Abstract
Central to the study of type III secretion systems is the availability of reporter systems to monitor bacterial protein translocation into host cells. We report here the development of a bacteriophage P1 Cre recombinase-based system to monitor the translocation of bacterial proteins into mammalian cells. Bacteriophage P1 Cre recombinase fused to the secretion and translocation signals of Salmonella enterica serovar Typhimurium of the type III secreted protein SopE was secreted in a type III secretion system-dependent fashion. More importantly, the SopE-Cre chimera was translocated into host cells via the type III secretion system and activated the expression of luciferase and green fluorescent protein reporters of Cre recombinase activity.
Many gram-negative bacteria pathogenic for humans, animals, and plants have evolved a specialized protein secretion system, generally known as “type III,” which mediates the direct transfer of bacterial proteins into host cells (3, 7). These proteins, known as “effectors,” have the capacity to modulate or interfere with a variety of cellular functions. Central to the function of these systems is an organelle known as the needle complex that serves as a conduit for the passage of the secreted proteins through the bacterial envelope (14). Proteins destined to travel the type III secretion (TTS) pathway posses specific signals, usually present at their amino terminus, which target them to the secretion machinery (17, 19, 22). Furthermore, a family of customized chaperones is thought to be crucial for the recognition of cognate secreted proteins by the secretion machinery (21, 28). These chaperones bind specific domains usually located between amino acids ∼20 and ∼100 of the secreted proteins. Once secreted from the bacterial cell, proteins must be translocated into host cells, a process mediated by a family of related type III secreted proteins that are thought to form a channel in the eukaryotic cell membrane (3).
The ability to monitor the transfer of type III secreted proteins into host cells is very important not only for the study of the mechanisms of TTS but also for the identification of potential TTS effector proteins. In general, the transfer of proteins by the TTS system has been monitored directly by biochemical and microscopy techniques (16, 23, 24) or indirectly with a variety of reporter systems. The most widely used reporter system is based on bacterial adenylate cyclase (Cya), an enzyme whose activity is strictly dependent on a cytosolic eukaryotic cell protein, calmodulin (27). Chimeric proteins composed of the secretion and translocation signals of type III secreted proteins fused to adenylate cyclase can be translocated into host cells by the TTS system (TTSS) and can be monitored by measuring the levels of cyclic AMP, the product of this enzyme (27). Another reporter system based on cellular activities relies on the incorporation of phosphorylation sites for intracellular kinases within the amino acid sequence of the protein whose translocation is to be monitored. The translocation of the protein of interest is then monitored by examining the phosphorylation state of the engineered site, usually with a phosphospecific antibody (4). Since these reporter systems rely on measurements of reversible and transient changes in host cells (e.g., levels of cyclic AMP or protein phosphorylation), these methods require previous knowledge of the time frame within which protein translocation will occur. Although powerful, the usefulness of these methods may be limited when this information is not available or cannot be predicted from the biology of the bacteria of interest. This limitation may also affect the performance of systems based on β-lactamase fusions, particularly if the half-life of the chimeric protein under examination is short (1).
In an attempt to overcome some of these limitations, we have developed a novel reporter system to monitor TTS-mediated protein translocation. The method is based on the use of the bacteriophage P1 Cre site-specific recombinase that catalyzes the recombination between two 34-bp sequences called loxP, thereby leading to the excision or inversion of intervening sequence. A similar approach has been previously described to monitor type IV protein secretion-mediated protein delivery into plant (29), mammalian (25), and bacterial cells (18). We have utilized this method to monitor protein transfer into mammalian cells mediated by the Salmonella enterica serovar Typhimurium (S. enterica serovar Typhimurium) TTS system encoded within its pathogenicity island 1 (SPI-1) (6). Furthermore, we have developed a transposon-based system to generate random fusion to Cre as a tool to identify type III secreted proteins.
MATERIALS AND METHODS
Plasmids.
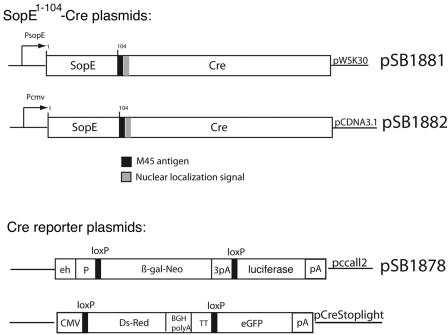
Diagrams of the relevant plasmids are shown in Fig. 1. Plasmid pSB1881 was constructed by fusing the bacteriophage P1 Cre recombinase in frame to the first 104 amino acids of the SPI-1 TTSS-secreted protein SopE, which contains its secretion and translocation signals (15) followed by an M45 epitope tag and the nuclear localization signal of the simian virus 40 (SV40) virus T antigen. The chimeric protein was then cloned into the bacterial expression vector pWSK30 (30). To express SopE1-104-Cre in mammalian cells, a DNA segment encoding the chimeric protein was amplified by PCR and cloned into the mammalian cell expression vector pCDNA3.1, yielding plasmid pSB1882. Plasmid pSB1878, which encodes a firefly luciferase gene preceded by a loxP site-flanked intervening sequence, was constructed by cloning the luciferase gene into plasmid pccall2 (20). The pStoplight plasmid is a mammalian expression vector that encodes the green fluorescent protein (GFP) preceded by a loxP-flanked intervening sequence (31). This vector also encodes the DsRed fluorescent protein.
FIG. 1.
Diagram of plasmids utilized in these studies. One hundred four amino acids of the N terminus of the S. enterica serovar Typhimurium effector protein SopE containing the domains required for secretion and translocation through the SPI-1 TTSS (signal sequence and chaperone binding domain) were fused in-frame with the M45 epitope tag, the nuclear localization signal of the SV40 large T antigen nuclear localization signal, and the full-length sequence of the P1 Cre recombinase. Expression of the chimeric protein was placed under the control of the native sopE promoter and cloned into the low-copy plasmid pWSK30, yielding plasmid pSB1881. The open reading frame of SopE-Cre, amplified by PCR, was cloned for eukaryotic expression in pCDNA3.1, yielding the plasmid pSB1882. A Cre reporter vector (pSB1878) was constructed by cloning the firefly luciferase gene in plasmid pccall2. A triple polyadenylation sequence (3pA) stops the transcription of firefly luciferase gene, which is only expressed after a recombination event that removes the lacZ gene and the transcriptional stop signals. β-Gal, β-galactosidase; CMV, cytomegalovirus; BGH, bovine growth hormone; eGFP, enhanced green fluorescent protein.
Bacterial strains.
All S. enterica serovar Typhimurium strains used in this work are derived from the wild-type strain SL1344 (10) and have been previously described (8, 9, 33). Bacterial strains were grown to maximize expression of the TTSS as previously described (2). Strains carrying the Δasd deletion mutation were grown in the presence of diaminopimelic acid (DAP) (50 μg/ml).
Construction of EZ::TN<cre-cat> and transposon mutagenesis.
To construct the EZ::TN<cre-cat> transposable element, a DNA segment encoding M45 epitope-tagged Cre containing the nuclear localization signal of the SV40 T antigen (SSDDEATADSQHSAPPKKKRKV) was amplified by PCR from the plasmid pSB2746. The resulting product was cloned into the ClaI-XbaI site of the vector pMOD-2<MCS>, a Tn5-based transposon construction vector (Epicenter). As a selection marker, a chloramphenicol resistance cassette isolated from pRY109 (32) was inserted downstream of cre into the PstI site of pMOD-2<cre>, resulting in EZ::TN<cre-cat>. A functional EZ::TN<cre-cat> transposon was isolated from the pMOD-2 vector by PCR amplification. To generate random SopE-Cre fusion proteins, the plasmid pSB1139, which encodes M45-tagged SopE, was subjected to in vitro transposon mutagenesis with EZ::TN<cre-cat> as described by the manufacturer (Epicenter).
Protein secretion assay.
The analysis of culture supernatant proteins was carried out as previously described (12).
SopE1-104-Cre translocation assay.
COS-2 cells were transfected with the Cre recombinase reporter plasmid pSB1878 or pStopLight using FUGENE-6 as indicated by the manufacturer. Four hours after transfection, cells were infected with the different S. enterica serovar Typhimurium strains at a multiplicity of infection of 25. After 45 min of infection, cells were thoroughly washed and noninternalized bacteria were killed by the addition of gentamicin (100 μg/ml in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum) for 2 h. Forty-eight hours after infection, the luciferase activity in cell lysates was measured using a luciferase assay system (Promega, Madison, WI) following the manufacturer's instructions. Alternatively, cells were observed under a fluorescence microscope to visualize GFP-expressing cells or processed for flow cytometry as follows. COS-2 cells were trypsinized, washed with RPMI containing 10% fetal bovine serum (FBS), and fixed with 4% paraformaldehyde in phosphate-buffered saline for 15 min. Cells were subsequently analyzed by flow cytometry in a FACScalibur flow cytometer (BD Biosciences, San Jose, CA). When indicated, the proteasome inhibitor MG132 was added at a final concentration of 1 μM 1 h before bacterial infection and kept throughout the experiment.
Mouse experiments.
The LacZ Rosa26 reporter mice (R26R) were obtained from Jackson Laboratories (26). These mice have a transgene in their Rosa26 locus in which the expression of lacZ is conditional to the removal of a loxP-flanked intervening sequence upon expression of the Cre recombinase. The Rosa26 locus allows expression in virtually all tissues. Eight-week-old mice were fasted for ∼6 h prior to oral (108 CFU) or intraperitoneal (107 CFU) infection with the Δasd S. enterica serovar Typhimurium strain carrying the plasmid pSB1881, which encodes SopE1-104-Cre. This strain undergoes very limited replication before undergoing DAP-less death. Five days after infection, animals were sacrificed and the organs and tissues were finely dispersed by passing them through a sterile steel mesh in RPMI containing 10% fetal bovine serum. The dispersed cells were washed and resuspended in the lysis buffer provided with the β-Galactosidase Reporter Gene Assay Chemiluminescence kit (Roche Applied Sciences). β-Galactosidase was then measured as indicated by the manufacturer.
RESULTS AND DISCUSSION
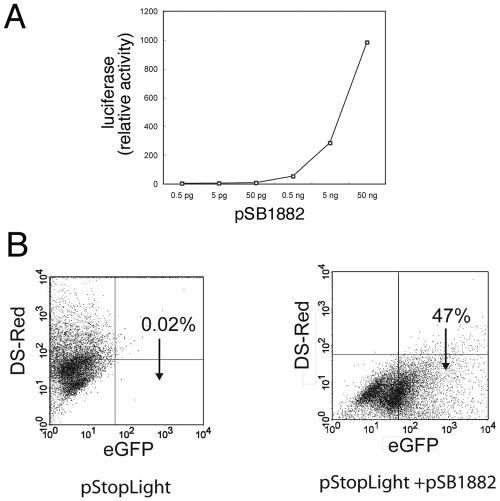
We constructed a chimeric protein consisting of the first 104 amino acids of SopE and the entire coding sequence of the bacteriophage P1 Cre recombinase (Fig. 1). SopE is a substrate of the S. enterica SPI-1-encoded TTSS, and its first 104 amino acids, which contain the secretion and translocation signals (15), have been previously used to deliver heterologous proteins into host cells for vaccine development (5). We first tested whether the chimeric protein retained recombinase activity. SopE1-104-Cre was cloned into a eukaryotic expression vector and cotransfected into COS-2 cells with a plasmid encoding the firefly luciferase or the green fluorescent protein so that their expression was dependent on the removal of a loxP-flanked intervening sequence. Expression of SopE1-104-Cre resulted in high levels of luciferase activity and a significant number of cells expressing GFP (Fig. 2A and B). No luciferase activity or GFP-expressing cells were observed when cells were cotransfected with the empty eukaryotic expression vector (data not shown). Both the level of luciferase activity and the number of GFP-expressing cells observed were directly correlated with the amount of plasmid DNA utilized in the transfections (Fig. 2A and data not shown). These results indicated that SopE1-104-Cre retains efficient recombinase activity.
FIG. 2.
SopE1-104-Cre exhibits Cre recombinase activity. A. Different amounts of plasmid pSB1882, which encodes SopE1-104-Cre, were cotransfected with the Cre reporter vector pSB1878, and firefly luciferase activity was determined 48 h after transfection. B. Plasmid pSB1882 was cotransfected with the reporter plasmid pStopLight, and 3 days after transfection cells were examined by flow cytometry for the presence of GFP-expressing cells, an indication of recombinase activity.
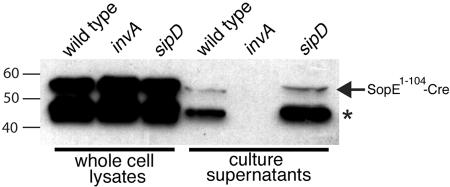
We then tested whether SopE1-104-Cre was secreted by the S. enterica serovar Typhimurium SPI-1 TTSS. A plasmid expressing SopE1-104-Cre from the psopE promoter was introduced into wild-type S. enterica serovar Typhimurium and their isogenic invA and sipD mutant derivatives. The invA mutant is defective for type III secretion (8) while the sipD mutant can secrete proteins from the bacteria through the TTSS, but it is defective for protein translocation into host cells (11). Whole-cell lysates and culture supernatants from these strains were then examined for the presence of SopE1-104-Cre. SopE1-104-Cre was observed in culture supernatants of the wild-type and sipD mutant strains but not of the invA S. enterica serovar Typhimurium strain, indicating that the chimeric protein could indeed be secreted by the type III secretion machinery (Fig. 3). Although secretion of the full-length SopE1-104-Cre protein was readily apparent, the most prominent secreted band had a molecular weight that was lower than that of the predicted full-length protein (Fig. 3). Since the epitope tag was present in the junction between SopE1-104 and Cre, most likely the lower-molecular-weight band is the result of proteolytic cleavage of its carboxy terminus.
FIG. 3.
SopE1-104-Cre is secreted by the S. enterica serovar Typhimurium SPI-1 type III secretion system. Whole-cell lysates and culture supernatants of wild-type S. enterica serovar Typhimurium and its isogenic derivatives invA and sipD mutant strains harboring the plasmid pSB1881, which encodes SopE1-104-Cre, were examined for the presence of SopE1-104-Cre by Western blot analysis. In addition to the full-length SopE1-104-Cre protein (arrow), a smaller polypeptide (*), presumably the result of proteolytic degradation, was also observed in both whole-cell lysates and culture supernatant samples.
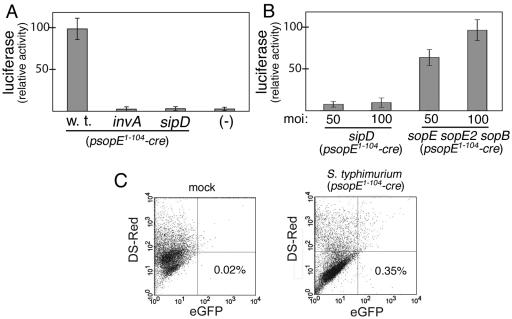
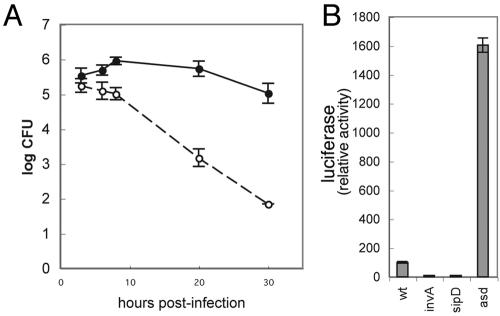
We then tested whether S. enterica serovar Typhimurium could deliver active SopE1-104-Cre into mammalian cells via the SPI-1 TTSS. COS-2 cells were transfected with plasmids encoding either the firefly luciferase or GFP reporter of Cre recombinase activity described above. Transfected cells were then infected with wild-type S. enterica serovar Typhimurium or derivatives carrying mutations in invA or sipD or simultaneously in sopE, sopE2, and sopB, all carrying a SopE1-104-Cre expression vector. The sopE sopE2 sopB triple mutant is competent for type III secretion and translocation, but it is defective for entry since these three effectors redundantly mediate this process (33). Forty-eight hours after infection with the various strains, cells were examined for the presence of luciferase activity or GFP, a measure of SopE1-104-Cre translocation. Cells infected with wild-type S. enterica serovar Typhimurium or the sopE sopE2 sopB triple mutant strains showed significant luciferase activity (Fig. 4A and B). In contrast, no significant luciferase activity was detected in cells infected with the secretion-defective invA or the translocation-defective sipD mutants. Furthermore, cells infected with wild-type S. enterica serovar Typhimurium expressing SopE1-104-Cre showed a 20-fold increase in the number of GFP-expressing cells over that of the control (Fig. 4C). These results indicated that S. enterica serovar Typhimurium can translocate SopE1-104-Cre via its SPI-1 TTSS and that translocation does not require bacterial internalization into host cells.
FIG. 4.
SopE1-104-Cre is translocated into cultured mammalian cells by the S. enterica serovar Typhimurium type III secretion system. (A and B) COS-2 cells transfected with the Cre recombinase luciferase reporter plasmid pSB1878 were infected with wild-type S. enterica serovar Typhimurium or its isogenic derivatives carrying mutations in invA, sipD, or simultaneously in sopE, sopE2, and sopB, all harboring the plasmid pSB1881, which encodes SopE1-104-Cre. The levels of translocated SopE1-104-Cre were measured by assaying the luciferase activity in infected cells. moi, multiplicity of infection. (C) COS-2 cells transfected with the Cre recombinase reporter plasmid pStoplight were mock treated (left panel) or infected with wild-type S. enterica serovar Typhimurium harboring pSB1881 (right panel). Cells expressing GFP, an indication of SopE1-104-Cre translocation, were quantified by flow cytometry. w. t., wild type; eGFP, enhanced green fluorescent protein.
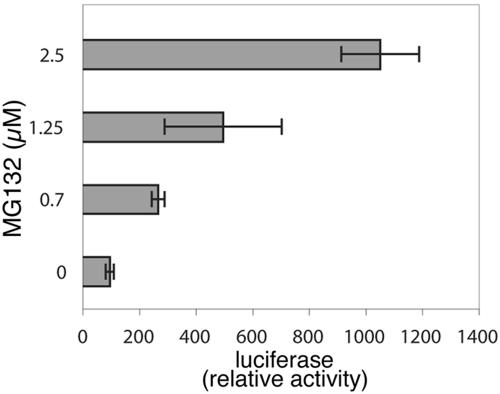
Previous studies have established that SopE is rapidly degraded upon translocation into host cells via the proteosome-mediated pathway (13). Furthermore, it was established that the signal for degradation is encoded within its secretion and translocation domain and that addition of this signal to other proteins also results in their rapid degradation (13). We hypothesized that if degradation of SopE1-104-Cre could be delayed, it might result in higher recombinase activity. We tested this hypothesis by infecting cells that had been transfected with a plasmid encoding the Cre recombinase luciferase reporter in the presence of the proteasome inhibitor MG132. Cells infected with wild-type S. enterica serovar Typhimurium expressing SopE1-104-Cre in the presence of different concentrations of the proteasome inhibitor exhibited up to a 10-fold increase in the luciferase activity (Fig. 5), indicating that preventing or retarding the degradation of the translocated protein results in higher recombinase activity. Since different translocated proteins exhibit different half-lives once inside cells (13), these results indicate that secretion and translocation signals from proteins with longer half-lives within the mammalian cell may serve as better surrogates of TTSS-mediated translocation.
FIG. 5.
Effect of proteasome inhibitors on the levels of translocated SopE1-104-Cre. COS-2 cells transfected with the Cre recombinase luciferase reporter plasmid pSB1878 were infected with wild-type S. enterica serovar Typhimurium harboring the plasmid pSB1881 in the presence or absence of the proteasome inhibitor MG132. The levels of translocated SopE1-104-Cre were measured by assaying the luciferase activity in infected cells.
Only a very small proportion of cells transfected with the GFP-based Cre recombinase reporter plasmid expressed GFP upon infection with wild-type S. enterica serovar Typhimurium expressing SopE1-104-Cre. Since expression of the reporter protein requires several hours of incubation, we hypothesized that the observed low efficiency of recombination might be due, at least in part, to the damage inflicted on the cell by the replicating bacteria. To test this hypothesis we introduced a plasmid expressing SopE1-104-Cre into an S. enterica serovar Typhimurium strain that carries a deletion in the asd gene. asd encodes aspartate semialdehyde dehydrogenase, an enzyme involved in the synthesis of diaminopimelic acid (DAP), an essential component of the cell wall (9). In the absence of DAP, this strain is unable to grow and undergoes DAP-less death. Since DAP is not synthesized by mammalian cells, S. enterica serovar Typhimurium Δasd is unable to grow within mammalian cells. As shown in Fig. 6A, in the absence of DAP, S. enterica serovar Typhimurium Δasd remained viable for up to 6 h after infection, although the number of viable bacteria declined sharply after that. Consistent with our hypothesis, cells transfected with the luciferase Cre recombinase reporter plasmid exhibited significantly higher (∼15 times) luciferase activity when infected with the Δasd S. enterica serovar Typhimurium strain expressing SopE1-104-Cre than when infected with the wild type carrying the same plasmid (Fig. 6B). These results suggest that bacterial toxicity to cells may hamper the efficiency of the recombinase-based type III secretion reporter systems.
FIG. 6.
Δasd mutation increases the levels of measurable translocated SopE1-104-Cre. A. Intracellular levels of wild-type (closed circles) and Δasd (open circles) S. enterica serovar Typhimurium strains at different times after infection. B. COS-2 cells transfected with the Cre recombinase luciferase reporter plasmid pSB1878 were infected with wild-type S. enterica serovar Typhimurium or its isogenic derivative invA, sipD, or Δasd mutant strains harboring the plasmid pSB1881, which encodes SopE1-104-Cre. The levels of translocated SopE1-104-Cre were measured by assaying the luciferase activity in infected cells. wt, wild type.
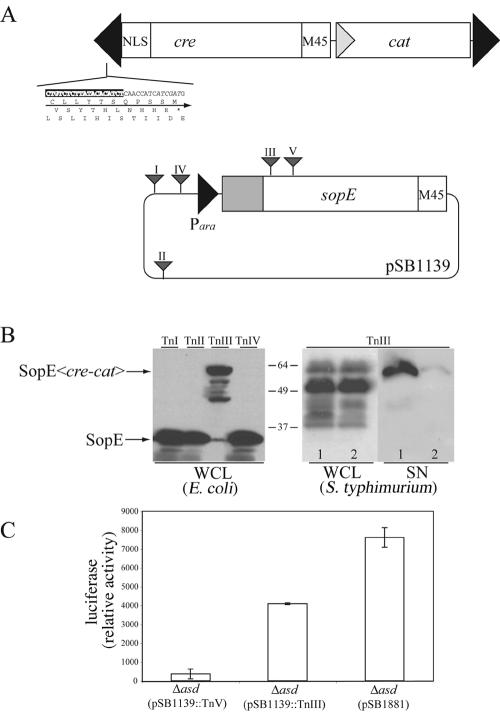
Since fusion of Cre to the secretion and translocation signals of SopE resulted in the translocation of the chimeric protein, it should be possible to use this reporter to identify potential type III secretion substrates or to delineate the minimal domain of a given type III secreted protein that is required for its translocation into host cells. To facilitate these applications, we constructed a transposon, EZ::Tn<cre-cat>, which is designed to create random translational fusions to cre (Fig. 7A). The resulting transposon was then tested by isolating random insertions in sopE and examining the resulting gene fusions for expression and secretion of the generated chimeric proteins. Random EZ::Tn<cre-cat> insertions in sopE generated after in vitro transposition were examined by restriction digestion and nucleotide sequencing. Clones with in-frame insertions in SopE were subsequently tested for expression and secretion of the SopE-Cre chimeras. Transposon insertions downstream of the secretion and translocation signals of SopE resulted in chimeric proteins that were secreted into culture supernatants in an SPI-1 TTSS-dependent manner (Fig. 7B). We also tested the ability of S. enterica serovar Typhimurium encoding sopE::EZ::Tn<cre-cat> to translocate the resulting chimeric protein into host mammalian cells by infecting COS-2 cells that had been transfected with a plasmid encoding the luciferase Cre recombinase reporter as indicated in Materials and Methods. Cells infected with S. enterica serovar Typhimurium expressing the SopE-Cre chimera exhibited significant luciferase activity, indicating that the fusion protein can be translocated into host cells (Fig. 7C). These results demonstrate that the EZ::Tn<cre-cat> transposon can be used to generate translocation-competent Cre fusions to type III secreted proteins.
FIG. 7.
Generation of SopE-Cre recombinase fusion proteins by transposon mutagenesis. A. (Upper panel) Diagram of the EZ::Tn<cre-cat> transposon. The inverted repeats of the transposable elements are shown by black arrowheads, and its nucleotide sequence as well as predicted amino acid sequences in the three reading frames are indicated. (Lower panel) EZ::Tn<cre-cat> insertions in pSB1139 are shown by triangles. The EZ::Tn<cre-cat> insertion III, located within the sopE open reading frame, generates an in-frame fusion between the first 167 amino acids of SopE and Cre. B. (Left panel) Whole-cell lysates of E. coli DH5α strains carrying the indicated EZ::Tn<cre-cat> insertions within plasmid pSB1139 were probed for the presence of SopE or the resulting SopE-Cre chimeric proteins. (Right panel) Plasmid pSB1139 carrying the EZ::Tn<cre-cat> insertion III, which generated the chimeric protein SopE1-167-Cre, was introduced into wild-type S. enterica serovar Typhimurium (lane 1) or its isogenic TTSS-defective invA mutant (lane 2). Whole-cell lysates (WCL) and cultured supernatants (SN) of these strains were probed for the presence of SopE1-167-Cre by Western blot analysis. C. COS-2 cells transfected with the Cre recombinase luciferase reporter plasmid pSB1878 were infected with S. enterica serovar Typhimurium Δasd carrying a plasmid that contains productive (TnIII) or nonproductive (TnV) EZ::Tn<cre-cat> insertions into sopE or the plasmid pSB1881, which encodes SopE1-104-Cre. The levels of the translocated SopE-Cre chimeras were measured by assaying the luciferase activity in infected cells. NLS, nuclear localization signal.
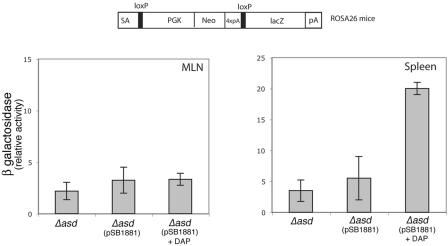
One of the main potential advantages of a Cre recombinase-based reporter system is the possibility of monitoring delivery of Cre recombinase in vivo. Mouse lines are available in which the expression of a reporter gene such as lacZ is conditional to the removal of loxP-flanked intervening sequence upon expression or delivery of the Cre recombinase (26). In particular, the Rosa26 strain carries such a reporter inserted within the Rosa26 locus, which can monitor Cre expression in virtually all tissues. We made use of this strain to attempt the monitoring of TTSS-mediated translocation of SopE1-104-Cre in vivo upon S. enterica serovar Typhimurium infection. Rosa26 mice were orally or intraperitoneally infected with different doses of wild-type or invA S. enterica serovar Typhimurium strains carrying a plasmid expressing SopE1-104-Cre. At different times after infection, the spleen, liver, intestine, and mesenteric lymph nodes were removed and lysed, and the β-galactosidase activity was measured as indicated in Materials and Methods. Even after several attempts using different bacterial doses and collecting samples at different times after infection, no β-galactosidase activity was detected in any of the tissues tested despite the presence of large number of bacteria in the tissues. We hypothesized that, similar to what was observed in vitro, it was possible that cells that received SopE1-104-Cre upon infection with wild-type S. enterica serovar Typhimurium may not survive long enough to allow for the expression of the Cre recombinase reporter. In an attempt to address this issue, mice were infected orally and intraperitoneally with various doses of a Δasd S. enterica serovar Typhimurium strain expressing SopE1-104-Cre, and the levels of β-galactosidase in different tissues were examined at different times after infection. No β-galactosidase activity was reproducibly detected in tissues, even when mice were infected with large doses of bacteria through either route of inoculation (Fig. 8). Since the Δasd S. enterica serovar Typhimurium strain undergoes only very limited replication in vivo, we attempted to increase the actual bacterial load in tissues by transiently administering DAP to the inoculated animals. Indeed, continued administration of DAP can effectively rescue the avirulence phenotype of this strain, suggesting that DAP is likely to be distributed throughout the different tissues (G. Briones and J. E. Galán, unpublished results). When DAP was administered once immediately after intraperitoneal inoculation of the Δasd S. enterica serovar Typhimurium strain, significant levels of β-galactosidase activity were observed in the spleen but not in other tissues of infected animals (Fig. 8). Attempts to identify the β-galactosidase-expressing cells in the spleen by flow cytometry were unsuccessful, suggesting that only a rather small number of cells were expressing the reporter gene.
FIG. 8.
In vivo detection of SopE1-104-Cre translocation into mouse cells. ROSA26 Cre recombinase reporter mice were orally infected with S. enterica serovar Typhimurium Δasd or the same strain carrying the plasmid pSB1881, which encodes SopE1-104-Cre, in the presence or absence of diaminopimelic acid (DAP) in the inoculum as indicated in Materials and Methods. Five days after infection, mice were euthanized and the levels of β-galactosidase activity in spleen and mesenteric lymph nodes (MLN) were measured as indicated in Materials and Methods. SA, splice acceptor; PGK, phosphoglycerate kinase 1 promoter.
We have developed a system to monitor type III secretion-mediated translocation of effector proteins into host cells both in vitro and potentially in vivo. The system should also be amenable to adaptation for high-throughput use, such as for the identification of inhibitors of type III secretion systems. In addition, we have constructed a transposon that should be useful for the identification of type III secreted proteins or the dissection of translocation signals of known effectors. Although the performance of the system in vitro was adequate for all experimental purposes, its performance in vivo was not efficient, at least with the type III secreted protein that was used as a surrogate. It is possible that the low in vivo efficiency of the reporter system is not due to the lack of sufficient delivery of the reporter enzyme but rather to aspects of the biology of Salmonella that may interfere with this application. It is conceivable that expression of the SPI-1 TTSS may eventually lead to cell death, not allowing enough time for expression of the reporter protein after recombination and its subsequent detection in the infected animal. It is therefore possible that under other conditions or in other bacteria less toxic for cells, the Cre recombinase system may be an efficient reporter of in vivo TTSS-mediated protein delivery.
Acknowledgments
We thank members of the Galán laboratory for critical reading of the manuscript.
This work was supported by Public Health Service Grants AI30492 and U54 AI0157158.
Editor: J. B. Bliska
REFERENCES
- 1.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, L. M., K. Kaniga, and J. E. Galán. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 4.Day, J. B., F. Ferracci, and G. V. Plano. 2003. Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol. Microbiol. 47:807-823. [DOI] [PubMed] [Google Scholar]
- 5.Evans, D. T., L.-M. Chen, J. Gillis, K.-C. Lin, B. Harty, G. P. Mazzara, R. O. Donis, K. G. Mansfield, J. D. Lifson, R. C. Desrosiers, J. E. Galan, and R. P. Johnson. 2003. Mucosal priming of simian immunodeficiency virus-specific cytotoxic T-lymphocyte responses in rhesus macaques by the salmonella type III secretion antigen delivery system. J. Virol. 77:2400-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galán, J. E. 2001. Salmonella interaction with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 7.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 8.Galán, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 17:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galán, J. E., K. Nakayama, and R. D. Curtiss. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94:29-35. [DOI] [PubMed] [Google Scholar]
- 10.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 11.Kaniga, K., D. Trollinger, and J. E. Galán. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaniga, K., S. C. Tucker, D. Trollinger, and J. E. Galán. 1995. Homologues of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubori, T., and J. E. Galan. 2003. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333-342. [DOI] [PubMed] [Google Scholar]
- 14.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galán, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. H., and J. E. Galan. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 16.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Z., and R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak, A., C. Guo, W. Yang, A. Nagy, and C. Lobe. 2000. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28:147-155. [PubMed] [Google Scholar]
- 21.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Ramamurthi, K. S., and O. Schneewind. 2003. Substrate recognition by the Yersinia type III protein secretion machinery. Mol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 23.Rosqvist, R., K. E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosqvist, R., C. Persson, S. Hakansson, R. Nordfeldt, and H. Wolf-Watz. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib. Microbiol. Immunol. 13:230-234. [PubMed] [Google Scholar]
- 25.Schulein, R., P. Guye, T. Rhomberg, M. Schmid, G. Schroder, A. Vergunst, I. Carena, and C. Dehio. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. USA 102:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 27.Sory, M.-P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 28.Stebbins, C. E., and J. E. Galan. 2003. Priming virulence factors for delivery into the host. Nat. Rev. Mol. Biol. 4:738-743. [DOI] [PubMed] [Google Scholar]
- 29.Vergunst, A., B. Schrammeijer, A. den Dulk-Ras, C. de Vlaam, T. Regensburg-Tuink, and P. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 30.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 31.Yang, Y., and T. Hughes. 2001. Cre stoplight: a red/green fluorescent reporter of Cre recombinase expression in living cells. BioTechniques 31:1036-1038. [DOI] [PubMed] [Google Scholar]
- 32.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galán. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]