Abstract
Cucumber mosaic virus (CMV) is an RNA plant virus with a tripartite genome and an extremely broad host range. Previous evolutionary analyses with the coat protein (CP) and 5′ nontranslated region (NTR) of RNA 3 suggested subdivision of the virus into three groups, subgroups IA, IB, and II. In this study 15 strains of CMV whose nucleotide sequences have been determined were used for a complete phylogenetic analysis of the virus. The trees estimated for open reading frames (ORFs) located on the different RNAs were not congruent and did not completely support the subgrouping indicated by the CP ORF, indicating that different RNAs had independent evolutionary histories. This is consistent with a reassortment mechanism playing an important role in the evolution of the virus. The evolutionary trees of the 1a and 3a ORFs were more compact and displayed more branching than did those of the 2a and CP ORFs. This may reflect more rigid host-interactive constraints exerted on the 1a and 3a ORFs. In addition, analysis of the 3′ NTR that is conserved among all RNAs indicated that evolutionary constraints on this region are specific to the RNA component rather than the virus isolate. This indicates that functions other than replication are encoded in the 3′ NTR. Reassortment may have led to the genetic diversity found among CMV strains and contributed to its enormous evolutionary success.
Cucumber mosaic virus (CMV) is the type species in the genus Cucumovirus, family Bromoviridae (20). CMV has the broadest host range of any known virus, infecting more than 1,000 species of plants, including monocots and dicots, herbaceous plants, shrubs, and trees (6). In the 85 years since its discovery (5, 15), CMV has been found in all parts of the world, and numerous strains have been characterized. Hence, CMV has been very successful in rapidly adapting to new hosts and new environments. Fifteen strains have complete nucleotide sequence data published or entered in GenBank, and more than 60 coat protein (CP) sequences are available.
CMV is a tripartite plus-sense RNA virus. Like most other plant viruses with divided genomes, the genomic RNAs are packaged in separate particles. This allows a larger genome to be packaged in a very simple virion but requires that multiple virus particles invade a single cell to initiate an infection. The cucumoviruses use vector transmission by aphids, which probably ensures a multiplicity of infection sufficient to reliably establish infection. This can also allow mixtures of virus particles to initiate infection in a new host during transmission from plants infected with more than one virus and can set the stage for genetic reassortment.
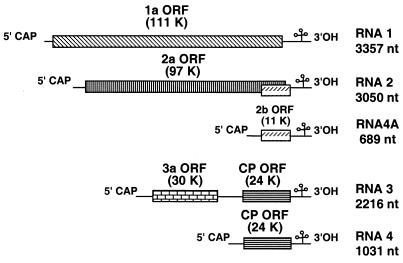
CMV contains five open reading frames (ORFs) (Fig. 1). The 1a and 2a ORFs, encoded on RNAs 1 and 2, respectively, are the viral components of the replicase. The 2b ORF, a gene overlapping the 2a ORF, is expressed from a subgenomic RNA, RNA 4A (4) and encodes a suppressor of posttranscriptional gene silencing (1). RNA 3 encodes the 3a protein, the viral movement protein, and the CP, expressed from subgenomic RNA 4 (for reviews, see references 6, 16, and 19).
FIG. 1.
Genome organization of CMV. Nucleotide (nt) numbers and the sizes for encoded proteins are given for the Fny strain. K, kilodalton.
Phylogenetic analyses by using maximum-parsimony methods are often used to resolve taxonomic questions in virology (13). However, these analyses can also reveal important information about the evolutionary history of a group of viruses. A phylogeny estimation of the species of the Cucumovirus genus showed noncongruent trees for the amino acid sequences of the different ORFs, indicating distinct evolutionary histories for each RNA and strongly supporting the occurrence of reassortment in the evolutionary history of the genus (24). Phylogeny estimations with the CP ORF, as well as rearrangements in the 5′ nontranslated region (NTR) of RNA 3, divided CMV strains into three subgroups: IA, IB, and II (21). We describe here a phylogenetic analysis of 15 CMV strains whose complete or nearly complete sequence information is available. This is the first study that shows a detailed analysis of the entire genome of this important species of plant viruses. Comparisons of the trees indicate that reassortment events were likely in the history of CMV evolution, that overprinting of the 2b ORF may have occurred more than once, and that conservation of the minus-strand replication signals is stronger for each RNA than it is within a strain. In addition, the CP and 2a ORFs exhibit both more rapid and radial evolution, in contrast to the evolutionary patterns of the 1a and 3a ORFs. This may result from a lack of constraint imposed by virus-host interactions.
MATERIALS AND METHODS
Sequence information.
Source of sequences are shown in Table 1. LS-CMV RNAs 1 and 2 were cloned as previously described (26). Plasmids pLS-1 and pLS-2 were sequenced by using a Taq DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). The products of the reaction were separated electrophoretically, and the data were processed by an ABI 373A automated DNA sequencer (Perkin-Elmer/Applied Biosystems). DNA sequences were assembled and edited by using the PC/Gene program ASSEMGEL (IntelliGenetics, Mountain View, Calif.). The sequences of LS-CMV RNAs 1 and 2 were deposited in GenBank under accession numbers AF416899 and AF416900, respectively.
TABLE 1.
Accession numbers for sequence data and country of origin
| Strain | Accession no.
|
Origina (state) | ||
|---|---|---|---|---|
| RNA 1 | RNA 2 | RNA 3 | ||
| Fny | D00356 | D00355 | D10538 | United States (N.Y.) |
| IA | AB042292 | AB042293 | AB042294 | Indonesia |
| Ix | U20220 | U20218 | U20219 | Philippines |
| Leg | D16403 | D16406 | D16405 | Japan |
| LS | AF416899 | AF416900 | AF127976 | United States (N.Y.) |
| Ly | AF198101 | AF198102 | AF198103 | Australia |
| Mf | AJ276479 | AJ276480 | AJ276481 | South Korea |
| Nt9 | D28778 | D28779 | D28780 | Taiwan |
| O | —b | D10209 | D00385 | Japan |
| Q | X02733 | X00985 | M21464 | Australia |
| S | Y10884 | Y10885 | U37227 and AF063610 | South Africa |
| SD | AF071551 | D86330 | AB008777 | [China] |
| SD | AF071551 | D86330 | AB008777 | [China] |
| Tfn | Y16924 | Y16925 | Y16926 | Italy |
| Trk7 | AJ007933 | AJ007934 | L15336 | Hungary |
| Y | D12537 | D12538 | D12499 | Japan |
| ER-PSV | U15728 | U15729 | U15730 | United States (Ky.) |
The country of origin is indicated where known. In cases where the country of origin is not indicated, the country where the sequence data was obtained is given in brackets.
—, see reference 11.
Multiple sequence alignments and phylogeny estimations.
Sequences were aligned by using the GCG package (version 10.0) program PILEUP, with the ER strain of Peanut stunt virus (ER-PSV) as an outgroup, and alignments were edited by using either the GCG Seqlab editing program or MacClade 4.0. Phylogeny estimations were done by using PAUP 4.0b5, maximum-parsimony setting, and a 100-replicate bootstrap search with either the heuristic or the branch-and-bound search option. Gaps were handled as a fifth character state. The length of each alignment and the number of informative characters used for analysis are shown in Table 2. All branches with <70% bootstrap support were judged inconclusive and were collapsed (7, 12). Branch lengths for all trees were normalized to 5% divergence, except for the 2bAA analysis, wherein branch lengths are compressed to represent twice as much divergence as in the other trees, in order to fit the figure into a similar space.
TABLE 2.
Details of alignments used for parsimony analyses
| ORFa | Typeb | Lengthc (no.) | No. of informative charactersd |
|---|---|---|---|
| 1a | nt | 3,045 | 860 |
| 2a | nt | 2,672 | 863 |
| 2b | nt | 372 | 154 |
| 2b | aa | 137 | 63 |
| 3a | nt | 866 | 197 |
| CP | nt | 788 | 212 |
| 3′ NTR | nt | 284 | 143 |
ORF used for analysis.
Type of sequence used: nt, nucleotide; aa, amino acid.
The length includes gaps in the alignment, which were considered as fifth-character states
The number of informative characters, e.g., for the 1a ORF, 860 of the 3,045 nucleotide positions in the alignment were parsimony informative.
RESULTS
Analysis of the RNA 3 ORFs.
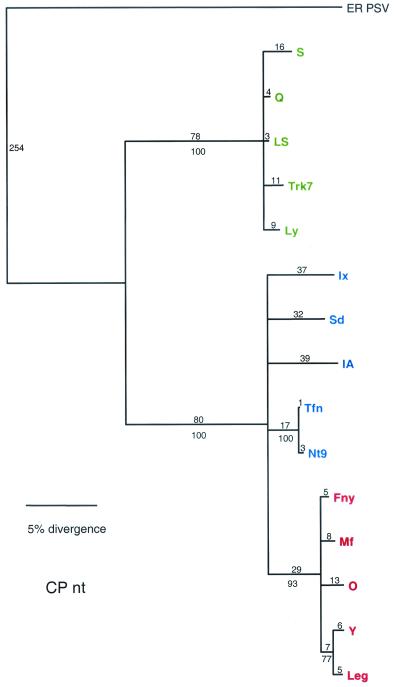
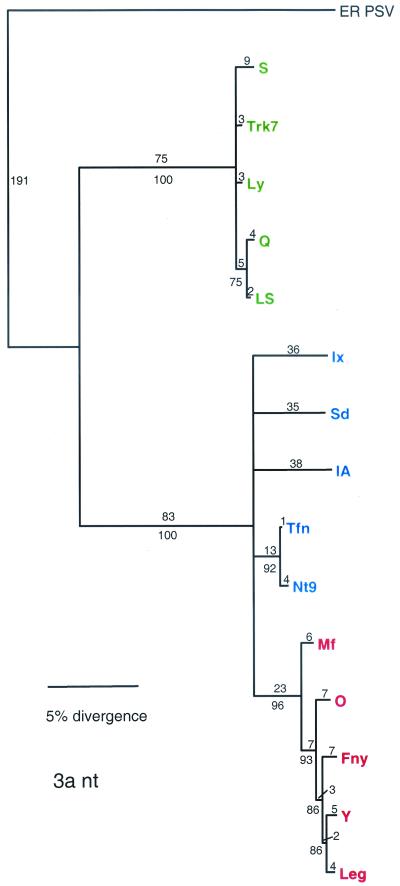
The CP gene for the 15 strains used in this study, encoded in the 3′ portion of RNA 3, shows a pattern of divergence similar to the earlier report with 53 strains (21) (Fig. 2). All of the subgroup IB strains used in the earlier report, however, originated in Asia. The Tfn strain, isolated from tomatoes in Italy, was included in this study and shows very close evolutionary ties to Nt9-CMV, a subgroup IB strain isolated in Taiwan. Also, as in the previous study, there is very little branching within the subgroup clades. In the 3a gene, however, there is more branching within the groups (Fig. 3). The number of informative characters for both ORFs is high (Table 2) but is greater in the CP ORF (212 of 788) than in the 3a ORF (197 of 866).
FIG. 2.
Phylogenetic analysis of the CP ORF, with aligned nucleotide sequences. Strains designated in green are from subgroup II, strains designated in blue are from subgroup IB, and strains designated in red are from subgroup IA. Bootstrap percentage values are shown below the branches, and distance, in number of nucleotide (nt) changes, is shown above the branch lines.
FIG. 3.
Phylogenetic analysis of the 3a ORF, with aligned nucleotide sequences. Strains designated in green are from subgroup II, strains designated in blue are from subgroup IB, and strains designated in red are from subgroup IA. Bootstrap percentage values are shown below the branches, and the distance, shown in number of nucleotide (nt) changes, is shown above the branch lines.
Analysis of the RNA 2 ORFs.
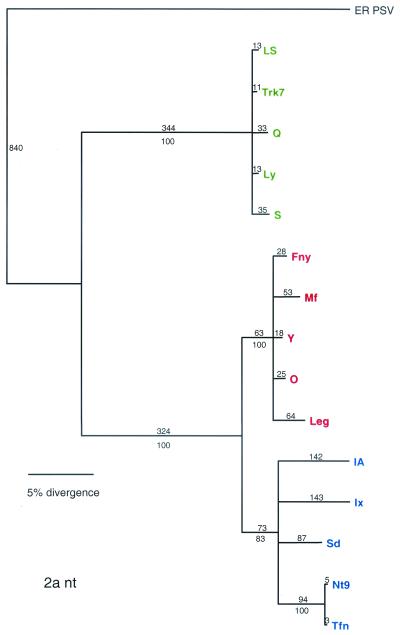
The 2a ORF can be clearly divided into three clades that correspond to subgroups IA, IB, and II. In addition, the pattern of sequence divergence in the 2a ORF is largely radial (Fig. 4), a finding similar to what is seen with the CP.
FIG. 4.
Phylogenetic analysis of the 2a ORF, with aligned nucleotide sequences. Strains designated in green are from subgroup II, strains designated in blue are from subgroup IB, and strains designated in red are from subgroup IA. Bootstrap percentage values are shown below the branches, and the distance, in number of nucleotide (nt) changes, is above below the branch lines.
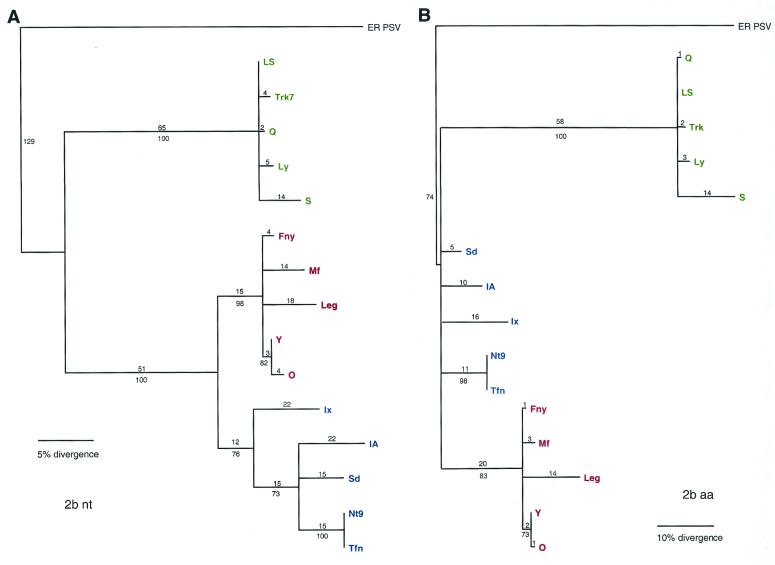
The nucleotide sequence divergence in the 2b ORF shows a pattern very similar to the 2a ORF (Fig. 5A) and is undoubtedly constrained by the 2a, since much of the ORF overlaps the 2a. The amino acid divergence pattern, however, differs dramatically in the length of the branches, particularly in the distance from the common ancestor of CMV to the subgroup II ancestor (Fig. 5B).
FIG. 5.
Phylogenetic analysis of the 2b ORF, with aligned nucleotide sequences (A) and aligned amino acid sequences (B). Strains designated in green are from subgroup II, strains designated in blue are from subgroup IB, and strains designated in red are from subgroup IA. Bootstrap percentage values are shown below the branches, and the distance, in number of nucleotide (nt) or amino acid (aa) changes, is shown above the branch lines.
Analysis of the 1a ORF.
The 1a ORF encodes the methyl transferase and helicase motifs conserved in many RNA viruses. The divergence of subgroups I and II is clearly seen in the 1a phylogeny estimation (Fig. 6); however, the further divergence of subgroup I into IA and IB is not as obvious and indicates an evolutionary history that is quite different from what is seen in the other RNAs.
FIG. 6.
Phylogenetic analysis of the 1a ORF, with aligned nucleotide sequences. Strains designated in green are from subgroup II, strains designated in blue are from subgroup IB, and strains designated in red are from subgroup IA. Bootstrap percentage values are shown below the branches, and the distance, in number of nucleotide (nt) changes, is shown above the branch lines.
3′ NTR.
The signals for virus replication are contained in the promoters at the 3′ ends of the plus and minus strands. The minus-strand promoter has been well defined and consists of a highly conserved region that can fold into two alternate structures of either a cruciform or a pseudoknot structure (16). This 3′ region was aligned for all three RNAs from each strain (strain S was not included in this analysis because not all of the NTR sequences are available). Care was taken not to include any sequences outside of the conserved 3′-end structure. When these sequences were analyzed phylogenetically, the 3′ regions grouped predominantly by RNA component rather than by virus strain, especially in the subgroup II isolates, indicating that constraints on this region are more complex than simply those imposed by the replicase (Fig. 7).
FIG. 7.
Phylogenetic analysis of the 3′ NTR, with aligned nucleotide sequences. RNAs designated in blue are from RNA 1, RNAs designated in green are from RNA 2, and RNAs designated in red are from RNA 3. Bootstrap percentage values are shown below the branches, and distance, in number of nucleotide (nt) changes, is shown above the branch lines.
DISCUSSION
In the CP and 3a phylogenies, the subgroup IB appears to be ancestral to the IA subgroup, i.e., the IA isolates probably radiated from a IB isolate. Tfn is the first non-Asian subgroup IB strain described to date and likely originated in Asia. The only apparent differences between the CP and 3a trees are in the degree of branching, which is somewhat higher in the 3a, and in the branch lengths, which are longer in the CP tree, especially the branches that lead to the subgroup I and II divisions. These differences probably reflect different constraints on the evolution of each ORF. As far as is known, the CP interacts predominantly with itself or with the viral RNA and has little interaction with the host. CP interactions with the aphid vector are also probably minimal and mostly nonspecific, since more than 85 species of aphids can transmit CMV (6). In contrast, the 3a protein is involved in virus movement and interacts with the host plasmadesmata. Hence, some of the evolutionary constraints on the 3a protein are imposed by the host, which may allow less radial divergence from the ancestral state and a more compact tree. A radial pattern in a phylogenetic tree can also indicate that there is not enough data to resolve the tree; however, the high level of informative characters in all of these analyses indicates that the trees are probably rigorous.
The 2a protein contains the GDD motif found in most RNA-dependent RNA polymerases. The 2a forms the viral replicase complex, together with the 1a protein and a number of host factors. Details of the interactions and formation of the complex are unknown but are presumed to be similar to the 1a-2a interactions of Brome mosaic virus (3, 17, 22). Upon comparison of the 2a tree with the 1a tree, it can be seen that there is a significant difference in the degree of branching and in overall branch lengths, a finding similar to what was seen in the CP and 3a trees. The 1a tree is almost completely resolved, in spite of the fact that there were less-informative characters than were used for the 2a ORF. This may indicate that the evolution of the 2a protein is not constrained by virus-host interactions and thus is not the replicase component that interacts with the host factors but rather that it interacts with the 1a and/or itself, as well as with the viral RNAs.
The 2b ORF is an overlapping ORF, found at the carboxy terminus of the 2a ORF (4). The region of overlap is apparently a highly flexible region of the 2a ORF. ORFs in this position occur in all members of the Cucumovirus genus but in only one other member of the Bromoviridae family: the Ilarvirus genus. All of these ORFs are found in the +1 reading frame, with reference to the 2a ORF (25). In all of the CMV isolates the 2b initiation codon is in a homologous position, but in the other members of the Cucumovirus genus initiation occurs in a different position. The level of 2b sequence similarity at the amino acid level is low among Cucumovirus species compared to the amino acid sequence similarity of other ORFs (data not shown). In addition, the identity at the nucleotide level (55 to 62%) is higher than the amino acid identity level (37 to 50%). Taken together, this indicates that the 2a is the ancestral ORF and may indicate that the cucumovirus 2b arose more than once and is found in a similar location because it is the most flexible portion of the viral genome. If the 2b ORFs did indeed arise more than once, the PSV and CMV amino acid sequences are not strictly homologous even though the nucleotides are homologous, and it may not be appropriate to use PSV as an outgroup. However, analysis of the 2b ORF by using an unrooted tree and deleting PSV did not change the relationships or branch lengths in the ingroup.
The 3′ NTR clearly contains the promoter for minus-strand synthesis of the viral RNA during replication. Its primary function involves interaction with the replicase (i.e., the 1a and 2a proteins, along with host factors). In both artificially generated and naturally occurring reassortants in the cucumoviruses, the 3′ regions of RNAs 3 have been exchanged with those of RNA 1 or RNA 2, presumably to better accommodate the replicase (8; M. J. Roossink, unpublished results). Hence, the grouping of the 3′ regions by RNA rather than by strain was surprising and indicates that RNA-specific functions are encoded in this region, in addition to the replicase functions. These functions may be related to controlling translation. In CMV, the 1a and 2a proteins are expressed at very low levels compared to the 3a protein and the CP. The 3′ region of RNA 3 of the related virus Brome mosaic virus is involved in regulating translation (10); hence, it seems likely that RNA-specific translational regulation would reside in this region.
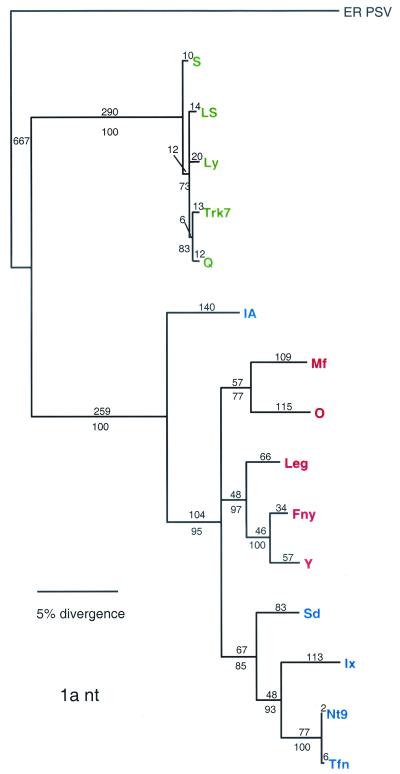
A closer look at each ORF in the subgroup I strains reveals that only the 3a and CP ORFs are congruent (with the exception of the divergence pattern: branched versus radial). In the RNA 3 ORFs the IA strains fall out as a clade within the IB strains. In the RNA 2 ORFs, subgroups IA and IB are found on separate clades, and in the RNA 1 ORF the IA strain is separated from the rest of the subgroup IB strains. The remainder of the subgroup I strains fall out as a three clades: one containing the rest of the IB subgroup, and two for the IA subgroup. Hence, each RNA appears to have a different evolutionary history. This implies that the extant strains were generated through reassortment events. Different selective constraints on the various RNAs is also consistent with these data, and such constraints could have facilitated the selection of reassortants over their parental variants.
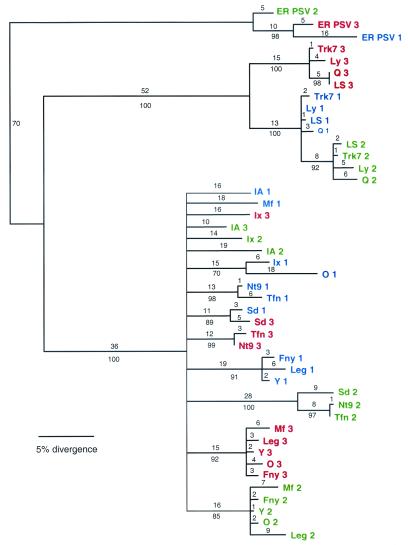
Reassortment of RNA viruses with divided genomes has been demonstrated for other viruses such as influenza virus (23) and bacteriophage ø6 (2), as well as for plant viruses such as tobraviruses (18) and cucumoviruses (24). Within-species reassortment has been demonstrated for PSV (14). Field surveys of CMV in Spain suggested that reassortment was a rare event (9), but these studies analyzed recent events of reassortment. In the present study events are analyzed that occurred over a much longer period of evolutionary time, although the actual amount of time that CMV has been diverging is unknown. Reassortment could have both positive and negative effects on a virus. The increased genetic diversity afforded by reassortment may allow for host range expansion and may allow for recovery of strains bearing deleterious mutations. However, if the reassortant event occurs between two strains that are already highly divergent, it may cause problems in protein-protein interactions, such as those required for a fully functional replicase, or in recognition of promoter sequences, RNA packaging signals, and other intra-RNA functions. In these cases the reassortment event may be common but the new viruses may be eliminated by strong negative selection. Clearly, CMV has been an evolutionarily successful virus, with both a very broad host range and a worldwide distribution. It is likely that reassortment events have played an important role in this success, and it is clear that reassortment is important in the evolution of new viruses.
Acknowledgments
I thank William Schneider and Gregory May for helpful suggestions on the manuscript.
This work was supported by the Samuel Roberts Noble Foundation.
REFERENCES
- 1.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Chao, L., T. Tran, and C. Matthews. 1992. Muller's ratchet and the advantage of sex in the RNA virus ø6. Evolution 46:289-299. [DOI] [PubMed] [Google Scholar]
- 3.Dinant, S., M. Janda, P. A. Kroner, and P. Ahlquist. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like virual RNA synthesis proteins. J. Virol. 67:7181-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding, S.-W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle, S. P. 1916. A new infectious mosaic disease of cucumber. Phytopathology 6:145-147. [Google Scholar]
- 6.Edwardson, J. R., and R. G. Christie. 1991. Cucumoviruses, p. 293-319. CRC Handbook of viruses infecting legumes. CRC Press, Boca Raton, Fla.
- 7.Efron, B., E. Halloran, and S. Holmes. 1996. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 93:7085-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Cuartero, B., J. Burgyán, M. A. Aranda, K. Salánki, E. Moriones, and F. García-Arenal. 1994. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203:373-377. [DOI] [PubMed] [Google Scholar]
- 9.Fraile, A., J. L. Alonso-Prados, M. A. Aranda, J. J. Bernal, J. M. Maplica, and F. García-Arenal. 1997. Genetic exchange by recombination or reassortment is infrequent in natural populations of a tripartite RNA plant virus. J. Virol. 71:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallie, D. R., and M. Kobayashi. 1994. The role of the 3-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 142:159-165. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa, T., M. Mizukami, I. Nakamura, and M. Suzuki. 1989. Cloning and sequencing of RNA-1 cDNA from cucumber mosaic virus strain O. Gene 85:533-540. [DOI] [PubMed] [Google Scholar]
- 12.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in plylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 13.Hillis, D. M., J. P. Huelsenbeck, and C. W. Cunningham. 1994. Application and accuracy of molecular phylogenies. Science 264:671-677. [DOI] [PubMed] [Google Scholar]
- 14.Hu, C.-C., and S. A. Ghabriel. 1998. Molecular evidence that strain BV-15 of peanut stunt cucumovirus is a reassortant between subgroup I and II strains. Phytopathology 88:92-97. [DOI] [PubMed] [Google Scholar]
- 15.Jagger, I. C. 1916. Experiments with the cucumber mosaic disease. Phytopathology 6:149-151. [Google Scholar]
- 16.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348. [DOI] [PubMed] [Google Scholar]
- 17.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson, D. J., W. D. O. Hamilton, B. D. Harrison, and D. C. Baulcombe. 1987. Two anomalous tobravirus isolates: evidence for RNA recombination in nature. J. Gen. Virol. 68:2551-2561. [Google Scholar]
- 19.Roossinck, M. J. 2001. Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 2:59-63. [DOI] [PubMed] [Google Scholar]
- 20.Roossinck, M. J., J. Bujarski, S. W. Ding, R. Hajimorad, K. Hanada, S. Scott, and M. Tousignant. 1999. Family Bromoviridae, p. 923-935. In M. H. V. vanRegenmortel, and C. M. Fauquet, and D. H. L. Bishop (ed.), Virus Taxonomy-Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 21.Roossinck, M. J., L. Zhang, and K.-H. Hellwald. 1999. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smirnyagina, E., N.-S. Lin, and P. Ahlquist. 1996. The polymerase-like core of brome mosaic virus 2a protein, lacking a region interacting with viral 1a protien in vitro, maintains activity and 1a selectivity in RNA replication. J. Virol. 70:4729-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster, R. G., W. J. Bean, and O. T. Gorman. 1995. Evolution of influenza viruses, p. 531-544. In A. J. Gibbs, and C. H. Calishe, and F. Garcia-Arenal (ed.), Molecular basis of viral evolution. Cambridge University Press, Cambridge, England.
- 24.White, P. S., F. J. Morales, and M. J. Roossinck. 1995. Interspecific reassortment in the evolution of a cucumovirus. Virology 207:334-337. [DOI] [PubMed] [Google Scholar]
- 25.Xin, H.-W., L.-H. Ji, S. W. Scott, R. H. Symons, and S.-W. Ding. 1998. Ilarviruses encode a cucumovirus-like 2b gene that is absent in other genera within the Bromoviridae. J. Virol. 72:6956-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, L., K. Hanada, and P. Palukaitis. 1994. Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 75:3185-3191. [DOI] [PubMed] [Google Scholar]