efficiently. Although the J-domain fragment T1-82 accumulated to levels equivalent to those of T1-708 (data not shown), it did not cooperate with Ras. These results suggested that a T-antigen segment containing the J domain (amino acids 1 to 82) and an Rb-binding motif (amino acids 106 to 114) was sufficient to cooperate with Ras. Constructs expressing full-length T antigen, T1-127, or T1-121 in the absence of small t antigen cooperated with Ras as efficiently as their counterparts expressing small t antigen.
FIG. 1.
Cooperation between T-antigen fragments and Ras. Each bar represents the average number of dense foci counted in duplicate flasks transfected with a ras oncogene expression plasmid and the DNA indicated. Different types of shading of the bars represent independent experiments. The results are expressed as a percentage of the number of foci produced by T1-708 and Ras. The average number of foci in flasks transfected with the T1-708 expression plasmid ranged from 81 to 142 in the individual experiments.
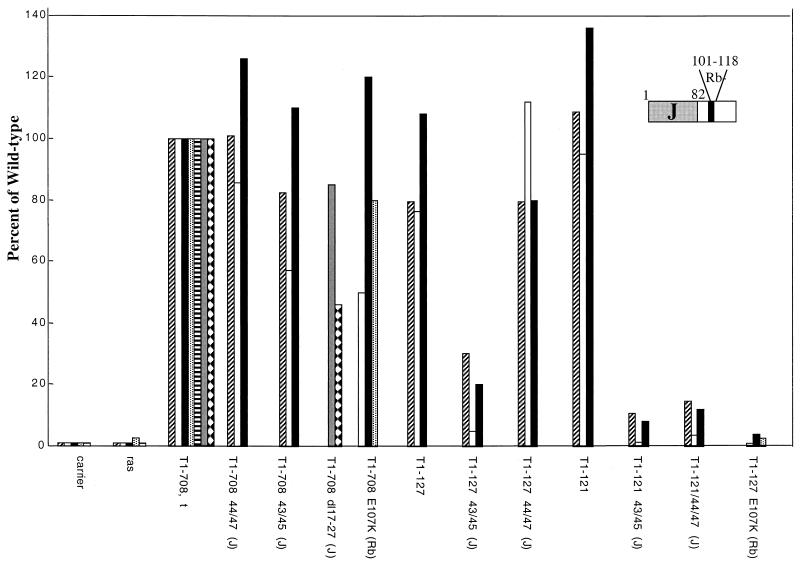
The roles of the J domain and the Rb-binding motif within the N-terminal segments then were examined. For this purpose, constructs that produce full-length T antigens with an amino acid substitution in the Rb-binding motif (E107K) or with double amino acid substitutions (D44E/G47R and P43L/K45N) or the deletion of amino acids 17 to 27 in the J domain were examined for the ability to cooperate with Ras. Figure 2 shows the results of these experiments. T antigens with mutations in the Rb-binding motif or in the J domain retained the ability to cooperate with Ras. This result was expected, as full-length T antigens contain the C-terminal Ras cooperation region.
FIG. 2.
Effect of J-domain and Rb-binding-region mutations on Ras cooperation. Each bar represents the average number of dense foci counted in duplicate flasks transfected with a ras oncogene expression plasmid and the DNA indicated. Different types of shading of the bars represent independent experiments. The results are expressed as a percentage of the number of foci produced by the T1-708,t expression vector and Ras. The average number of foci in flasks transfected with the T1-708,t expression vector ranged from 50 to 103 in the individual experiments.
Introduction of the E107K mutation into T1-127 (T1-127E107K) abrogated Ras cooperation, suggesting that cooperation between Ras and the N-terminal T-antigen segment was dependent on the integrity of the Rb-binding motif. Similarly, introduction of the P43L/K45N substitutions into either T1-127 or T1-121 abrogated the ability to cooperate with Ras in transforming REF. The effects of the D44E/G47R mutations in the two N-terminal T-antigen fragments differed, however. Whereas Ras cooperation was inhibited in the context of the T1-121 protein, cooperation was retained with the T1-127D44E/G47R protein. This result suggested two possibilities. First, Ras cooperation may not depend on a J-domain activity but instead may depend on a required activity contained within a neighboring region that is disturbed by the P43L/K45N mutations. Second, Ras cooperation may depend on a J-domain activity that is disrupted by the P43L/K45N mutations but is not disturbed by the D44E/G47R mutations unless they occur in the context of the shorter protein. To distinguish between these possibilities, it was necessary to determine whether the double amino acid substitution mutations affected J-domain activities similarly. Previously, Porras et al. (45) showed that these mutations in the context of small t antigen failed to promote anchorage-independent growth of human diploid fibroblasts, an activity that depends on an intact J domain.
The mutant T antigens were examined for the ability to transactivate the cyclin A promoter and to overcome the growth-suppressive effects of Rb overexpression. These activities were selected because they examine a requirement for cell cycle progression (cyclin A is expressed at the G1-S border and is required for maintenance of the S phase) and because they may be involved in overcoming natural or ras-induced senescence, respectively. ras-induced senescence depends, in part, on promoting the growth-inhibitory activities of Rb through the p16 pathway (29, 50)
Transactivation of the cyclin A promoter.
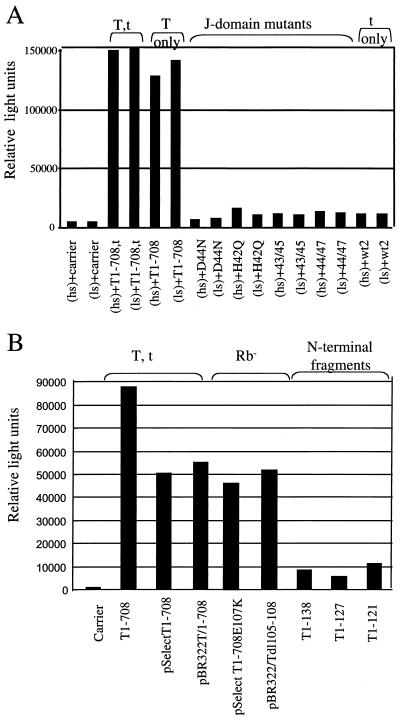
Both SV40 large T and small t antigens transactivate the cyclin A promoter (45). With small t antigen, transactivation is J-domain dependent (45). Similarly, polyomavirus large T antigen (PyT) transactivates this promoter. Sheng et al. (51) recently showed that PyT-mediated transactivation of the cyclin A promoter is only partially dependent on Rb-binding and J-domain functions in cells under normal growth conditions and is Rb-binding dependent and J-domain independent in serum-starved cells. Therefore, the abilities of the T-antigen J-domain mutants with the P43L/K45N, D44E/G47R, H42Q, and D44N mutations and the Rb-binding-defective T1-708E107K and T dl105-108 mutants to transactivate the cyclin A promoter were examined with cells in each condition. The results are shown in Fig. 3.
FIG. 3.
Transactivation of the cyclin A promoter is a J-domain-dependent, Rb-binding-independent T-antigen activity. TC7 cells were transfected with the reporter pAluc and the DNA indicated. (A) Transient transfection assays performed with medium containing high or low concentrations of serum (hs or ls, respectively). (B) Results obtained with hs. Since transactivation levels are influenced by the plasmid backbone of constructs expressing T antigen (7), the mutants in each experiment were examined in relation to wild-type T antigen expressed from the same plasmid backbone.
As expected (Fig. 3A), T1-708,t and T1-708 efficiently (31.9- and 27.3-fold, respectively) transactivated the cyclin A promoter. Small t antigen (wt2) transactivated it 2.5-fold. Transactivation was equivalent in growing and serum-starved cells. The J-domain mutants with the P43L/K45N, D44E/G47R, H42Q, and D44N mutations transactivated the cyclin A promoter between 1.4- and 3.5-fold. These results show that for large T antigen, as shown previously for small t antigen by using a small t antigen containing the P43L/K45N mutations (45), transactivation of the cyclin A promoter requires an intact J domain. The Rb-binding-defective T1-708E107K and T dl105-108 mutants (Fig. 3B), however, transactivated efficiently in these transient transfection assays. Thus, transactivation of the cyclin A promoter is J-domain dependent and Rb-binding independent.
The N-terminal T-antigen fragments T1-138, T1-127, and T1-121 (Fig. 3B) transactivated the cyclin A promoter between 5.6- and 11-fold. In contrast, in the same experiment, plasmids expressing full-length T antigen (pPVU0, pSelectESV, and pBR322/T1-708) transactivated between 45.6- and 86-fold. The N-terminal fragments are expressed at levels equal to or higher than that of T1-708 (data not shown). The inability of T1-127 and T1-121 to transactivate the cyclin A promoter is not simply due to the absence of a nuclear localization signal, as the nuclear localization signal is present in T1-138. All of the plasmids used to generate the data in Fig. 3B express small t antigen. Thus, the differences observed in transactivation reflect the contribution of large T antigen or fragments thereof to the process. These results indicated that one or more activities located between or dependent on amino acids 128 to 708 cooperate with the N terminus of T antigen in transactivating the cyclin A promoter.
Inhibition of Rb-mediated growth arrest or senescence.
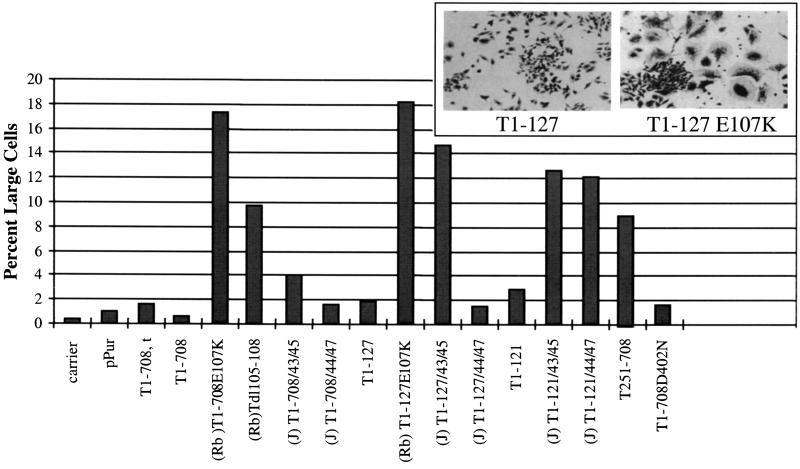
The T antigens were tested for the ability to inactivate the growth-inhibiting or senescence-inducing activity of Rb. Overexpression of Rb causes certain cells to arrest growth and adopt the morphological and biochemical properties of senescent cells. Specifically, the cells stop dividing, assume a large flat shape (63), and express the biochemical marker for senescence, the senescence-associated β-galactosidase (SAβgal) (20). Rb-deficient SAOS-2 cells were transfected with constructs expressing Rb and a puromycin resistance gene along with either the wild type or one of the mutant T-antigen constructs. After 9 days of puromycin selection, the cultures were fixed and stained with crystal violet, and the numbers of large flat cells were determined. Figure 4 displays the morphologies of cells expressing a T antigen (T1-127) that can bind Rb and of cells expressing T1-127E107K as well as quantitative results. Cells with a large flat shape were rare in cultures transfected with the puromycin resistance plasmid alone or in cells expressing Rb and T1-708 or Rb and T1-708,t. Similarly, T1-121 and T1-127 effectively prevented Rb-mediated growth arrest or senescence. As expected, T1-708E107K, T dl105-108, and T1-127E107K, each of which contains a mutation in the Rb-binding motif, could not prevent the appearance of flat cells. In the context of T1-708, the J-domain mutations had no effect on T-antigen-mediated inactivation or growth-suppressive properties of Rb.
FIG. 4.
Ability of T antigens to overcome Rb-induced growth arrest. In each of duplicate dishes, the number of large cells in a total of 1,000 cells was determined. The insert shows the morphology of cells transfected with Rb and T1-127 (left) and Rb and T1-127E107K (right) expression plasmids.
Introduction of the P43L/K45N mutations into T1-121 or T1-127 prevented the truncated protein from inhibiting Rb-mediated growth arrest or senescence. Introduction of the D44E/G47R mutations prevented T1-121 but not T1-127 from overcoming Rb-induced growth arrest or senescence. As expected, a C-terminal T-antigen fragment (T251-708) which does not contain either a J domain or an Rb-binding motif could not override growth arrest. A mutant with a defect in the p53-binding region (T1-708D402N) but intact J domain and Rb-binding site showed wild-type T-antigen activity in this assay. Examination of parallel cultures of transfected cells for the expression of SAβgal (data not shown) produced the same profile of results. The inability of the D44N and H42Q mutations to overcome Rb-mediated growth arrest was demonstrated previously (70). Thus, the same mutations that inactivate J-domain or Rb-binding-dependent functions involved in overriding Rb-mediated growth arrest abrogate Ras cooperation.
Role of p53 binding in Ras cooperation.
It was shown previously that the C-terminal T251-708 fragment cooperated with Ras whereas a fragment containing amino acids 300 to 708 did not (6). To further define the limits of the T-antigen region involved, a mutant T antigen containing amino acids 272 to 708 (T272-708) was examined for the ability to cooperate with Ras. The results are shown in Fig. 1. Although the level of accumulated T272-708 protein following transfection of plasmid pT272-708 was equivalent to that of full-length T antigen (data not shown), T272-708 did not cooperate with Ras. This result indicates that amino acids between residues 252 and 271 are essential either to preserve a function dependent on that region or to maintain a conformation-dependent function elsewhere within T251-708.
Previously, evidence was provided that the dl400 mutation, which encodes three nonauthentic T-antigen amino acids in place of amino acid 400, prevented Ras cooperation by T251-708 without limiting p53 binding (6). This result suggested two possibilities. The insertion of three amino acids could affect a T-antigen function that is either adjacent to or overlaps the p53-binding region or, alternatively, the binding of p53 may not be equivalent to inactivation of its functions. In order to probe more fully the impact of amino acid substitutions in the region that surrounds residue 400, we inserted a series of single amino acid substitution mutations between codons 382 and 411 into T251-708 and examined the ability of the resulting proteins to cooperate with Ras. The point mutations were characterized previously by Lin and Simmons (30) for their abilities to cause p53 binding and to immortalize human and rodent cells. The results are shown in Fig. 5. Nine of the mutant T251-708 proteins (carrying mutations C41lR, F408V, D402N, D402H, D402E, P399L, L397W, C396S/L397W, and A382P) displayed a substantially decreased ability to cooperate with Ras. Mutant proteins containing mutations K410R, D407L, S403A, M401T, K400R, K400E, L398V, L397V, L397M, C396S, H395N, L394V, G390A, and M388L cooperated with Ras at levels similar to those obtained with parental T251-708.
FIG. 5.
Effect of mutations in the p53-binding region on Ras cooperation. (Top) Positions of p53-binding regions and amino acid substitutions examined. For each mutant tested, the amino acid substitution and position within T antigen are indicated. (Bottom) Ability of a mutant to cooperate with Ras, indicated by a plus or a minus sign and quantitatively as a percentage of the wild-type value; values are expressed as the average of three or more experiments. The percentage of the wild-type value for the T251-708 segment (49%) is the average value obtained from six experiments. The average number of foci in flasks transfected with the T1-708 expression plasmid ranged from 80 to 168 in the individual experiments.
In order to exclude the possibility that these amino acid substitutions globally distorted T antigen, each substitution was examined in the context of T1-708 for Ras cooperation (data not shown). Retention of the ability to cooperate with Ras would indicate that the N-terminal Ras cooperation region was not compromised by the mutation in the C-terminal Ras cooperation region. Only one of the amino acid substitutions, P399L, abrogated Ras cooperation in the context of T1-708. With the remainder, N-terminal Ras cooperation activity remained intact. These results confirmed the requirement for amino acids surrounding residue 400 in Ras cooperation by T251-708.
Ability of mutant T251-708 proteins to inactivate p53.
The amino acid substitutions between amino acids 382 and 411 are located within one of two regions (amino acids 351 to 450 and amino acids 533 to 650) known to be involved in binding p53 (25). The ability of T251-708 proteins containing the substitutions to bind p53 was examined for two reasons. The first was to determine whether p53 binding correlated with Ras cooperation. The second was to provide an additional indication of whether gross structural alterations had occurred as a result of introducing the substitutions into the T-antigen fragment. Each T251-708 mutant was tested first in transient transfection assays. Accordingly, TC7 cells were cotransfected with a mouse p53 expression vector and a plasmid expressing one of the T251-708 mutants. The ability to bind p53 was monitored by coimmunoprecipitation and Western blot analyses with monoclonal antibody PAb421 or PAb901 for the immunoprecipitation of p53 or T antigen, respectively. Immunoblots were probed with a mixture of PAb421 and PAb901. Reduced levels of T-antigen proteins were observed in PAb421 immunoprecipitates from cells transfected with T251-708 proteins containing the amino acid substitutions D402N, D402E, P399L, and L397V (data not shown).
Previously, Lin and Simmons (30) showed that wild-type monkey p53 has a weaker association with T antigen than does wild-type mouse p53. p53 tetramers formed in transiently transfected TC7 cells might contain both monkey p53 and mouse p53. Such a combination might have adversely affected the binding or detection of p53-mutant T-antigen complexes. Therefore, Rat-2 cell lines expressing T251-708D402N, T251-708D402E, T251-708P399L, or T251-708L397V as well as cell lines expressing T251-708D402H, T251-708C411, T251-708 dl400, or T251-708P399L were generated. The results of coimmunoprecipitation assays showed that the mutant proteins bound rat p53 at levels similar to T251-708 when taking into consideration the relative amounts of T antigen in the samples (data not shown)
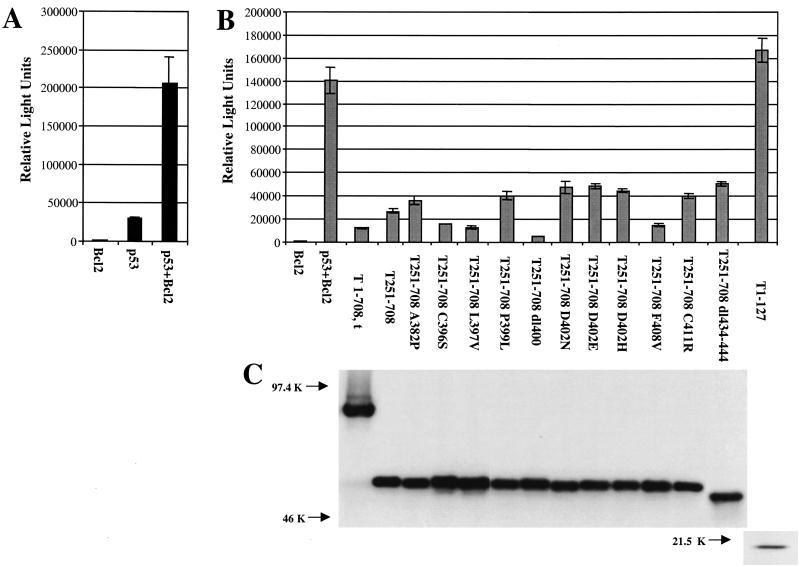
Next, mutant T251-708 proteins that failed to cooperate with Ras were examined for the ability to inactivate p53 as a transcriptional transactivator. Various mutant T antigens accumulate to different levels in transiently transfected cells. Therefore, prior to examination of the T antigens for the ability to inactivate p53, the level of protein produced by each construct relative to the amount of expression plasmid was determined by Western blot analysis of transfected cells. The amounts of mutant plasmids required to produce levels of protein equivalent to those produced in cells transfected with pPVU0 (T1-708,t) were cotransfected along with a wild-type p53 expression plasmid and a p53-responsive reporter (p50-2luc) into p53-null (SAOS-2) cells. The introduction of wild-type p53 into SAOS-2 cells results in apoptosis (2, 28). To avoid this potential complication, a plasmid that expresses the antiapoptosis protein Bcl2 was added to all transfection mixtures for testing T-antigen mutants. The results shown in Fig. 6A indicate that p53 transactivated the reporter, but Bcl2 did not. The presence of p53 and Bcl2, however, resulted in levels of luciferase substantially higher than those observed when p53 alone was transfected along with the reporter. This difference is presumed to result from the abrogation of apoptosis. Figure 6B shows that T1-708, T251-708, T251-708C396S, and T251-708L397V, all of which cooperated efficiently with Ras, prevented p53-mediated transactivation. The T251-708 proteins containing amino acid substitution A382P, P399L, D402N, D402E, D402H, F408V, or C411R or deletion dl400 or dl434-444 (6), which failed to cooperate with Ras, also retained the ability to inhibit p53-mediated transactivation. As expected, T1-127, which is missing the p53-binding region, did not prevent p53-mediated transactivation. Figure 6C shows that the T1-708 and T251-708 proteins were expressed at equivalent levels during the transient transfection assays. Thus, T251-708 proteins containing amino acid substitutions that prevent Ras cooperation retain the ability to inactivate p53 as a transcription factor in this assay. These results suggest that at least one activity in the C-terminal T-antigen segment other than the ability to inactivate this p53 activity is required for Ras cooperation.
FIG. 6.
T251-708 proteins with amino acid substitutions in the p53-binding region that do not cooperate with Ras retain the ability to inhibit p53 as a transcriptional transactivator. (A) Effect of adding Bcl2 to transient transfection-luciferase assays involving p53. Error bars indicate standard deviations. (B) Luciferase assays of cells transfected with the mutant indicated, Bcl2, wild-type p53, and the reporter construct. Narrow bars indicate standard deviations. (C) Immunoprecipitation and Western blot analysis of the T-antigen levels in parallel cultures of transfected cells. Equal amounts of protein were immunoprecipitated with PAb901, which recognizes an epitope in the C terminus of T antigen in all cases except T1-127. T1-127 protein was immunoprecipitated with PAb416, which recognizes an epitope in the N terminus of T antigen adjacent to but not including the region common to T antigen and t antigen; this antibody does not recognize small t antigen.
DISCUSSION
Expression of a ras oncogene in primary cells is initially mitogenic; however, prolonged expression induces premature senescence (29, 50). Cells expressing SV40 large T antigen and Ras bypass senescence and become transformed. It was shown previously (6) that either the N-terminal T-antigen fragment T1-147 or the C-terminal fragment T251-708 cooperated with Ras to bypass senescence and form dense foci on monolayers of REF. Large T antigen shares its first 131 amino acids with the 17,000-molecular-weight (17K) T antigen. The 17K T antigen contains three additional amino acids derived from an alternative reading frame. The presence of the 17K T protein has been demonstrated for both lytically infected and transformed cells (72). The potential activities of the 17K T antigen can be inferred from the activities of N-terminal T-antigen fragments. Thus, the transforming activities in N-terminal T-antigen fragments are potentially relevant to transformation by SV40. It was also shown that the C-terminal fragment T251-708 cooperated with Ras in the transformation of primary cells. The analyses reported here further defined the limits of the Ras cooperation regions. The results indicated that the N-terminal Ras cooperation activities are contained within the first 121 amino acids of T antigen. Within the C terminus, further truncation of T251-708 by 21 amino acids abrogated Ras cooperation. This result indicated that amino acids 252 to 271 must be retained to maintain transforming activity.
Analysis of the activities in the N-terminal Ras cooperation fragment focused on the Rb-binding motif and the J domain. Introduction of the E107K mutation, which is known to prevent Rb binding and T-antigen functions dependent on binding (Fig. 2), into T1-127 abrogated Ras cooperation, indicating that Rb binding is required.
Mutational analysis of the J domain suggests a complex relationship among the activities of the region and the protein context in which they are located. Various J-domain-dependent activities are differentially affected by specific mutations. For instance, both of the mutation sets P43L/K45N and D44E/G47R inactivate the ability of T1-708 to transactivate the cyclin A promoter, yet neither set prevents T1-708 from overcoming Rb-mediated growth arrest. In addition, as indicated by others and confirmed here (16, 58), certain mutations, such as P43L/K45N and D44E/G47R, inactivate a J-domain function only in the absence of the C terminus, suggesting that the C terminus of T antigen compensates for the loss of J-domain activity. Recently, Sullivan et al. (62) showed that N-terminal T-antigen fragments form only transient complexes with Hsc70. A stable interaction requires one or more regions between amino acids 137 and 708. It is possible, therefore, that activities that appear J-domain dependent in N-terminal T-antigen fragments but not full-length T antigen represent activities that require stable Hsc70 binding. The introduction of either set of mutations into T1-121 abrogated Ras cooperation. The reasons for the differences in the responses of the N-terminal fragments to mutations in the J domain are not known; however, the differences emphasize the influence of context on the mutant phenotype in the J domain.
The effects of T-antigen Rb-binding and J-domain mutations on transactivation of the cyclin A promoter were investigated also. Cyclin A is a downstream target of Rb. Hypophosphorylated Rb represses the cyclin A promoter (26). The ability of T antigen to bind and inactivate hypophosphorylated Rb could be involved in transactivation of the cyclin A gene. The resulting increase in cyclin A levels could further limit Rb activity by contributing to Rb phosphorylation. Cyclin A-cdk2 complexes, such as the G1-phase cyclins and their kinase partners, phosphorylate and thereby inactivate Rb (1). T-antigen-mediated transactivation of the cyclin A promoter, therefore, would be expected to decrease the level of active Rb and contribute to overcoming its growth-suppressive effects. Since the J domain is required for T antigen to overcome the growth-suppressive effects of Rb, it was reasonable to examine whether J-domain mutants retained or lost the ability to transactivate the cyclin A promoter. The results showed that both an intact J domain and one or more activities C terminal to amino acid 139 were required. However, transactivation did not require an intact Rb-binding motif.
One concern was that many cell lines contain mutations in the p16 gene. It is known that TC7 cells, the cells with which the transactivation experiments were performed, contain Rb. However, the p16 status of the cells has not been determined. If p16 were absent or mutated in the cells, an excess of hyperphosphorylated Rb would accumulate. Since hyperphosphorylated Rb does not readily bind to T antigen, the absence of p16 could mask a requirement for Rb binding by T antigen. Therefore, we repeated the transactivation experiments with a p16 expression plasmid. In the presence of exogenously added p16, wild-type T antigen and T1-708E107K transactivated the cyclin A promoter to similar extents (data not shown).
The activities of SV40 T antigen involved in transactivation of the cyclin A promoter differ in some respects from those observed by Sheng et al. (51) for mouse PyT. In serum-starved cells, Rb binding was necessary for PyT to activate the cyclin A promoter, whereas J-domain function was not. In growing cells, both Rb and J-domain mutants retained substantial transactivating potential. In addition, the J-domain mutation H42Q did not affect the transactivation of the cyclin A promoter by PyT, yet this same mutation, as well as all other J-domain mutations examined, eliminated transactivation by SV40 T antigen.
The promoter region used in the present study contained a 7-kb region used to first define the cyclin A promoter (19), to demonstrate the ability of small t antigen to transactivate the promoter, and to demonstrate a defect in transactivation when the J-domain P43L/K45N mutations were introduced into small t antigen (45). Sheng et al. (51) used the region from −89 to +11. Thus, differences in the results may reflect the extent of the cyclin A promoter used. These authors showed that this promoter region, which contains a serum-responsive E2F site (−37 to −33), also contains uncharacterized targets for PyT-mediated transactivation of the cyclin A promoter that operate independently of Rb binding, indicating that both PyT and SV40 T antigen can transactivate the cyclin A promoter independently of Rb binding.
Alternatively, as suggested by others (51), the differences may reflect divergence in the interactions of the two T antigens with components of the transcription complex. The SV40 N-terminal T-antigen fragment T1-138 is sufficient to bind a number of transcription factors, including TEF-1, Sp-1, transcription factor IIB, and TATA-binding protein (21) and to transactivate the SV40 late promoter and the Rous sarcoma virus long terminal repeat. Whether the J domain is needed in order for this N-terminal fragment to bind the transcription factors was not determined. However, a higher-affinity binding site for the same transcription factors is located between amino acids 101 and 249, and a segment of T antigen containing only those amino acids is sufficient for binding (21). It is possible that the inability of the N-terminal T-antigen segments tested here to transactivate the cyclin A promoter relates to the absence of amino acids in this higher-affinity binding site. The finding that all of the SV40 T-antigen J-domain mutants failed to transactivate the cyclin A promoter indicated that all of the mutations inactivated one J-domain-dependent activity. The finding that individual J-domain mutants differed in their ability to cooperate with Ras and to overcome Rb-induced growth arrest in the context of an N-terminal T-antigen fragment raises the possibility that the J domain may contain overlapping activities or structurally dependent, separable activities.
Clearly, regions of T antigen that are sufficient to cooperate with Ras to transform primary cells did not transactivate the cyclin A promoter. Thus, Ras cooperation with the N-terminal T-antigen regions does not depend on this T-antigen activity. Cooperation more closely correlates with the override of Rb-mediated growth arrest.
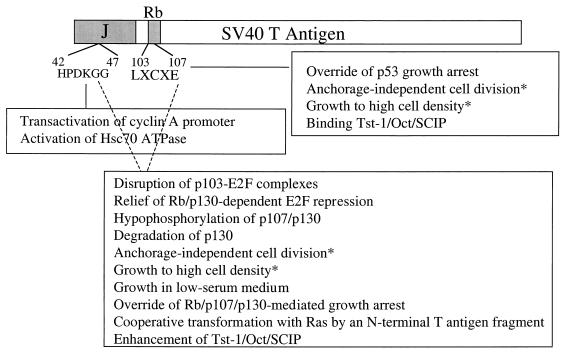
Both the T-antigen J domain and Rb-binding functions affect multiple T-antigen activities. Some T-antigen functions depend on both an intact J domain and an Rb-binding motif. Other activities are dependent on the J domain and independent of Rb binding, and yet other activities are independent of the J domain and dependent on Rb binding. These activities and their dependence on the J domain or the Rb-binding motif are shown in Fig. 7. Cotransformation by the N-terminal Ras cooperation region joins the Rb-dependent, J-domain-dependent activities, whereas transactivation of the cyclin A promoter is a J-domain-dependent, Rb-binding-independent T-antigen activity.
FIG. 7.
Diagram indicating T-antigen activities that are dependent on the J domain, the Rb-binding motif, or both. Data were obtained from the following sources: transactivation of cyclin A (this study); activation of Hsc70 ATPase (58); override of p53 growth arrest (16, 46); anchorage-independent cell division, growth in medium containing low serum concentrations, and growth to high cell density (61, 65); binding and enhancement of Tst-1/Oct/SCIP (55); disruption of p103-E2F complexes, relief of Rb/p130-dependent E2F repression, and override of Rb/p107/p130-mediated growth arrest (70); hypophosphorylation of p107/p130 (61); degradation of p130 (60); and cooperative transformation with Ras by an N-terminal fragment of T antigen (6). Asterisks indicate divergence in results from different studies.
Lin et al. (29) showed that activation of the mitogen-activated protein kinase pathway following binding of Raf to Ras was sufficient to induce cell senescence in REF. Senescence was abrogated by inactivation of either p53 or p16 and possibly p19ARF (29). Our results showing that the ability of an N-terminal fragment of T antigen to cooperate with Ras in transforming primary cells is dependent on binding and inactivating the growth-suppressive effect of Rb are consistent with those findings. Since T antigen binds underphosphorylated Rb, accumulation of this active form of the tumor suppressor in response to a Ras-induced increase in p16 levels would be counteracted. However, interruption of the p53 pathway does not appear to be sufficient to overcome ras-induced senescence in T-antigen-expressing cells.
The C-terminal Ras cooperation segment T251-708 binds p53 (6) and prevents p53 from activating a p53-responsive promoter (Fig. 6). However, binding and inactivation of p53 as a transcriptional transactivator were not sufficient for Ras cooperation. Seven mutants containing amino acid substitution mutations A382P, P399L, D402E, D402H, D402N, F408V, and C411R failed to cooperate with Ras. Each, like T1-708 dl400, retained the ability to inhibit p53-mediated transactivation to approximately the same extent as did T251-708. A mutant T antigen lacking the p53-binding domain (T1-127) did not inhibit p53-mediated transactivation. The lack of correlation between Ras cooperation and p53 inactivation was not anticipated. Ras readily transforms p53-null cells (29, 50). In addition, Ras cooperates with MDM2 (14), the negative regulator of p53, or dominant-negative forms of p53 (15) to transform REF. In each of these cases, the transcriptional transactivating capacity of p53 is inhibited (12, 21, 35, 37, 39). Therefore, we anticipated that T251-708 would cooperate with Ras by virtue of binding p53 and rendering p53 inactive as a transcriptional transactivator. Based on the results presented, it seems likely that either T251-708 provides an additional activity needed to cooperate with Ras or Ras cooperation depends on abrogating a p53 activity other than or in addition to transactivation. Further studies are needed to distinguish between the possibilities.
The ras oncogene activates two additional signaling pathways (reviewed in references 34 and 52) by binding to the effector protein phosphatidylinositol 3-kinase (PI3K) or RaIGDS (a GTP-GDP exchange factor). Interaction of Ras with PI3K stimulates its lipid kinase activity and the subsequent activation of Akt/PKB on the one hand and Rac on the other. Both of the resulting cascades limit apoptosis. Ras interaction with RaIGDS results in the inhibition of Cd42/Rac and the generation of phosphatidic acid. Which of the three Ras effector pathways is involved in cooperation with T antigen and segments thereof in transforming rodent cells remains to be determined.
ras-induced senescence and natural senescence share many properties. In both cases, senescent cells display similar morphological and biochemical changes. During both natural senescence and ras-induced senescence, cells at subconfluent densities stop proliferating, flatten, increase in volume, and express senescence-associated biochemical markers such as SAβgal (10). Similarly, following prolonged Ras expression or during natural senescence, cells express increased levels of p53 and p16 (reviewed in reference 50). The involvement of these genes in senescence was confirmed by the observation that p16- or p53-null cells readily bypassed both natural senescence (24) and ras-induced senescence (51, 52, 65). Also, the p16 and p53 pathways appear to act cooperatively in ras-induced senescence (29). Similarity between natural senescence and ras-induced senescence is indicated further by the absence of a role for p21. p21 levels increase during natural senescence. However, p21-null murine fibroblasts still undergo senescence, and they are resistant to transformation by ras (40).
However, the T-antigen activities needed to overcome the two processes differ. T-antigen-mediated bypass of natural senescence is Rb and J-domain independent and p53-binding dependent (25, 66). In contrast, Ras cooperation is Rb dependent and J-domain dependent and either is p53 inactivation dependent or requires activities in addition to inactivation of p53 transcriptional transactivating capacity. Further studies, particularly those testing T-antigen fragments with specific mutations for the ability to extend cell life span and to cooperate with C-terminal segments of T antigen in bypassing natural senescence, are required to elucidate similarities and differences in the bypass of natural senescence and ras-induced senescence.
Acknowledgments
We thank Holly Lacko for excellent technical assistance.
This work was supported by grant CA24694 from the National Cancer Institute of the National Institutes of Health. T.M.B. was supported by training grant 5-T23-CA60395 (Viruses and Cancer) from the National Cancer Institute of the National Institutes of Health.
REFERENCES
- 1.Adams, P. D., X. Li, W. R. Sellers, K. B. Baker, X. Leng, J. W. Harper, Y. Taya, and W. G. Kaelin, Jr. 1999. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol. Cell. Biol. 19:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Barbacid, M. 1987. ras genes. Annu. Rev. Biochem. 56:779-827. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky, J. L., and J. M. Pipas. 1998. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol. 72:5329-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campisi, J. 1997. The biology of replicative senescence. Eur. J. Cancer 33:703-709. [DOI] [PubMed] [Google Scholar]
- 6.Cavender, J. F., A. Conn, M. Epler, H. Lacko, and M. J. Tevethia. 1995. Simian virus 40 large T antigen contains two independent activities that cooperate with a ras oncogene to transform rat embryo fibroblasts. J. Virol. 69:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavender, J. F., C. Mummert, and M. J. Tevethia. 1999. Transactivation of a ribosomal gene by simian virus 40 large-T antigen requires at least three activities of the protein. J. Virol. 73:214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeCaprio, J. A. 1999. The role of the J domain of SV40 large T in cellular transformation. Biologicals 27:23-28. [DOI] [PubMed] [Google Scholar]
- 9.Diller, L., J. Kassel, C. E. Nelson, M. A. Gryka, G. Litwak, M. Gebhardt, B. Bressac, M. Ozturk, S. J. Baker, B. Vogelstein, et al. 1990. p53 functions as a cell cycle control protein in osteosarcomas. Mol. Cell. Biol. 10:5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliyahu, D., D. Michalovitz, and M. Oren. 1985. Overproduction of p53 antigen makes established cells highly tumorigenic. Nature 316:158-160. [DOI] [PubMed] [Google Scholar]
- 12.Farmer, G., J. Bargonetti, H. Zhu, P. Friedman, R. Prywes, and C. Prives. 1992. Wild-type p53 activates transcription in vitro. Nature 358:83-86. [DOI] [PubMed] [Google Scholar]
- 13.Filmus, J., A. I. Robles, W. Shi, M. J. Wong, L. L. Colombo, and C. J. Conti. 1994. Induction of cyclin D1 overexpression by activated ras. Oncogene 9:3627-3633. [PubMed] [Google Scholar]
- 14.Finlay, C. A. 1993. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol. Cell. Biol. 13:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finlay, C. A., P. W. Hinds, and A. J. Levine. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57:1083-1093. [DOI] [PubMed] [Google Scholar]
- 16.Gjoerup, O., H. Chao, J. A. DeCaprio, and T. M. Roberts. 2000. pRB-dependent, J domain-independent function of simian virus 40 large T antigen in override of p53 growth suppression. J. Virol. 74:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldfarb, M., K. Shimizu, M. Perucho, and M. Wigler. 1982. Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature 296:404-409. [DOI] [PubMed] [Google Scholar]
- 18.Haupt, Y., and M. Oren. 1996. p53-mediated apoptosis: mechanisms and regulation. Behring Inst. Mitt. 97:32-59. [PubMed]
- 19.Henglein, B., X. Chenivesse, J. Wang, D. Eick, and C. Brechot. 1994. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc. Natl. Acad. Sci. USA 91:5490-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, D., A. Srinivasan, G. Lozano, and P. D. Robbins. 1993. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene 8:2805-2812. [PubMed] [Google Scholar]
- 21a.Johnston, S. D., X.-M. Yu, and J. E. Mertz. 1996. The major transcriptional transactivation domain of simian virus large T antigen associates noncurrently with multiple components of the transcriptional preinitiation complex. J. Virol. 70:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaelin, W. G., Jr. 1999. Functions of the retinoblastoma protein. Bioessays 21:950-958. [DOI] [PubMed] [Google Scholar]
- 23.Kalderon, D., and A. E. Smith. 1984. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology 139:109-137. [DOI] [PubMed] [Google Scholar]
- 24.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 25.Kierstead, T. D., and M. J. Tevethia. 1993. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol. 67:1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen, K. E., A. F. Fribourg, M. W. Strobeck, J. M. Blanchard, and E. S. Knudsen. 1999. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J. Biol. Chem. 274:27632-27641. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 28.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 29.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J. Y., and D. T. Simmons. 1991. Stable T-p53 complexes are not required for replication of simian virus 40 in culture or for enhanced phosphorylation of T antigen and p53. J. Virol. 65:2066-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludlow, J. W., J. A. DeCaprio, C. M. Huang, W. H. Lee, E. Paucha, and D. M. Livingston. 1989. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell 56:57-65. [DOI] [PubMed] [Google Scholar]
- 32.Ludlow, J. W., J. Shon, J. M. Pipas, D. M. Livingston, and J. A. DeCaprio. 1990. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell 60:387-396. [DOI] [PubMed] [Google Scholar]
- 33.Manfredi, J. J., and C. Prives. 1994. The transforming activity of simian virus 40 large tumor antigen. Biochim. Biophys. Acta 1198:65-83. [DOI] [PubMed] [Google Scholar]
- 34.McCormick, F. 1999. Signalling networks that cause cancer. Trends Cell Biol. 9:M53-M56. [PubMed] [Google Scholar]
- 35.Mietz, J. A., T. Unger, J. M. Huibregtse, and P. M. Howley. 1992. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 11:5013-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 37.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 38.Muller, H., and K. Helin. 2000. The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta 1470:M1-M12. [DOI] [PubMed] [Google Scholar]
- 39.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 40.Pantoja, C., and M. Serrano. 1999. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 18:4974-4982. [DOI] [PubMed] [Google Scholar]
- 41.Peeper, D. S., T. M. Upton, M. H. Ladha, E. Neuman, J. Zalvide, R. Bernards, J. A. DeCaprio, and M. E. Ewen. 1997. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386:177-181. [DOI] [PubMed] [Google Scholar]
- 42.Phillips, B., and K. Rundell. 1988. Failure of simian virus 40 small t antigen to disorganize actin cables in nonpermissive cell lines. J. Virol. 62:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pipas, J. M. 1998. Molecular chaperone function of the SV40 large T antigen. Dev. Biol. Stand. 94:313-319. [PubMed] [Google Scholar]
- 44.Pipas, J. M., K. W. Peden, and D. Nathans. 1983. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol. Cell. Biol. 3:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porras, A., J. Bennett, A. Howe, K. Tokos, N. Bouck, B. Henglein, S. Sathyamangalam, B. Thimmapaya, and K. Rundell. 1996. A novel simian virus 40 early-region domain mediates transactivation of the cyclin A promoter by small-t antigen and is required for transformation in small-t antigen-dependent assays. J. Virol. 70:6902-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quartin, R. S., C. N. Cole, J. M. Pipas, and A. J. Levine. 1994. The amino-terminal functions of the simian virus 40 large T antigen are required to overcome wild-type p53-mediated growth arrest of cells. J. Virol. 68:1334-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy, V. B., S. S. Tevethia, M. J. Tevethia, and S. M. Weissman. 1982. Nonselective expression of simian virus 40 large tumor antigen fragments in mouse cells. Proc. Natl. Acad. Sci. USA 79:2064-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ries, S., C. Biederer, D. Woods, O. Shifman, S. Shirasawa, T. Sasazuki, M. McMahon, M. Oren, and F. McCormick. 2000. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103:321-330. [DOI] [PubMed] [Google Scholar]
- 49.Serrano, M. 1997. The tumor suppressor protein p16INK4a. Exp. Cell Res. 237:7-13. [DOI] [PubMed] [Google Scholar]
- 50.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 51.Sheng, Q., T. M. Love, and B. Schaffhausen. 2000. J domain-independent regulation of the Rb family by polyomavirus large T antigen. J. Virol. 74:5280-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shields, J. M., K. Pruitt, A. McFall, A. Shaub, and C. J. Der. 2000. Understanding Ras: "it ain't over 'til it's over.' Trends Cell Biol. 10:147-154. [DOI] [PubMed] [Google Scholar]
- 53.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 54.Sleigh, M. J., W. C. Topp, R. Hanich, and J. F. Sambrook. 1978. Mutants of SV40 with an altered small t protein are reduced in their ability to transform cells. Cell 14:79-88. [DOI] [PubMed] [Google Scholar]
- 55.Sock, E., J. Enderich, and M. Wegner. 1999. The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol. Cell. Biol. 19:2455-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sompayrac, L., and K. J. Danna. 1983. Simian virus 40 deletion mutants that transform with reduced efficiency. Mol. Cell. Biol. 3:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sompayrac, L., and K. J. Danna. 1989. An SV40 mutant oncoprotein has a nuclear location. Virology 171:267-270. [DOI] [PubMed] [Google Scholar]
- 58.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stein, G. H., L. F. Drullinger, A. Soulard, and V. Dulic. 1999. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 19:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stubdal, H., J. Zalvide, and J. A. DeCaprio. 1996. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J. Virol. 70:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan, C. S., S. P. Gilbert, and J. M. Pipas. 2001. ATP-dependent simian virus 40 T-antigen-Hsc70 complex formation. J. Virol. 75:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Templeton, D. J., S. H. Park, L. Lanier, and R. A. Weinberg. 1991. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc. Natl. Acad. Sci. USA 88:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tevethia, M. J., R. H. Bonneau, J. W. Griffith, and L. Mylin. 1997. A simian virus 40 large T-antigen segment containing amino acids 1 to 127 and expressed under the control of the rat elastase-1 promoter produces pancreatic acinar carcinomas in transgenic mice. J. Virol. 71:8157-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tevethia, M. J., H. A. Lacko, T. D. Kierstead, and D. L. Thompson. 1997. Adding an Rb-binding site to an N-terminally truncated simian virus 40 T antigen restores growth to high cell density, and the T common region in trans provides anchorage-independent growth and rapid growth in low serum concentrations. J. Virol. 71:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson, D. L., D. Kalderon, A. E. Smith, and M. J. Tevethia. 1990. Dissociation of Rb-binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virology 178:15-34. [DOI] [PubMed] [Google Scholar]
- 67.Tornow, J., and C. N. Cole. 1983. Intracistronic complementation in the simian virus 40 A gene. Proc. Natl. Acad. Sci. USA 80:6312-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wynford-Thomas, D. 1999. Cellular senescence and cancer. J. Pathol. 187:100-111. [DOI] [PubMed] [Google Scholar]
- 69.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 70.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zambetti, G. P., J. Bargonetti, K. Walker, C. Prives, and A. J. Levine. 1992. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 6:1143-1152. [DOI] [PubMed] [Google Scholar]
- 72.Zerrahn, J., U. Knippschild, T. Winkler, and W. Deppert. 1993. Independent expression of the transforming amino-terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 12:4739-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, J., P. W. Rice, L. Gorsch, M. Abate, and C. N. Cole. 1992. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J. Virol. 66:2780-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]