Abstract
Oral delivery of toxin-negative derivatives of enterotoxigenic Escherichia coli (ETEC) that express colonization factor antigens (CFA) with deletions of the aroC, ompC, ompF, and toxin genes may be an effective approach to vaccination against ETEC-associated diarrhea. We describe the creation and characterization of an attenuated CFA/I-expressing ETEC vaccine candidate, ACAM2010, from a virulent isolate in which the heat-stable enterotoxin (ST) and CFA/I genes were closely linked and on the same virulence plasmid as the enteroaggregative E. coli heat-stable toxin (EAST1) gene. A new suicide vector (pJCB12) was constructed and used to delete the ST and EAST1 genes and to introduce defined deletion mutations into the aroC, ompC, and ompF chromosomal genes. A phase I trial, consisting of an open-label dose escalation phase in 18 adult outpatient volunteers followed by a placebo-controlled double-blind phase in an additional 31 volunteers, was conducted. The vaccine was administered in two formulations, fresh culture and frozen suspension. These were both well tolerated, with no evidence of significant adverse events related to vaccination. Immunoglobulin A (IgA) and IgG antibody-secreting cells specific for CFA/I were assayed by ELISPOT. Positive responses (greater than twofold increase) were seen in 27 of 37 (73%) subjects who received the highest dose level of vaccine (nominally 5 × 109 CFU). Twenty-nine of these volunteers were secreting culturable vaccine organisms at day 3 following vaccination; five were still positive on day 7, with a single isolation on day 13. This live attenuated bacterial vaccine is safe and immunogenic in healthy adult volunteers.
Enterotoxigenic Escherichia coli (ETEC) causes diarrhea in both humans and animals. Worldwide, it is estimated to cause 210 million cases of human diarrhea per year, leading to the deaths of 380,000 children under the age of 5 years, mostly in developing countries (46). It also causes debilitating illness in adults who lack preexisting immunity and presents a significant problem for travelers and the military operating in regions where it is endemic.
The major virulence factors of ETEC strains include colonization factor antigens (CFA) and toxins (reviewed in reference 25). The CFA are usually fimbriae that promote adherence of the ETEC strain to the epithelium of the small intestine of the host. CFA-mediated adherence is thought to allow secreted toxins to exert their effect on the epithelial cells, inducing fluid and electrolyte secretion, causing diarrhea. There are many known CFA fimbriae, but some studies suggest that up to 70% of strains that cause diarrhea in humans express CFA/I, CFA/II, or CFA/IV (27, 34, 45). This, however, is likely to vary according to location and time (16, 26). In many areas of endemicity CFA/I is one of the most common CFA expressed by ETEC and so represents an important component of any vaccine.
In addition to colonization factors ETEC strains express at least one of two enterotoxins, the heat-stable (ST) and the heat-labile (LT) toxin. ETEC strains that express CFA/I almost always express ST. Derivatives of CFA/I ETEC strains that have lost ST frequently become concurrently CFA/I negative, and the genetic loci for these virulence factors have been found to be linked on a single plasmid for a number of ETEC strains (8, 19, 20, 24, 37, 44). STIh (ST) is encoded by the estA gene and is a monomeric polypeptide of 19 amino acids (25). LT is encoded by the eltAB operon and is heteromeric, consisting of a toxic A subunit (LTA) and a pentamer of receptor-binding B subunits (LTB) similar to cholera toxin (reviewed in reference 29). A significant proportion of ETEC strains also harbor the enteroaggregative heat-stable enterotoxin (EAST1) gene, astA (31, 33, 47).
Protection against ETEC disease has been associated with a humoral immune response to both the CFA and the toxins (4, 5, 10, 39, 40, 41). We have previously described the derivation of an oral live attenuated vaccine strain, PTL003 (42), from the ETEC strain E1392/75-2A. E1392/75-2A is a spontaneous toxin-negative derivative of strain E1392/75 which expresses CFA/II, ST, and LT. Human volunteer studies using E1392/75-2A as an oral live attenuated vaccine indicated that it gave 75% protection against challenge with a virulent ST+ LT+ ETEC strain expressing the same CFA but a different O:H serotype. However, it also caused some mild but unacceptable diarrhea (reviewed in reference 40). To overcome these side effects of strain E1392/75-2A, defined deletion mutations were introduced into the aroC gene required for biosynthesis of aromatic amino acids and the ompC and ompF genes that code for outer membrane porins. The resulting strain, PTL003, was tested in human volunteers and found to be safe and immunogenic (21, 42). Based on this successful approach we are continuing to develop a vaccine that protects against a diverse range of ETEC strains, by raising an immune response to a number of CS antigens (CFA/I and CS1 to CS6), as well as to LTB. In this study we describe the attenuation of a CFA/I ETEC clinical isolate to generate a candidate vaccine strain called ACAM2010. Furthermore, we have demonstrated in a phase I trial in healthy adult volunteers that ACAM2010 is well tolerated and immunogenic and therefore represents a significant milestone in the development of a live attenuated ETEC vaccine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. ETEC strain WS-1858B was a generous gift from S. Savarino (Naval Medical Research Center, Maryland). Suicide vector pDM4 (23) was kindly provided by P. Barrow, Institute for Animal Health, Compton, United Kingdom. Suicide vector plasmids with the replication origin derived from plasmid R6K and their derivatives were maintained in E. coli SY327λpir (22) and then transferred into E. coli SM10λpir (36) for conjugation to ETEC strains. Plasmid pACYC-Tc is a derivative of pACYC184 (3) in which the chloramphenicol resistance determinant was inactivated by deletion of a DNA fragment using restriction endonuclease BsmBI (New England BioLabs, Inc.). Plasmids used as covalently closed circular DNA (cccDNA) molecular size standards were RP4 (60 kb), pColVIk94 (127 kb), and the virulence plasmid from Salmonella enterica serovar Typhimurium strain F98 (∼90 kb); the strains harboring these plasmids were obtained from D. Pickard (Imperial College, London, United Kingdom). Conjugation for the transfer of suicide vector derivatives into ETEC strains was performed as described previously (42) except that transconjugants were selected using chloramphenicol and tetracycline. Bacteria were cultured in LB broth (Sigma) or in LB broth prepared from soy peptone A3 (10 g/liter; Organotechnie), yeast extract (5 g/liter; Sigma), and NaCl (10 g/liter; BDH). Solid medium was LB broth supplemented with 15 g/liter Bacto Agar (Difco). Strains were grown routinely at 37°C. For identification of antibiotic-sensitive derivatives, dilutions of an overnight culture in LB broth were spread onto LB agar and incubated until colonies appeared. These colonies were then replica plated onto LB agar supplemented with appropriate antibiotics, and then all plates were returned to incubation. When maximal expression of CFA/I was required, ETEC strains were grown on CFA agar (10 g/liter Casamino Acids [Difco], 20 g/liter Noble agar [Difco], 1.5 g/liter yeast extract [Sigma], 0.05 g/liter MgSO4 [Sigma], 0.005 g/liter MnCl2 [Sigma]) (9). When required, antibiotics were used at the indicated concentrations: tetracycline (Sigma), 15 μg/ml; streptomycin (Sigma), 20 μg/ml; chloramphenicol (Sigma), 10 μg/ml; nalidixic acid (Sigma), 50 μg/ml. In counterselection against the sacB gene, NaCl was omitted from LB medium and 5% sucrose (Sigma) was added (18).
TABLE 1.
Bacterial strains and plasmids
| Name | Relevant genotype, phenotype, or description | Source/referencea |
|---|---|---|
| Strains | ||
| DH5αλpir | pir Nalr | IAH |
| SY327αλpir | pir Nalr | IAH/22 |
| SM10λpir | pir traRP4 | IAH/36 |
| WS-4437A | O128:H12 CFA/I ST EAST1 LT (wild-type ETEC) | NMRC/this work |
| WS-1858B | O71:H− CFA/I ST EAST1 Apr Tpr Sur (wild-type ETEC) | NMRC/this work |
| A18-34 | O71:H− CFA/I ΔestA EAST1 Apr Tpr Sur | This work |
| A18-34Aps | O71:H− CFA/I ΔestA EAST1 Tpr Sur | This work |
| A18-34ApsTps | O71:H− CFA/I ΔestA EAST1 | This work |
| A18-34ApsTps ΔastA | O71:H− CFA/I ΔestA ΔastA | This work |
| ACAM2010(pSTREP) | O71:H− CFA/I ΔestA ΔastA ΔaroC ΔompC ΔompF Smr | This work |
| E1392/75 | O6:H16 CS1 CS3 ST LT Sur Smr | 40 |
| E1392/75-2A | O6:H16 CS1 CS3 Sur Smr | 40 |
| PTL003 | O6:H16 CS1 CS3 Sur Smr ΔaroC ΔompC ΔompF | 42 |
| Plasmids | ||
| RP4 | 60-kb plasmid used as cccDNA size standard | IC/23 |
| pvirF98 | ∼90-kb virulence plasmid from serovar Typhimurium strain F98 used as cccDNA size standard | IC |
| ColVIk94 | ∼127-kb plasmid used as cccDNA size standard | IC |
| pACYC-Tc | ori15A Tcr | This work |
| pDM4 | oriR6K mobRP4 sacB cat | IAH/31 |
| pDM4A7 | oriR6K mobRP4 cat | This work |
| pDM4A7ΔEco | oriR6K mobRP4 cat | This work |
| pJCB10 | oriR6K mobRP4 cat | This work |
| pJCB12 | oriR6K mobRP4 sacB cat | This work |
| pJCB12-estA | 345-bp fragment, including the whole estA gene, in pJCB12 | This work |
| pJCB12-ΔastA | 286-bp fragment, incorporating a 62-bp defined deletion in astA gene, in pJCB12 | This work |
| pJCB12-ΔaroC345 | 748-bp fragment, incorporating a 345-bp defined deletion in aroC, in pJCB12 | This work |
| pJCB12-ΔompC | 1,016-bp fragment, incorporating a defined deletion of the whole ompC gene, in pJCB12 | This work |
| pJCB12-ΔompF | 943-bp fragment, incorporating a defined deletion of the whole ompF gene, in pJCB12 | This work |
| pJCB12-ori15A | pACYC-Tc replication origin in pJCB12 | This work |
IAH, provided by P. Barrow, Institute for Animal Health, Compton, United Kingdom; NMRC, provided by S. Savarino, Naval Medical Research Center; IC, provided by D. Pickard, Imperial College, London, United Kingdom.
DNA manipulations.
DNA manipulations were performed using standard procedures. Chromosomal and plasmid DNA was prepared, and DNA fragments were isolated from agarose gels using the appropriate kits from QIAGEN (Crawley, United Kingdom). PCR was performed using Taq DNA polymerase (Invitrogen) except during the construction of recombinant DNA molecules, when Pfu Turbo DNA polymerase (Stratagene) was used. Bacterial transformation by electroporation was performed as described previously (7). Southern hybridizations were performed using the ECL random prime labeling and detection system (version II), the ECL direct nucleic acid labeling and detection system, or the alkaline phosphatase direct labeling kit with chemiluminescent detection using the CDP-Star detection reagent (Amersham Pharmacia). Southern hybridization probes were prepared by PCR using plasmid DNA from strain WS-1858B as template and the following oligonucleotides (Table 2): 47100 and 47101 for the estA (ST) gene, 4749 and 4752 for the astA (EAST1) gene, 47104 and 4728 for the cfaABC genes, and 4785 and 4786 for the cfaD gene.
TABLE 2.
Oligonucleotides
| Name | Nucleotide sequence | Target |
|---|---|---|
| 4727 | GCCGCATGCATTAATTCCATATATAGGGG | cfaC |
| 4728 | GCCGTCGACTGCCATAAGGTAAACGAGC | cfaC |
| 4749 | GGCGTCGACGAAAATGAAGGGGCGAAGTTC | astA (EAST1 gene) |
| 4750 | ATGACACGAATGTTGATGGCATCCGGGAAGC | astA (EAST1 gene) |
| 4751 | GCCATCAACATTCGTGTCATGGAAGGACTAC | astA (EAST1 gene) |
| 4752 | GGCGCATGCAAGATTCGGCCAGTTAGCC | astA (EAST1 gene) |
| 4764 | AATATTACTATGCTCTTCGTAGCGG | estA (ST gene) |
| 4765 | ATTAATAGCACCCGGTACAAGCAGG | estA (ST gene) |
| 4785 | TATGGATATATATTCAGAAGAAGAG | cfaD (CFA/I regulator gene) |
| 4786 | AATAAGACGCACTGGAAATTCC | cfaD (CFA/I regulator gene) |
| 47100 | CCGGCATGCGATGCCCTGCAGATGG | estA (ST gene) |
| 47101 | GCCGTCGACTATGCTCTTCGTAGCGGAG | estA (ST gene) |
| 47104 | TTATTGATGGAAGCTCAGGAGG | cfaA |
| 47116 | GCGTCTAGACACAACAATAACGGAGCCGTG | aroC |
| 47117 | GGCGAGCTCGGAATATCAGTCTTCACATCGG | aroC |
| 47118 | CCACGCCTTTCACCCCACCGCCGCGATAATCGC | aroC |
| 47119 | CGCGGCGGTGGGGTGAAAGGCGTGGAAATTGGC | aroC |
| APSrev | GGGAATTCTTAATAGCACCCGGTACAAGCAGGTTTACAACAC | estA (ST gene) |
Construction of suicide vector pJCB12.
A new suicide vector was derived from pDM4 in which more than 50% of the nonfunctional DNA was removed. The resultant plasmid, pJCB12, retains the oriR6K origin, mobRP4 mobilization origin, multiple cloning site, and cat and sacB genes of pDM4 but is only 3.1 kb in size. In our hands this modified vector was found to be inserted into the desired location with much greater efficiency than pDM4.
Construction of pJCB12 derivatives.
The ompC and ompF defined deletion mutations have been described previously (42), and the deletion gene fragments were ligated to pJCB12 for incorporation into WS-1858B. For targeting of the ST gene the pJCB12-estA plasmid derivative was constructed by amplification of the estA (ST) gene with oligonucleotides 4764 and 4765. The fragment was ligated with pJCB12 digested with restriction endonuclease SmaI. The ΔaroC345 and ΔastA defined deletion fragments were generated by overlap extension PCR. For the ΔaroC345 fragment, which incorporates a deletion of 345 bp from the middle of the aroC gene, oligonucleotides 47116, 47117, 47118, and 47119 were used and for ΔastA 4749, 4750, 4751, and 4752 were used (43). The 62-bp astA deletion does not span the whole of the astA gene but leaves intact the first four codons, while the remaining 13 complete codons at the 3′ end of the gene are out of translational reading frame. Fragments were ligated to pJCB12 using appropriate restriction enzyme sites to give derivatives pJCB12-ΔaroC345 and pJCB12-ΔastA. Plasmid pJCB12-ori15A incorporates the replication origin from plasmid pACYC-Tc.
Incorporation of mutations into WS-1858B using the pJCB12-derived targeting vectors.
Plasmid pACYC-Tc was introduced into WS-1858B to provide a tetracycline resistance determinant for selection during manipulation of the strain. Following the transfer of pJCB12 derivatives into WS-1858B, transconjugant colonies were analyzed by PCR to identify those in which recombination had occurred at the correct locus. Appropriate transconjugants were then grown in medium supplemented with 5% sucrose to select for derivatives in which the suicide vector had been lost (18, 30). These derivatives were screened by PCR using appropriate oligonucleotides in order to identify those in which the correct mutation had been incorporated. Upon completion of the genetic manipulations, pACYC-Tc was specifically cured from the strain by introducing pJCB12-ori15A and selecting for chloramphenicol resistance. Tetracycline-sensitive derivatives were identified, and these were then grown on 5% sucrose medium to select derivatives from which pJCB12-ori15A had been lost.
Introduction of streptomycin resistance gene marker to facilitate detection of vaccine strain.
In our previous work with CFA/II ETEC vaccine strain PTL003 (21, 42), we utilized the streptomycin resistance genes carried by this strain on a natural plasmid to allow us to easily isolate and quantitate it in fecal samples. This plasmid, called pSTREP, was isolated from PTL003, purified, and introduced into ACAM2010 by electroporation to create a derivative (strictly called ACAM2010pSTREP but abbreviated to ACAM2010) which was used in the clinical studies described here to facilitate the qualitative and quantitative detection of vaccine organisms in the stool of volunteers.
Antibiotic resistance profiles were determined using Sensidisks from Oxoid.
Preparation of CFA/I.
For demonstration of expression of CFA/I, bacteria were grown overnight by being spread on a CFA agar plate and harvested into 0.5 ml phosphate-buffered saline (PBS), a volume of cell suspension equivalent to 1 × 109 CFU was centrifuged, and the cell pellet was resuspended in 10 μl PBS. This sample was incubated at 65°C for 10 min to release the CFA and centrifuged for 5 min at 15,000 × g, and the supernatant was mixed with an equal volume of Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye (Invitrogen) supplemented with 0.1 M dithiothreitol. CFA/I was visualized by SDS-PAGE on precast 12% polyacrylamide gels (Invitrogen) stained with SimplyBlue SafeStain (Invitrogen).
For use in immunoassays purified CFA/I was prepared from a second strain (WS-4437A, serotype O128:H12) as follows. Bacteria cultured overnight on CFA agar were harvested into PBS and incubated at 65°C for 25 min to release pili (17). The suspension was centrifuged for 15 min at 11,500 rpm in a Sorvall SS34 rotor at 4°C, and the supernatant was harvested and centrifuged at 18,500 rpm in a Sorvall SS34 rotor at 4°C for 2 h to remove membrane debris. The pili were harvested from the supernatant by being pelleted for 2 h at 43,000 rpm in a Beckman 70.1Ti rotor at 4°C. The pellet was resuspended in PBS, CsCl was added to 0.5 g/ml, and the resulting solution was centrifuged at 55,000 rpm for 16 to 20 h in a Beckman 70.1Ti rotor. The resulting CFA/I band was removed and dialyzed against PBS.
Toxin assays.
Three different toxin assays were employed, namely, enzyme-linked immunosorbent assay (ELISA) (GM1 binding for LT [38] and a commercial kit for ST [Oxoid]), Y1 adrenal cell toxicity assays (6), and transepithelial electrical resistance (TEER) of CaCo-2 cells, which can detect both ST and LT. CaCo-2 cells were grown on collagen-coated transwells (12-mm diameter, 0.4-μm mesh) for about 18 days. Bacterial supernatants or purified toxins as positive controls were added to the medium inside the transwell, and the change in TEER was monitored using “chopstick” electrodes (WPI, Stevenage, United Kingdom).
Preparation of vaccines for administration to volunteers.
The vaccine lot was produced by a standardized, aseptic, fully enclosed process in which cells from a characterized research master cell bank were expanded in a 2-liter fermentor, washed free from growth media, concentrated, and finally suspended in PBS-20% glycerol at >1010 CFU/ml, and used to fill a large number of cryovials for storage at −80°C. Representative vials were tested for potency, microbiological purity, and absence of bacteriophage with full QC and QA audit by Bioreliance (Glasgow, United Kingdom). The vaccine strain was administered to half of the volunteers by thawing these vials, diluting the strain in PBS to provide the desired dose of organisms in 1 ml of suspension, adding 1 ml to 200 ml of CeraVacx buffer (Cera Products Inc., Jessup, MD; rice solids, 7.0 g; sodium bicarbonate, 2 g; trisodium citrate, 0.5 g, in 200 ml of water), and having them drink it (“frozen vaccine”). To compare this vaccine with studies done previously (21, 42), the remaining volunteers were given “fresh vaccine” in which a freshly prepared lawn of bacteria was grown by inoculating CFA agar plates with 100 μl of the thawed frozen suspension and incubating the plates at 35 to 37°C for 18 h. The lawn was harvested in sterile PBS; based on optical density at 600 nm, the concentration of bacterial cells was adjusted to give the appropriate number of CFU per ml, and then 1 ml was diluted into CeraVacx and administered as doses as above.
Immunization of volunteers and assessment of vaccine safety and immunogenicity.
The study was conducted in two phases. Phase A consisted of an open-label dose escalation in which groups of six subjects, aged 18 to 50 years, received a single dose of vaccine at 5 × 107 CFU, 5 × 108 CFU, and 5 × 109 CFU, three receiving fresh and three receiving frozen vaccine. Phase B was conducted as a two-dose, placebo-controlled, double-blind protocol in 31 subjects who were randomized to one of six groups to receive vaccine or placebo on days 0 and 10. Fifteen subjects were randomized to receive two doses of vaccine (fresh/fresh or frozen/frozen, n = 7 and 8, respectively), and 16 were randomized to receive one dose of vaccine (fresh/placebo, frozen/placebo, placebo/fresh, or placebo/frozen; n = 4 per group). Subjects fasted for 90 min before and after dosing. Volunteers were immunized by drinking 200 ml of CeraVacx containing 1 ml of the appropriate vaccine suspension. Placebo recipients received 200 ml of CeraVacx buffer alone. For 7 days after each dose, subjects recorded symptoms on diary cards. Peripheral blood mononuclear cells (PBMC) were obtained on days 0, 7, and 10 in phase A and days 0, 7, 10, 17, and 20 in phase B to assess local and systemic immune responses. Fecal samples were obtained for culture on days 0, 3, 7, 10, and 13 in phase A and on days 0, 3, 7, 10, 13, 17, and 20 in phase B.
Isolation of vaccine strain from stools.
Samples were plated onto MacConkey agar and on MacConkey agar containing streptomycin (25 μg/ml). Ten colonies were taken from the streptomycin-containing plates and plated onto Davies minimal medium, which lacks aromatic metabolites, and onto Luria agar (42). Colonies growing on Luria agar but not on minimal agar were confirmed to be the vaccine strain by specific PCR. The detection limit for vaccine strains is estimated to be 30 CFU/g stool. Three days following each vaccination stool samples were analyzed quantitatively to estimate the levels of vaccine shedding. Weighed stool samples (approximately 1 g) were suspended thoroughly in PBS and diluted serially in the same medium. One-hundred-microliter aliquots of appropriate dilutions were plated onto MacConkey agar plus streptomycin as above. Colonies which looked like ACAM2010 were counted on an appropriate dilution plate, and 10 were plated onto minimal and Luria agars as above to confirm their phenotype. The level of vaccine organisms was then calculated as (number counted × dilution factor × proportion aro dependent/weight of stool sample).
Detection of anti-CFA/I ASCs.
PBMCs were isolated from peripheral blood samples by Ficoll-Hypaque gradient centrifugation, and anti-CFA/I-specific immunoglobulin A (IgA) and IgG antibody-secreting cells (ASCs) were determined using the enzyme-linked immunospot assay (ELISPOT) as described previously (1, 11). A positive ASC response was defined as a ≥2-fold increase over the baseline (day 0) value when the baseline value was ≥0.5/106 PBMC. If the baseline value was <0.5/106 PBMC, a response was defined as ≥2 spots/106 PBMC.
RESULTS
The CFA/I ETEC strain WS-1858B harbors the genes coding for ST and the EAST1 toxins and antibiotic resistance determinants.
ETEC strain WS-1858B (Table 1) was isolated in Cairo, Egypt, from a child with diarrhea and was characterized previously as serotype O71:H negative, CFA/I and STIh positive, but LT negative (S. Savarino, personal communication). SDS-PAGE analysis confirmed that the strain expressed CFA/I. Toxin ELISAs and Y1 adrenal cell toxicity assays showed that it expressed ST but not LT (data not shown), as confirmed by TEER analysis (Fig. 1). Results of PCRs using suitable primers (Table 2) were consistent with WS-1858B being ST positive and LT negative and, in addition, indicated that it harbored the astA gene coding for the EAST1 toxin (data not shown).
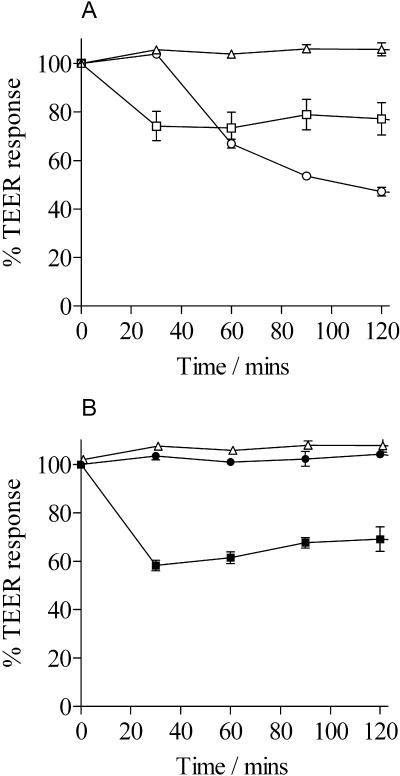
FIG. 1.
TEER responses of CaCo-2 cells to ST, LT, and culture supernatants. Electrical resistance is measured in ohms and plotted as a percentage of the level at time zero. (A) TEER responses to ST and LT. Open triangles, LB broth; open squares, LB broth supplemented with ST at 100 ng/ml; open circles, LB broth supplemented with LT at 100 ng/ml. (B) TEER responses to WS-1858B and ACAM2010. Open triangles, LB broth; filled squares, WS-1858B culture supernatants; filled circles, ACAM2010 culture supernatants. Each experiment was performed in triplicate, and the mean values were plotted, with error bars equal to the standard deviation.
For release into the environment a live attenuated bacterium must carry no antibiotic resistance determinants which may spread to other bacterial species. Therefore, the antibiotic resistance profile of WS-1858B was determined using disk diffusion. The strain was sensitive to nalidixic acid, kanamycin, chloramphenicol, tetracycline, streptomycin, furazolidone, and ciprofloxacin but resistant to ampicillin, trimethoprim, and sulfamethoxazole.
The gene coding for ST and those coding for CFA/I are 1.9 kb apart and divergently transcribed in WS-1858B.
CFA/I ETEC strains frequently express ST, and closer analysis of some strains has confirmed that the respective genes are located on a high-molecular-weight plasmid (20, 27, 44). Moreover, loss of the ST gene has usually been associated with loss of CFA/I expression (8, 19, 24, 37). Southern hybridization demonstrated that the estA (Fig. 2) and astA (data not shown) genes in WS-1858B are carried on a large plasmid of ca. 100 kb. In order to characterize the linkage of the ST and CFA/I loci, plasmid DNA from strain WS-1858B was digested with a number of restriction enzymes followed by Southern hybridization, which revealed that both loci were on an 8-kb SphI fragment (not shown). Furthermore, PCR analysis using oligonucleotides APSrev and 4728 indicated that the ST gene is ∼2 kb upstream of the cfaA open reading frame and that the ST and CFA/I genes are divergently transcribed. The nucleotide sequence of this ∼2-kb intergenic region was determined for strain WS-1858B (EMBL database accession number AJ868113) and confirmed that 1,876 bp separate the estA and cfaA open reading frames. Within this region were 794 nucleotides that were 94% identical to part of the 1.3-kb IS629 insertion sequence from Shigella sonnei.
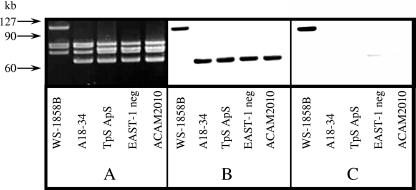
FIG. 2.
Southern hybridization analysis of covalently closed circular plasmid DNA from ETEC strain WS-1858B and its derivatives. A. Plasmid DNA was electrophoresed through 0.6% agarose and stained with SYBR-Gold (Cambridge Bioscience). The position of large-molecular-size cccDNA standards is indicated on the left of the figure, and derived strains are indicated below each lane. B and C. Southern hybridization results obtained using cfa (B) and estA (C) gene-specific probes are shown. Results obtained using the EAST1 gene probe gave results identical to those obtained with estA. Each strain possessed small-molecular-size plasmids of about 5.1 and 4.2 kb, but only that portion of the gel which includes the large-molecular-size plasmids is shown.
Removal of toxin genes and antibiotic resistance determinants from WS-1858B.
Toxin-negative strains of WS-1858B were generated using pJCB12. After each stage the derivatives were tested by PCR using appropriate oligonucleotides (Table 2) to identify those that continued to harbor the cfa genes and then by SDS-PAGE to confirm that they remained capable of expressing CFA/I at levels similar to WS-1858B. Removal of the ST toxin gene was achieved using the suicide vector derivative pJCB12-estA, which incorporated a fragment of the ST gene. Once recombinants of strain WS-1858B with pJCB12 at the estA gene had been identified by PCR, sucrose selection allowed identification of derivatives from which the suicide vector had been lost. Screening of these sucrose-resistant derivatives by PCR using oligonucleotides specific for estA identified 19 potentially toxin-negative derivatives. One of these ST-negative derivatives, called A18-34, was found to retain wild-type levels of CFA/I expression (data not shown) and was used for further manipulation. Southern hybridization demonstrated that the loss of estA had occurred via the deletion of a large fragment from the 100-kb plasmid that had also, by chance, removed the astA gene (Fig. 2).
To isolate a derivative of A18-34 sensitive to ampicillin, trimethoprim, and sulfamethoxazole, approximately 20,000 colonies of A18-34 were sequentially replica plated onto LB plates supplemented with these antibiotics. An ampicillin-sensitive derivative was first identified. By subsequent screening with trimethoprim, a derivative sensitive to this and to sulfamethoxazole was found, suggesting that the genes for these latter two phenotypes are linked. When approximately109 CFU of the antibiotic-sensitive derivative was plated separately onto LB supplemented with ampicillin, trimethoprim, or sulfamethoxazole, no colonies were obtained, confirming that reversion to resistance was unlikely. This derivative continued to express CFA/I and was called A18-34ApsTps.
Plasmid DNA from A18-34ApsTps no longer hybridized to ST- or EAST1-specific probes on Southern blots; however, PCR analysis revealed that it continued to harbor the EAST1 gene, astA, presumably in a chromosomal location. This was again targeted for deletion using pJCB12-ΔastA. The sucrose-resistant derivatives were completely negative by PCR analysis using primers flanking the expected deletion site, indicating that the entire EAST1 gene locus had been lost through undefined deletion events. The toxin-negative, antibiotic-sensitive derivative identified was finally tested to confirm its continued ability to express CFA/I and was called A18-34ApsTpsΔastA.
Introduction of deletion mutations into aroC, ompC, and ompF, to generate vaccine candidate strain ACAM2010.
The deletions previously demonstrated to attenuate PTL003 (42) were introduced into these genes in strain A18-34ApsTpsΔastA using pJCB12. The deletions were confirmed by PCR of the relevant loci and nucleotide sequencing of the PCR products obtained. Southern hybridization analysis performed as previously (42) also confirmed that these three loci were deleted (not shown). Following the introduction of these mutations, the expression of CFA/I and the lipopolysaccharide was analyzed and shown to be indistinguishable from that seen in the ancestral wild-type strain (not shown). The aroC, ompC, and ompF mutant was called ACAM2010.
Confirmation of the loss of ST activity from ACAM2010 using TEER.
Following its isolation, strain ACAM2010 was confirmed to no longer express ST by using an inhibition ELISA, while the parent strain had already been determined to lack LT as described above. The toxin status of the final strain was confirmed by measuring TEER across CaCo-2 cell monolayers with a “chopstick” electrode. This assay can detect and differentiate between the ST and LT toxins. Following the addition of LB broth supplemented with ST at 100 ng/ml, the TEER of the CaCo-2 cell monolayer showed an immediate sharp decrease over a period of about 0.5 h by 20 to 30%. Over the next 1.5 h electrical resistance recovered slightly. When LB broth supplemented with 100 ng/ml LT was added, the TEER decreased after a delay of about 0.5 h and continued to do so over the following 1.5 h (Fig. 1A). When a culture supernatant from strain WS-1858B was used, a TEER response typical for ST was observed. In contrast, a culture supernatant from ACAM2010 gave no TEER response, confirming that no ST or LT was present in the culture supernatant (Fig. 1B).
Analysis of plasmids in ETEC strain WS-1858B and its derivatives.
In order to define more fully the nature of the toxin and antibiotic resistance gene deletions that occurred during the construction of ACAM2010, plasmid DNA from this strain, its wild-type parent, and selected intermediate strains was examined by agarose gel electrophoresis and Southern hybridization (Fig. 2). Each strain had two plasmids of 4 to 5 kb and at least four large-molecular-size (60- to 100-kb) plasmids (Fig. 2A). Probes specific for the cfa operon (Fig. 2B) and cfaD (data not shown) both hybridized with a ∼100-kb plasmid in WS-1858B. Deletion of the toxin genes resulted in the ∼100-kb plasmid being replaced by one of ∼70 kb to which the cfa and cfaD probes hybridized. Probes for estA (Fig. 2C) and astA (not shown) hybridized only to the ∼100-kb plasmid in WS-1858B and not the ∼70-kb plasmid which replaced it in A18-34 and subsequent derivatives. These results indicate that the 100-kb plasmid in WS-1858B codes for the major ETEC virulence factors and that loss of ST resulted from the deletion of ∼30 kb from this plasmid which was associated with the loss of a copy of the astA gene also.
Although the astA gene probe did not hybridize to plasmid DNA from strain A18-34 or its antibiotic-sensitive derivatives, these strains remained positive for the astA gene by PCR, suggesting that a second copy of this EAST1 toxin gene was present on the WS-1858B chromosome. This second copy of astA was subsequently deleted using pJBC12-ΔastA (see above) as confirmed by Southern hybridization analysis. There was no observable difference in the plasmid profiles of the strains prior to and after removal of this second copy of astA (Fig. 2A).
Loss of the antibiotic resistance determinants also coincided with a small increase in the mobility of another high-molecular-weight plasmid (Fig. 2A), suggesting that loss of antibiotic resistance was due to deletion of part of a, rather than a whole, plasmid; loss of all three resistance determinants coincided with visible changes to only one plasmid.
Immunization of volunteers in phase A: dose escalation study.
Vaccines were successfully administered to all 18 subjects, proceeding to the higher dose levels after confirmation of the safety of the previous level. No serious adverse events (AEs) were noted, and the treatment-related AEs were mostly gastrointestinal (GI) symptoms as usually observed following administration of such vaccines in buffered suspension. All were mild with the exception of two moderate incidents of abdominal pain and one of constipation. In the absence of limiting toxicity the maximum tolerable dose was considered to be at least 5 × 109 CFU, so this dose was selected for the double-blind phase. There was no correlation between dose level and frequency or severity of AEs.
Immunization of volunteers in phase B: double-blind randomization and administration of vaccine.
Thirty-one subjects were randomly assigned to one of six groups to receive vaccine (fresh, frozen, or placebo) on days 0 and 10. All volunteers received both doses of vaccine or placebo. The 15 subjects who received one or two doses of fresh ACAM2010 had a mean age of 25.6 years; 11 were female. The 16 recipients of frozen ACAM2010 had a mean age of 26.8 years; 12 were female.
Vaccine strain ACAM2010 was well tolerated in either formulation.
In phase A, no subject experienced diarrhea, but one recipient of 5 × 108 CFU of frozen vaccine vomited. Two subjects experienced headache after receiving frozen vaccine (at 5 × 108 and 5 × 109 CFU). In phase B the rates of symptoms during the 7 days following ingestion of fresh and frozen ACAM2010 were not significantly different from those following ingestion of placebo (Table 3). There was a higher frequency of AEs in recipients of fresh vaccine than in recipients of either frozen vaccine or placebo. The incidence of abdominal pain and combined GI symptoms (abdominal pain, diarrhea, and vomiting) was significantly higher in recipients of fresh than in those of frozen vaccine, although not significantly different from placebo due to the lower number in this group.
TABLE 3.
Incidence of gastrointestinal and general symptoms reported by subjects during the 7 days following ingestion of fresh or frozen ACAM2010 or placebo on days 0 and 10a in phase B
| Symptom | No. of subjects (%)
|
P (fresh/placebo; frozen/placebo; fresh/frozen)e | ||
|---|---|---|---|---|
| Fresh (n = 22)b | Frozen (n = 24)c | Placebo (n = 16)d | ||
| Loose stools | 6 (27) | 6 (25) | 6 (38) | 0.37; 0.30; 0.56 |
| Diarrheaf | 3 (14)g | 0 (0) | 1 (6) | 0.43; 0.40; 0.10 |
| Vomiting | 2 (9)g | 1 (4) | 1 (6)g | 0.62; 0.65; 0.47 |
| Abdominal pain | 7 (32)g | 2 (8) | 2 (13) | 0.16; 0.53; 0.05 |
| Any of three GI symptomsh | 12 (55) | 3 (13) | 4 (25) | 0.06; 0.27; 0.01 |
| Fever (temp >38°C) | 0 (0) | 0 (0) | 0 (0) | 1.0; 1.0; 1.0 |
| Headache | 13 (59)g | 8 (33) | 7 (44)g | 0.27; 0.37; 0.07 |
Volunteers were orally administered ACAM2010 on day 0 and/or day 10 in 200 ml of CeraVacx buffer. Placebo was CeraVacx buffer alone.
Comprises eight volunteers receiving a single dose and seven receiving two doses of vaccine.
Comprises eight volunteers receiving a single dose and eight receiving two doses of vaccine.
Comprises four groups of four receiving placebo either before or after fresh or frozen vaccine.
By Fisher's exact test. Vaccine formulations were compared to the placebo group and each other.
Diarrhea was defined as ≥3 unformed stools in a 24-h period.
All symptoms were rated as mild by the volunteers except one diarrhea following placebo, one vomiting following fresh vaccine, one vomiting following placebo, four headaches following fresh vaccine, one headache following placebo, and one abdominal pain following fresh vaccine, all of which were rated moderate.
Sum of incidences of diarrhea, vomiting, and abdominal pain.
Fecal shedding of vaccine strain ACAM2010 was short-lived and equivalent following ingestion of either formulation.
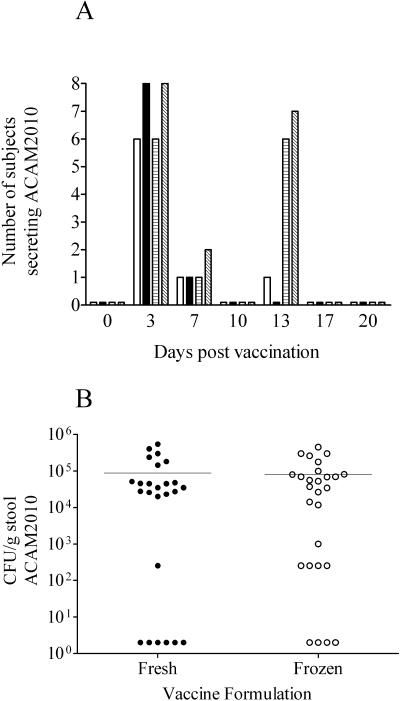
Fecal samples collected from volunteers following vaccination were analyzed for the presence of ACAM2010; the frequency with which vaccine organisms were isolated from the stool of subjects receiving either one or two doses at the highest dose level is shown in Fig. 3A. ACAM2010 was recovered from the stool of 13/18 volunteers at least once following dosing with fresh vaccine and from the stool of 16/19 volunteers at least once following dosing with frozen vaccine. The majority of subjects were shedding on day 3 following any dose, with only a few still positive on day 7. The longest documented shedding was in asingle volunteer who received fresh vaccine on day 0 and placebo on day 10 and who gave a positive stool culture on day 13. Antibiotics were not required to abolish shedding in any volunteers before the end of the study period.
FIG. 3.
Shedding of vaccine strains following ingestion of one or two doses of fresh or frozen ACAM2010. Stool samples obtained from subjects were cultured for the presence of ACAM2010. Graph A shows the number of subjects at each time point following vaccination from which the vaccine strain was recovered: open bars, one dose of fresh vaccine; filled bars, one dose of frozen vaccine; hatched bars, two doses of fresh vaccine; diagonally hatched bars, two doses of frozen vaccine. Graph B shows the levels of shedding in CFU per gram of stool estimated from samples taken 3 days following ingestion of a first or second dose of ACAM2010. Negative stool samples are plotted as 2 CFU. Horizontal bars indicate the mean response in each group.
Quantitative estimates were made of the level of shedding in the stool of volunteers 3 days following receipt of vaccine (Fig. 3B). The majority of positive samples contained between 104 and 106 CFU of ACAM2010 per gram, and the total estimated excretion per volunteer is estimated to be less than the dose administered (5 × 109 CFU).
ACAM2010 induces systemic and mucosal anti-CFA/I immune responses.
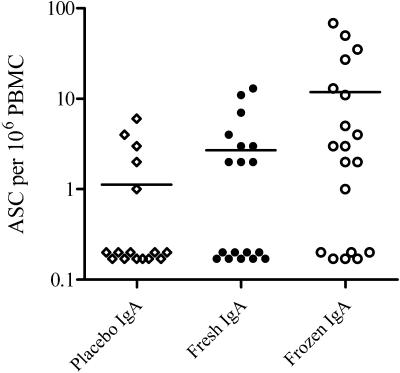
In keeping with other authors in evaluating ASC responses following oral vaccination and the definitions in the clinical protocol, a positive response is defined as a twofold or higher increase in the number of spots observed on day 7 or day 10 compared to the prevaccination level on day 0 or else a value of ≥2 spots per 106 PBMC in those volunteers whose baseline value was zero. The majority of volunteers had significant ELISPOT responses to oral vaccine irrespective of the method of vaccine preparation (Table 4). Altogether, 27 of 37 (73%) recipients of vaccine showed an ELISPOT response compared to 7 of 16 (44%) recipients of placebo (P = 0.04). Although the proportion of recipients of placebo developing an ELISPOT response looks high, these responses were small, with mean ASC counts below 2 per million PBMCs. Responses were quantitatively higher in recipients of vaccine (Table 4). The mean number of IgA plus IgG ASCs generated by vaccination with frozen ACAM2010 is significantly greater than that generated by vaccination with fresh ACAM2010 (t test, P < 0.02), although the frequencies of positive responses are not significantly different (Table 4 and Fig. 4). There was a low level of background responses to CFA/I measured in prevaccination samples and also seemingly induced following placebo dosing in some volunteers, although the frequency and magnitude were lower than those in vaccinees. The difference in mean response (for both IgA and IgG) in recipients of frozen vaccine from controls was highly significant (P = 0.01, t test with Welch's correction), whereas the corresponding differences for the fresh vaccine were not significantly different from controls (P = 0.07, 0.09).
TABLE 4.
ASC responses to CFA/I measured following ingestion of fresh or frozen ETEC vaccine strain ACAM2010 or placeboa
| Isotype of response | Vaccine administered | No. of respondersb | Mean ASC/106 PBMCc | Pd |
|---|---|---|---|---|
| IgA | Fresh | 8/18 (44) | 2.6 | 0.33 |
| Frozen | 13/19 (68) | 11.8 | 0.03 | |
| Placebo | 5/16 (31) | 1.0 | NA | |
| IgG | Fresh | 10/18 (56) | 9.0 | 0.24 |
| Frozen | 13/19 (68) | 9.2 | 0.07 | |
| Placebo | 6/16 (38) | 0.8 | NA | |
| IgA or IgG | Fresh | 12/18 (67) | 11.6 | 0.16 |
| Frozen | 15/19 (79) | 21.1 | 0.04 | |
| Placebo | 7/16 (44) | 1.8 | NA |
A positive response in the ASC assay is a ≥2-fold increase over baseline and ≥2 spots/106 PBMC, measured at day 7 following ingestion of a first dose of vaccine or placebo.
Responders/total number of subjects in group (percent); vaccine groups include recipients of highest dose in phase A; the placebo group comprises recipients of placebo before or after a single dose of either vaccine formulation in phase B. (Two additional ACAM2010 recipients in phase B who received a second dose, one fresh and one frozen, responded positively to the second but not the first dose.)
Peak response at day 7 or 10 following either dose of vaccine, calculated as frequency of ASC at peak − frequency of ASC on day of vaccination.
Fisher's exact test comparing the number of responders to each formulation of vaccine with the number of responders to placebo. NA, not applicable.
FIG. 4.
IgA CFA/I-specific ASC responses in recipients of one or two doses of ACAM2010. Shown is a dot plot showing the maximal response seen at day 7 or day 10 in individual subjects following ingestion of one or two doses of ACAM2010 at the highest dose level (third cohort in phase A and all subjects in phase B). Responses are plotted after subtraction of the response on the day of the appropriate vaccination; zero or negative responses are plotted as 0.2. Horizontal bars indicate the mean response in each group.
DISCUSSION
A number of different strategies are being employed to develop a vaccine against ETEC infections including oral killed whole-cell preparations (14, 15, 28, 32) and subunit vaccines (12, 17, 48). In addition, live attenuated vaccines against ETEC that are in development include those based on Shigella (1, 2) or Salmonella (11, 13) strains expressing CFA or LTB and attenuated ETEC derivatives (21, 42). Use of orally delivered live attenuated vaccines may be more advantageous for enteric diseases than killed or subunit vaccines as the antigens are delivered to the intestinal mucosal surface where immunity is required. Moreover, while oral killed or subunit vaccines present antigen only during their brief passage through the alimentary tract, where they are prone to digestive degradation, the aim of live attenuated vaccines is to achieve transient colonization of the host. This colonization is expected to result in expression of relevant antigens in situ, achieving a more appropriate presentation of antigens for a prolonged period to the immune system. A live attenuated vaccine based on a number of diverse natural ETEC strains may have the added advantage of presenting ETEC antigens in addition to CFA, which may add to the protection elicited.
Immune responses to the CFA are important for protection against ETEC infection (5, 10, 39, 40, 41). Since different ETEC strains express a number of CFA, to protect against the majority of ETEC strains a vaccine will need to consist of several strains expressing the most common CFA. It was demonstrated previously that vaccine candidate PTL003, a toxin-negative derivative of ETEC strain E1392/75 with mutations introduced into the aroC, ompC, and ompF chromosomal genes and which expresses CFA/II, was well tolerated and immunogenic when administered to volunteers (21, 42).
One of the more common CFA is CFA/I (35, 45), which is expressed by ETEC strain WS-1858B. Using the same basis of attenuation as for PTL003, this strain was rendered toxin negative, and aroC, ompC, and ompF deletion mutations were introduced into the chromosome. Antibiotic-sensitive derivatives were identified, and the resulting strain, ACAM2010, now represents an important component of a live attenuated ETEC vaccine.
Southern hybridization analysis performed on the parent strain of ACAM2010, WS-1858B, revealed that the genes associated with virulence (CFA/I, ST, and EAST1) are all carried by the same plasmid. Likewise, loss of the antibiotic resistance determinants coincided with a visible change in mobility to only one other plasmid, suggesting that these determinants are encoded by a resistance plasmid. In WS-1858B the region between the ST and cfaA open reading frames is only 1,876 bp. This intragenic region includes nucleotide sequences that are 94% identical to half of IS629 from Shigella sonnei. For two other CFA/I- and ST-expressing ETEC strains, WS-4437A and WS-6117A (S. Savarino), the nucleotide sequence between the ST and cfaA open reading frames was almost identical to that in strain WS-1858B (A. K. Turner, unpublished data). These three strains express different O:H serotypes (O128:H12; O153:H45, and O71:H−, respectively) and so are not clonally related. This suggests that the CFA/I and ST loci initially became tightly linked by transposition and have been subsequently transferred together to a variety of different strains. Linkage of ETEC virulence factors is of clear evolutionary significance as it facilitates this simultaneous transfer. Spontaneous loss of toxin expression has been observed previously in ETEC strains (37), and the pJCB12 approach used here was designed to permit selection for and easy isolation of the desired derivative. The estA gene fragment in pJCB12-estA did not incorporate a deletion but rather was used simply to insert a counterselectable marker into the locus. The loss of estA in the ST-negative derivatives occurred by spontaneous recombination resulting in deletion of approximately 30 kb from the plasmid. Similarly, for the second copy of the astA gene, presumed to be on the chromosome, the deletion construct was not incorporated, but rather the whole astA gene and proximal flanking sequences were lost. The results for both estA and astA in WS-1858B are consistent with the hypothesis that these enterotoxins are encoded by unstable genetic loci that are prone to spontaneous recombination.
Previous phase I trials of live attenuated ETEC vaccine candidates (21, 42) have been carried out using vaccines formulated from freshly grown overnight cultures on solid medium, which maximizes expression of the CFA. While this provides an acceptable procedure for initial clinical proof-of-concept studies, it clearly bears little or no relation to a process by which a final vaccine product will be manufactured. An aim of the study with ACAM2010 was to compare this “fresh vaccine” approach with an alternative formulation which represents a step towards a scalable manufacturing process. Accordingly half of the subjects in this study received vaccine which had been produced by culturing ACAM2010 in a fermentor; washing and concentrating it and filling cryovials with it in a closed aseptic manner; and storing it at −80°C until use. Under these conditions no effort is made to maximize expression of CFA in vitro, relying on their expression by the live vaccine strains following their ingestion. The phase I trial described here thus represents the first administration to human volunteers of a new candidate ETEC vaccine strain expressing CFA/I as well as addressing the issue of formulation to move towards a process which can be carried out on a commercial scale.
ACAM2010 has been tested in a phase I clinical trial, with a total of 37 volunteers receiving nominal doses of 5 × 109 CFU, 19 doses prepared directly from a frozen suspension and 18 from a fresh overnight culture. Both formulations were generally well tolerated with no AEs occurring at frequencies significantly higher in vaccinees than in placebo recipients. Lower numbers of AEs were observed in recipients of the frozen than in those of the fresh formulation, although this difference was significant only for abdominal pain and the three aggregated GI symptoms (Table 3). This is an encouraging result because as product development proceeds and vaccine is manufactured by large-scale fermentation for later-stage clinical trials it will be more similar to the frozen formulation than to freshly cultured organisms. The data presented here suggest that we can expect the safety and tolerability of larger-scale lots of vaccine to be similarly good. Equally encouraging is the observation that, although the frequencies of response were not significantly different, the magnitude of the immune responses to the CFA/I antigen following vaccination were significantly higher in recipients of the frozen formulation than in those ingesting the freshly grown vaccine.
These studies with ACAM2010 therefore represent an important milestone in the development of a broadly protective ETEC vaccine, providing a safe and immunogenic CFA/I-expressing strain and demonstrating that it is not necessary to maximize expression of CFA/I during growth of live vaccine strains in order to ensure optimal immunogenicity, the frozen vaccine being at least as immunogenic as vaccine freshly grown on CFA agar. The lower reactogenicity and higher immunogenicity of the frozen formulation, together with the greater ease of dose preparation and improved level of characterization possible, make this the protocol of choice for future clinical studies prior to full-scale cGMP manufacture of the final product.
Acknowledgments
We are grateful to the United States Naval Medical Research Center, Silver Spring, MD, and Unit 3, Cairo, Egypt, for providing us with ETEC strains.
This work was funded in part by a grant from the Dual Use Science and Technology program from the U.S. Army Medical Research and Materiel Command, Fort Detrick, Maryland.
Editor: A. D. O'Brien
REFERENCES
- 1.Altboum, Z., M. M. Levine, J. E. Galen, and E. M. Barry. 2003. Genetic characterization and immunogenicity of coli surface antigen 4 from enterotoxigenic Escherichia coli when it is expressed in a Shigella live-vector strain. Infect. Immun. 71:1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, E. M., Z. Altboum, G. Losonsky, and M. M. Levine. 2003. Immune responses elicited against multiple enterotoxigenic Escherichia coli fimbriae and mutant LT expressed in attenuated Shigella vaccine strains. Vaccine 21:333-340. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemens, J. D., D. A. Sack, J. R. Harris, J. Chakraborty, P. K. Neogy, B. Stanton, N. Huda, M. U. Khan, B. A. Kay, M. R. Khan, et al. 1988. Cross-protection by B subunit-whole cell cholera vaccine against diarrhoea associated with heat-labile toxin-producing enterotoxigenic Escherichia coli: results of a large-scale field trial. J. Infect. Dis. 158:372-377. [DOI] [PubMed] [Google Scholar]
- 5.Cravioto, A., R. E. Reyes, F. Trujillo, F. Uribe, A. Navarro, J. M. De La Roca, J. M. Hernandez, G. Perez, and V. Vazquez. 1990. Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens. Am. J. Epidemiol. 131:886-904. [DOI] [PubMed] [Google Scholar]
- 6.Donta, S. T., H. W. Moon, and S. C. Whipp. 1974. Detection of heat-labile Escherichia coli enterotoxin with the use of adrenal cells in tissue culture. Science 183:334-336. [DOI] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echeverria, P., J. Seriwatana, D. N. Taylor, S. Changchawalit, C. J. Smyth, J. Twohig, and B. Rowe. 1986. Plasmids coding for colonization factor antigens I and II, heat-labile enterotoxin, and heat-stable enterotoxin A2 in Escherichia coli. Infect. Immun. 51:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. G., D. J. Evans, Jr., S. Clegg, and J. A. Pauley. 1979. Purification and characterization of the CFA/I antigen of enterotoxigenic Escherichia coli. Infect. Immun. 25:738-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman, D. J., C. O. Tacket, A. Delehanty, D. R. Maneval, J. Nataro, and J. H. Crabb. 1998. Milk immunoglobulin with specific activity against purified colonization factor antigens can protect against oral challenge with enterotoxigenic Escherichia coli. J. Infect. Dis. 177:662-667. [DOI] [PubMed] [Google Scholar]
- 11.Giron, J. A., J. G. Xu, C. R. Gonzalez, D. Hone, J. B. Kaper, and M. M. Levine. 1995. Simultaneous expression of CFA/I and CS3 colonization factor antigens of enterotoxigenic Escherichia coli by ΔaroC, ΔaroD Salmonella typhi vaccine strain CVD 908. Vaccine 13:939-946. [DOI] [PubMed] [Google Scholar]
- 12.Guerena-Burgueno, F., E. R. Hall, D. N. Taylor, F. J. Cassels, D. A. Scott, M. K. Wolf, Z. J. Roberts, G. V. Nesterova, C. R. Alving, and G. M. Glenn. 2002. Safety and immunogenicity of a prototype enterotoxigenic Escherichia coli vaccine administered transcutaneously. Infect. Immun. 70:1874-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillobel, H. C., J. I. Carinhanha, L. Cardenas, J. D. Clements, D. F. de Almeida, and L. C. Ferreira. 2000. Adjuvant activity of a nontoxic mutant of Escherichia coli heat-labile enterotoxin on systemic and mucosal immune responses elicited against a heterologous antigen carried by a live Salmonella enterica serovar Typhimurium vaccine strain. Infect. Immun. 68:4349-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, E. R., T. F. Wierzba, C. Ahren, M. R. Rao, S. Bassily, W. Francis, F. Y. Girgis, M. Safwat, Y. J. Lee, A. M. Svennerholm, J. D. Clemens, and S. J. Savarino. 2001. Induction of systemic antifimbria and antitoxin antibody responses in Egyptian children and adults by an oral, killed enterotoxigenic Escherichia coli plus cholera toxin B subunit vaccine. Infect. Immun. 69:2853-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jertborn, M., C. Ahren, and A. M. Svennerholm. 2001. Dose-dependent circulating immunoglobulin A antibody-secreting cell and serum antibody responses in Swedish volunteers to an oral inactivated enterotoxigenic Escherichia coli vaccine. Clin. Diagn. Lab. Immunol. 8:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Z. D., B. Lowe, M. P. Verenkar, D. Ashley, R. Steffen, N. Tornieporth, F. von Sonnenburg, P. Waiyaki, and H. L. DuPont. 2002. Prevalence of enteric pathogens among international travelers with diarrhoea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay). J. Infect. Dis. 185:497-502. (First published online 22 January 2002.) [DOI] [PubMed] [Google Scholar]
- 17.Katz, D. E., A. J. DeLorimier, M. K. Wolf, E. R. Hall, F. J. Cassels, J. E. van Hamont, R. L. Newcomer, M. A. Davachi, D. N. Taylor, and C. E. McQueen. 2003. Oral immunization of adult volunteers with microencapsulated enterotoxigenic Escherichia coli (ETEC) CS6 antigen. Vaccine 21:341-346. [DOI] [PubMed] [Google Scholar]
- 18.Lawes, M., and S. Maloy. 1995. MudSacI, a transposon with strong selectable and counterselectable markers: use for rapid mapping of chromosomal mutations in Salmonella typhimurium. J. Bacteriol. 177:1383-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, M. M., P. Ristaino, R. B. Sack, J. B. Kaper, F. Orskov, and I. Orskov. 1983. Colonization factor antigens I and II and type 1 somatic pili in enterotoxigenic Escherichia coli: relation to enterotoxin type. Infect. Immun. 39:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConnell, M. M., H. R. Smith, G. A. Willshaw, A. M. Field, and B. Rowe. 1981. Plasmids coding for colonization factor antigen I and heat-stable enterotoxin production isolated from enterotoxigenic Escherichia coli: comparison of their properties. Infect. Immun. 32:927-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenzie, R., A. L. Bourgeois, F. Engstrom, E. Hall, H. S. Chang, J. G. Gomes, J. L. Kyle, F. Cassels, A. K. Turner, R. Randall, M. J. Darsley, C. Lee, P. Bedford, J. Shimko, and D. A. Sack. 2006. Comparative safety and immunogenicity of two attenuated enterotoxigenic Escherichia coli vaccine strains in healthy adults. Infect. Immun. 74:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. USA 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, B. E., D. J. Evans, Jr., M. E. Penaranda, and D. G. Evans. 1983. CFA/I-ST plasmids: comparison of enterotoxigenic Escherichia coli (ETEC) of serogroups O25, O63, O78, and O128 and mobilization from an R factor-containing epidemic ETEC isolate. J. Bacteriol. 153:566-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oyofo, B. A., D. S. Subekti, A. M. Svennerholm, N. N. Machpud, P. Tjaniadi, T. S. Komalarini, B. Setiawan, J. R. Campbell, A. L. Corwin, and M. Lesmana. 2001. Toxins and colonization factor antigens of enterotoxigenic Escherichia coli among residents of Jakarta, Indonesia. Am. J. Trop. Med. Hyg. 65:120-124. [DOI] [PubMed] [Google Scholar]
- 27.Qadri, F., S. K. Das, A. S. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack, and A. M. Svennerholm. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 38:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qadri, F., C. Wenneras, F. Ahmed, M. Asaduzzaman, D. Saha, M. J. Albert, R. B. Sack, and A. Svennerholm. 2000. Safety and immunogenicity of an oral, inactivated enterotoxigenic Escherichia coli plus cholera toxin B subunit vaccine in Bangladeshi adults and children. Vaccine 18:2704-2712. [DOI] [PubMed] [Google Scholar]
- 29.Rappuoli, R., M. Pizza, G. Douce, and G. Dougan. 1999. Structure and mucosal adjuvanticity of cholera and Escherichia coli heat-labile enterotoxins. Immunol. Today 20:493-500. [DOI] [PubMed] [Google Scholar]
- 30.Ried, J. L., and A. Collmer. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 31.Savarino, S. J., A. Fasano, J. Watson, B. M. Martin, M. M. Levine, S. Guandalini, and P. Guerry. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc. Natl. Acad. Sci. USA 90:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savarino, S. J., E. R. Hall, S. Bassily, T. F. Wierzba, F. G. Youssef, L. F. Peruski, Jr., R. Abu-Elyazeed, M. Rao, W. M. Francis, H. El Mohamady, M. Safwat, A. B. Naficy, A. M. Svennerholm, M. Jertborn, Y. J. Lee, and J. D. Clemens. 2002. Introductory evaluation of an oral, killed whole cell enterotoxigenic Escherichia coli plus cholera toxin B subunit vaccine in Egyptian infants. Pediatr. Infect. Dis. J. 21:322-330. [DOI] [PubMed] [Google Scholar]
- 33.Savarino, S. J., A. McVeigh, J. Watson, A. Cravioto, J. Molina, P. Echeverria, M. K. Bhan, M. M. Levine, and A. Fasano. 1996. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173:1019-1022. [DOI] [PubMed] [Google Scholar]
- 34.Shaheen, H. I., K. A. Kamal, M. O. Wasfy, N. M. El-Ghorab, B. Lowe, R. Steffen, N. Kodkani, L. Amsler, P. Waiyaki, J. C. David, S. B. Khalil, and L. F. Peruski. 2003. Phenotypic diversity of enterotoxigenic Escherichia coli (ETEC) isolated from cases of travelers' diarrhoea in Kenya. Int. J. Infect. Dis. 7:35-38. [DOI] [PubMed] [Google Scholar]
- 35.Shaheen, H. I., S. B. Khalil, M. R. Rao, R. Abu Elyazeed, T. F. Wierzba, L. F. Peruski, Jr., S. Putnam, A. Navarro, B. Z. Morsy, A. Cravioto, J. D. Clemens, A. M. Svennerholm, and S. J. Savarino. 2004. Phenotypic profiles of enterotoxigenic Escherichia coli associated with early childhood diarrhea in rural Egypt. J. Clin. Microbiol. 42:5588-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 37.Sommerfelt, H., B. I. Haukanes, K. H. Kalland, A. M. Svennerholm, J. Sanchez, and B. Bjorvatn. 1989. Mechanism of spontaneous loss of heat-stable toxin (STa) production in enterotoxigenic Escherichia coli. APMIS 97:436-440. [DOI] [PubMed] [Google Scholar]
- 38.Svennerholm, A.-M., and J. Holmgren. 1978. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM-1 ELISA) procedure. Curr. Microbiol. 1:19-23. [Google Scholar]
- 39.Svennerholm, A. M., C. Wenneras, J. Holmgren, M. M. McConnell, and B. Rowe. 1990. Roles of different coli surface antigens of colonization factor antigen II in colonization by and protective immunogenicity of enterotoxigenic Escherichia coli in rabbits. Infect. Immun. 58:341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tacket, C. O., and M. M. Levine 1997. Vaccines against enterotoxigenic Escherichia coli infections, p. 875-883. In M. M. Levine, G. C. Woodrow, J. B. Kaper, and G. S. Cobon (ed.), New generation vaccines, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 41.Tacket, C. O., R. H. Reid, E. C. Boedeker, G. Losonsky, J. P. Nataro, H. Bhagat, and R. Edelman. 1994. Enteral immunization and challenge of volunteers given enterotoxigenic E. coli CFA/II encapsulated in biodegradable microspheres. Vaccine 12:1270-1274. [DOI] [PubMed] [Google Scholar]
- 42.Turner, A. K., T. D. Terry, D. A. Sack, P. Londono-Arcila, and M. J. Darsley. 2001. Construction and characterization of genetically defined aro omp mutants of enterotoxigenic Escherichia coli and preliminary studies of safety and immunogenicity in humans. Infect. Immun. 69:4969-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, A. K., J. Greenwood, J. C. Stephens, J. C. Beavis, and M. J. Darsley. September. 2002. Attenuated bacteria useful in vaccines. European patent number EP1,425,038.
- 44.Willshaw, G. A., H. R. Smith, M. M. McConnell, E. A. Barclay, J. Krnjulac, and B. Rowe. 1982. Genetic and molecular studies of plasmids coding for colonization factor antigen I and heat-stable enterotoxin in several Escherichia coli serotypes. Infect. Immun. 37:858-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf, M. K. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization. 1999. New frontiers in the development of vaccines against enterotoxigenic (ETEC) and enterohaemorrhagic (EHEC) E. coli infections. Wkly. Epidemiol. Rec. 74:98-101. [PubMed] [Google Scholar]
- 47.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, J., F. Cassels, T. Scharton-Kersten, S. A. Hammond, A. Hartman, E. Angov, B. Corthesy, C. Alving, and G. Glenn. 2002. Transcutaneous immunization using colonization factor and heat-labile enterotoxin induces correlates of protective immunity for enterotoxigenic Escherichia coli. Infect. Immun. 70:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]