Abstract
This is the first report of a Cryptococcus neoformans var. gattii strain (serotype B) that switches reversibly between its parent mucoid (NP1-MC) colony morphology and a smooth (NP1-SM) colony morphology. Similar to C. neoformans var. grubii and C. neoformans var. neoformans strains, the switch is associated with changes in the polysaccharide capsule and virulence in animal models. In murine infection models, NP1-MC is significantly more virulent than NP1-SM (P < 0.021). In contrast to the serotype A and D strains, the serotype B strain switches in vivo reversibly between both colony morphologies. The polysaccharide of NP1-MC exhibits a thicker capsule, and thus NP1-MC exhibits enhanced intracellular survival in macrophages. Consistent with this finding, switching to the mucoid variant is observed in pulmonary infection with NP1-SM. In contrast, the thin polysaccharide capsule of NP1-SM permits better crossing of the blood-brain barrier. In this regard, only smooth colonies were grown from brain homogenates of NP1-MC-infected mice. Our findings have important implications for the pathogenesis of cryptococcosis and suggest that phenotypic switching affects host-pathogen interactions in the local microenvironment. This altered interaction then selects for specific colony variants to arise in a pathogen population.
Cryptococcus neoformans is a human fungal pathogen that exists as three distinct varieties, specifically C. neoformans var. grubii (serotype A), C. neoformans var. neoformans (serotype D), and C. neoformans var. gattii (serotypes B and C). The majority of disease is due to C. neoformans var. grubii, followed by C. neoformans var. neoformans, and these varieties are found worldwide (29, 33). C. neoformans var. gattii is predominately restricted to tropical regions; however, a recent outbreak of C. neoformans var. gattii infections on Vancouver Island in Canada (19, 23) has raised the level of interest in this pathogen. Except for the recently described high prevalence of serotype C infections in AIDS patients living in sub-Saharan Africa (26), the majority of cryptococcosis due to serotype B and C strains occurs in immunologically normal individuals. Disease with serotype A and D strains occurs more often in immunosuppressed hosts (7) but has also been described for immunocompetent patients (20). The biological and genetic differences between the serotype A and D strains and C. neoformans var. gattii strains are so substantial that it has been proposed that C. neoformans var. gattii should be recognized as a separate species (7, 10, 24, 41, 42, 46).
We have previously shown that serotype A and D strains can undergo phenotypic switching from a smooth parent variant to more virulent mucoid or wrinkled colony variants (17). Furthermore, switches to a mucoid colony variant during chronic murine infections were associated with poor outcomes (16). Phenotypic switching only occurs in a subpopulation, and therefore a switch variant has to be selected by the host to become dominant (14, 16). In the setting of infection with a switching strain, antifungal interventions can select for the mucoid variant (13). This is relevant because frequent treatment failures in immunocompromised hosts generally are the result of persistence of the initial strain despite standard therapy (1, 2, 34). The relevance of phenotypic switching in infection with C. neoformans var. gattii is not known, but these infections often require more prolonged antifungal therapy and are associated with neurological sequelae and a higher frequency of neurosurgical interventions than those due to serotype A and D isolates (42).
Incoming mycological clinical specimens at All India Institute of Medical Sciences were typed by molecular typing methods and screened for colony variation (20). Five of 57 clinical isolates were found to be serotype B. One of these serotype B strains exhibited both smooth and mucoid colonies on the original plate that was inoculated with a patient's cerebrospinal fluid (CSF). We now present findings on this C. neoformans var. gattii strain that reversibly switches between its mucoid parent variant and a smooth colony morphology. Similar to the case with serotype A and D strains, the switch is associated with changes in the polysaccharide capsule and the cell wall. Most importantly, in murine infection models, the phenotypic switch to a smooth colony permits dissemination to the central nervous system (CNS), which has important implications for pathogenesis. In contrast to the serotype A and D strains, the serotype B strain switches in vivo reversibly between the mucoid and smooth variants. The colony variant that dominates the pathogen population is dependent on the local microenvironment promoting selection.
(Part of the work presented herein is part of the Ph.D. thesis work of N.J. at Jiwaji University, and All India Institute of Medical Sciences.)
MATERIALS AND METHODS
Strain.
Strain NP1 was isolated from the CSF of a patient with cryptococcal meningitis. The patient was not infected with human immunodeficiency virus and had no other obvious predisposing factors for infections (22). Colonies with smooth (NP1-SM) and mucoid (NP1-MC) morphologies were isolated from the first culture of the CSF specimen. The varietal status was determined by the use of canavanine-glycine-bromothymol blue medium and glycine-cycloheximide-phenol red medium. This identification was confirmed by rabbit serum antigenic factor typing with a commercial kit (Iatron, Japan). In addition, other C. neoformans var. gattii strains were screened for colony morphology. They included 11 environmental isolates from Australia (AS2554, AS2553, AS2552, AS2543, AS2542, AS2546, AS2551, AS2555, AS2556, AS2544, and AS2617), 5 clinical isolates from Canada (CN2615, CN2611, CN2613, CN2618, and CN2617), and 1 clinical serotype C isolate from New York (NY1343C) (a generous gift from Tom Mitchell).
Molecular characterization of NP1-SM and -MC.
Random amplified polymorphic DNA (RAPD) analysis was done by a PCR-based method using the mini-satellite-specific core sequence of the wild-type phage M13 as previously described (28). Restriction fragment length polymorphism (RFLP) analysis was performed as described previously, with a C. neoformans-specific transposon sequence (TCN-1) (20, 21). The TCN-1 probe (634 bp) was amplified from cryptococcal DNA with specific primers (TCN1.F.2 [5′-TCATGTCAGGTCCTTCCACTCGTAG-3′] and TCN1.R.2 [5′-CATAAACTTGGGCTGGGGATCG-3′]). The amplicon was cloned, purified, and labeled with [α-32P]dCTP, using a High Prime DNA labeling kit (Boehringer, Mannheim, Germany). For karyotype analysis, chromosomal DNA plugs were prepared from cultures derived from single colonies as described previously (12).
Phenotypic characterization.
Doubling times were determined by optical density measurements and manual counting of diluted suspensions in Sabouraud dextrose (SD) broth and in cell culture medium (Dulbecco's modified Eagle's medium [DMEM] with 5% fetal calf serum [FCS]). Melanization was determined with Sabouraud dextrose agar (SDA) plates supplemented with 1 μM l-dopa. MICs of fluconazole and amphotericin B for both variants were determined by microdilution and E tests per standard laboratory protocols. Capsule size was measured with C. neoformans cells from 24-h cultures in SD broth or with cells in lung homogenates 24 h after intratracheal (i.t.) injection or 3 h after intravenous (i.v.) injection in brain homogenates. Yeast cells were suspended in India ink (Becton Dickinson, NJ) and visualized at a magnification of ×1,000 with an Olympus AX70 microscope. Images were captured with a QImaging Retiga 1300 digital camera using QCapture suite V2.46 software (QImaging, Burnaby, BC, Canada). Capsule measurements were made with 25 randomly chosen cells from each strain, using Adobe Photoshop 7.0 for Windows, and the capsule thickness was calculated using the conversion of 45 pixels to 1 micrometer. The capsules of the NP1-SM and -MC yeast cells were stained with the monoclonal antibody (MAb) 18B7 to glucuronoxylomannan (GXM) and visualized with a fluorescein isothiocyanate-labeled sheep antibody to mouse immunoglobulin G as previously described (32). The viscosity of GXM dissolved in water and phosphate-buffered saline (PBS) were measured using a capillary viscometer (Technical Glass Products, Inc., Dover, NJ).
Resistance to temperature, osmotic stresses, and lysing enzyme.
NP1-SM and -MC were grown for 2 days at 30°C, suspended in PBS at 5 × 103 cells per ml, and either frozen overnight at −20°C or heated to 45°C for 30 min, 45 min, and 1 h or 50°C for 5 min. Ten microliters of suspension was plated on SDA plates to calculate the survival fraction relative to that of a control suspension. To compare the abilities of cells to tolerate high osmotic pressure, NP1-SM and -MC were grown on SDA plates supplemented with 1 M NaCl, 1 M sorbitol, or 10 mM glycerol. Sensitivities to lysing enzyme were determined by incubating NP1-SM and NP1-MC cells in sorbitol-sodium citrate buffer with different concentrations (96 μg/ml to 0.35 μg/ml) of lysing enzyme (from Trichoderma harzianum; Sigma Aldrich, St. Louis, Mo.) at 37°C. Every 15 min, cells were exposed to 2% sodium dodecyl sulfate to monitor the lysis of yeast cells under magnification (×400). The concentration of enzyme at which >80% of cells were lysed compared to control cells (incubated without lysing enzyme) was determined.
Analysis of GXM.
GXM purification and determination of its chemotype were done as previously described (8, 9). Prior to nuclear magnetic resonance (NMR) spectroscopy, GXM was sonicated and exchanged in 99.99% D2O (Cambridge Isotope Laboratories, Andover, MA). 1H chemical shifts were measured relative to the methyl groups of sodium 4,4-demethyl-4-silapentane-1-sulfonate taken at 0.00 ppm. Spectroscopy was done with a Bruker DRX600 NMR spectrometer (600 MHz) in our NMR facility. The viscosity of GXM dissolved in water and PBS was measured by using a capillary viscometer (Technical Glass Products, Inc., Dover, NJ) and determining the time the liquid took to fall a specific distance.
Animal studies.
BALB/c and SCID/BALB/c mice (male, 6 to 12 weeks old) were obtained from the National Cancer Institute (Bethesda, MD). Mice (n = 5 to 7 per group) were infected i.t. or i.v. via the tail vein. NP1-SM and NP1-MC were grown for 24 h in SD broth washed twice with PBS. For i.t. infections, mice were anesthetized and injected in the trachea with 5 × 106 C. neoformans cells in 50 μl sterile PBS using a 26-gauge needle. For i.v. infections, mice were infected with 1 × 106 to 5 × 106 cells in 100 μl PBS. Dilutions of the inoculum were plated onto SDA to ensure that comparable numbers of yeast cells were injected and that the colony phenotypes were stable. Mice were observed daily for signs of disease. Mice were killed by cervical dislocation after anesthesia, and organ CFU were determined at 14 days postinfection by homogenizing lung and brain tissue in PBS and plating 100 μl of different dilutions on SDA. Colonies were counted after 72 to 96 h. For histology and immunohistochemistry, mice were anesthetized and perfused with 4% paraformaldehyde in PBS for fixation under constant pressure, and tissue sections were stained with hematoxylin and eosin or mucicarmine.
Determination of in vivo switching of SM to MC phenotype.
For calculations of switching frequencies, a single colony of NP1-SM or -MC that was grown overnight in SD broth or organ homogenates were plated on SDA plates (200 to 300 colonies per plate). The in vitro switching rate of NP1-SM to the MC colony phenotype or vice versa was determined by visually scoring 104 colonies after growth on SDA plates. For in vivo switching rates, the percentages of colonies of the total CFU with MC relative to SM morphology were determined.
Phagocytosis and killing assays.
In vitro killing assays were performed with the murine macrophage-like cell line J774.16 (ATCC, Rockville, MD) (30, 45). C. neoformans var. gattii cells were added at a 1:5 ratio to cells with MAb 18B7 and coincubated with macrophages in DMEM supplemented with 5% FCS at 37°C for 2 h. The cell layers were then washed with PBS to remove nonadherent yeast cells and further incubated with cell culture medium or stained with Giemsa Wright stain after fixation with methanol. The starting phagocytosis index (PI) was determined by microscopy as follows: PI = number of yeast in macrophages/number of macrophages. The medium was replaced with medium that contained no MAb to avoid further phagocytosis, which was not observed with complement only. The mixture was incubated for 36 h at 37°C in 10% CO2. The ability of NP1-SM and -MC to replicate in J774 cells was determined by counting the CFU of C. neoformans var. gattii after it was cocultured with macrophages in the presence (intracellular growth) and absence (extracellular growth) of MAb 18B7. After 16 and 36 h, the supernatants of the wells were removed and separated in sterile tubes. Cells were lysed by incubation in sterile water and repeated vigorous aspiration with a pipette to complete the disruption. Supernatants and lysates were plated in different dilutions on SDA to determine the CFU and percent killing relative to yeast cells that grew extracellularly in the same medium.
Transmigration of blood-brain barrier.
A murine model of i.v. infection was used to study crossing of the blood-brain barrier by NP1-SM and -MC. The cells were grown in SD broth at 37°C for 20 h. Organisms were washed and counted, and NP1-SM cells were sonicated twice for 15 s each time on ice to disperse clumps. BALB/c mice (seven per group) were infected i.v. with 5 × 106 cells. The inoculum size and colony morphology were verified by backplating dilutions of the inoculum onto SDA plates. After 3 h, the mice were anesthetized and perfused by injecting 30 ml sterile PBS into the left ventricle. The right atrium was cut open to allow drainage during the procedure. The brains were removed and homogenized in sterile PBS on SDA plates to determine the CFU per brain.
Statistical analysis.
The student t test and the Kruskal-Wallis test were used to compare cell sizes, capsule sizes, and log-transformed fungal burdens (with the statistical program Primer). SPSS, version 8.0 (SPSS Inc., Chicago IL), was used to generate Kaplan-Meier survival curves.
RESULTS
NP1 phenotypic switching system.
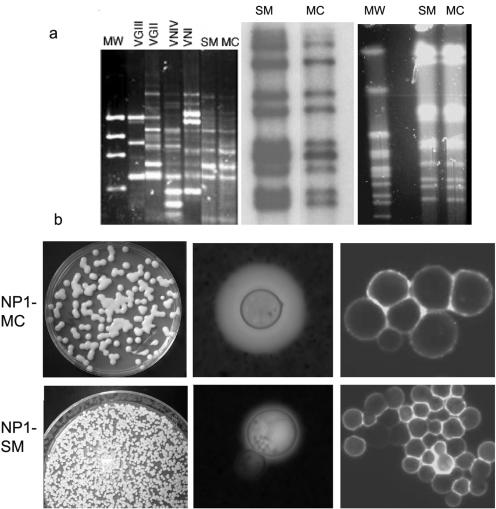
NP1 is a C. neoformans var. gattii strain that undergoes reversible phenotypic switching from a mucoid (NP1-MC) to a smooth (NP1-SM) colony phenotype (Table 1). The NP1-MC-to-NP1-SM switching rate was higher than that of NP1-SM to NP1-MC switching. RFLP and karyotype analyses confirmed that NP1-SM and NP1-MC colonies were the same C. neoformans var. gattii strain. M13 RAPD typing yielded a VGII group pattern, which is consistent with the classification of this strain as a C. neoformans var. gattii strain (Fig. 1a). Analysis of the average GXM structure by NMR yielded similar serotype-B-specific reporter group structures for both NP1-SM and -MC GXM (Table 1). This was confirmed by polyclonal antibody typing (data not shown). Because the majority of C. neoformans var. gattii strains (95% of our collection of >20 strains) exhibit a mucoid colony morphology, we concluded that NP1-MC represents the parent colony and NP1-SM the switch variant. Among 18 additional C. neoformans var. gattii strains tested, we found one other strain that exhibited a smooth morphology (AS2617). For this strain, 30,000 smooth colonies were plated to screen for switching to a mucoid colony morphology. No switching was detected, but it may happen at a lower frequency (<1 in 30,000). NP1-SM colonies exhibited a smooth domed surface and smooth edges, whereas NP1-MC colonies exhibited a shiny domed surface (Fig. 1b). Under high osmotic pressure conditions, NP1-MC consistently switched to the smooth colony phenotype, whereas NP1-SM remained smooth. In DMEM under 10% CO2, the doubling times of the variants were comparable, but in SD broth, NP1-SM grew slower than NP1-MC (Table 1).
TABLE 1.
Phenotypic characteristics of NP1-SM and NP1-MC
| Parameter | Value for colony type
|
|
|---|---|---|
| NP1-SM | NP1-MC | |
| Spontaneous in vitro switching rate | SM→MC, 7 × 10−5 | MC→SM, 5 × 10−4 |
| Doubling time at 37°C in SD broth (h) | 13.9 | 6.6 |
| Doubling time at 37°C in DMEM (h) | 7.3 | 6.3 |
| Cell size at 37°C (μm) | 4.7 ± 0.2 | 5.4 ± 0.4 |
| Capsule size (μm) | ||
| 20 h of growth in SD broth at 37°C | 0.63 ± 0.4 | 1.5 ± 0.25 |
| 24 h of growth in DMEM-FCS in 5% CO2 | 0.63 ± .025 | 3.6 ± 0.6 |
| In vivo 24 h after i.t. injection | 1.4 ± 0.11 | 5.3 ± 0.6 |
| At time of death in lung homogenates | 2.7 ± 0.47 | 6.0 ± 1.6 |
| Melanization | Yes | Yes |
| MIC of amphotericin B (μg/ml) | 0.25 | 0.25 |
| MIC of fluconazole (μg/ml) | 16 | 16 |
| GXM triad structure, as determined by NMR | M3, M1, M6 | M3, M1, M6 |
| Viscosity of GXM | MC >> SM | |
| Heat resistance at 45°C | Death after 30 min | Death after 60 min |
| Cold resistance | No difference | |
| Osmotic resistance (colony morphology on different plates) | SM >> MC | |
| 1 M NaCl | SM | SM |
| 1 M sorbitol | SM | SM |
| 10 mM glycerol | SM | SM |
| Resistance to lysing enzyme | SM >> MC | |
| Concn required for complete lysis (μg/ml) | 48 | 12 |
FIG. 1.
(a) Molecular characterization demonstrates that NP1-SM and -MC are switch variants of the same C. neoformans var. gattii strain (NP1). The left panel shows by RAPD analysis that both variants are VGII, consistent with their varietal status. The middle panel demonstrates identical RFLP patterns, and the right panel shows identical karyotype patterns for the SM and MC variants of NP1. (b) NP1-MC (upper panels) and NP1-SM (lower panels) colonies. From the left, the panels show the following: first column, comparison of colony morphologies; second column, capsule sizes in vivo; and third column, staining patterns with a GXM-specific MAb (18B7).
In SD broth at 37°C, NP1-SM cells exhibited some flocculence (clumps). Microscopic analysis of the NP1 phenotypes revealed striking differences in capsule size. The NP1-MC capsule was larger than the NP1-SM capsule. This was also observed during murine infection (Fig. 1b). Immunofluorescence staining with a MAb to the capsule demonstrated the presence of a GXM-like epitope(s) on both variants. Thus, the primary differences between the phenotypes were the capsule and cell sizes, cell wall stabilities, and doubling times (Fig. 1b).
Virulence studies.
The virulence levels of the NP1-SM and -MC variants were compared in BALB/c mice in i.t. and i.v. infection models. The measures of virulence were organ CFU and survival. In both infection models, the NP1-MC variant was more virulent (Fig. 2a and b). Both routes of infection were lethal with the NP1-MC isolate. Mice infected i.t. with NP1-SM survived >100 days, and at that time, no CFU could be detected in the lungs or brains of the i.t. infected mice (Fig. 2a). Half of the mice infected i.v. with NP1-SM died of disseminated cryptococcosis (Fig. 2b). Consistent with the survival difference, the lung CFU on day 14 were significantly lower in NP1-SM- than in NP1-MC-infected mice (i.t. and i.v.) (Fig. 2c). The brain CFU were not different for NP1-SM- and -MC-infected mice (Fig. 2c). SCID mice infected i.t. with the NP1-SM or -MC variant also died and had comparable median survival rates. Interestingly, NP1-SM-infected SCID mice cleared the infection from the lungs but eventually died from disseminated CNS disease (data not shown).
FIG. 2.
Comparison of virulence of NP1-SM and NP1-MC in murine animal models. These studies show that NP1-SM-infected BALB/c mice live significantly longer than NP1-MC-infected BALB/c mice in a pulmonary infection model (P = 0.021) (a) as well as an intravenous infection model (P = 0.008) (b). (c) Consistent with these findings, significantly lower (P ≤ 0.03) CFU were found in the lungs of NP1-SM-infected (i.t. and i.v.) BALB/c mice on day 14.
Inflammatory response to switch variants.
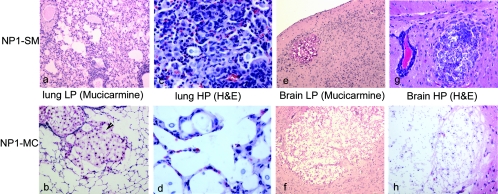
The extent of the inflammatory response directly correlated with the clearance of fungal burden. The histology of the lung sections demonstrated an intense inflammatory response in lung tissue infected with NP1-SM (Fig. 3a), characterized by perivascular cuffing, hyperplasia of the peribronchial lymphoid tissue, and granuloma formation. The mononuclear inflammation was composed of lymphocytes and macrophages (Fig. 3c). In NP1-MC-infected mice, lung histology revealed minimal inflammation and the presence of large cryptococcomas (Fig. 3b and d). For mice infected either i.v. or i.t. with NP1-SM or -MC, multiple cryptococcomas were seen in CNS tissue (Fig. 3e and f). The cryptococcomas in NP1-SM-infected mice (Fig. 3g) appeared smaller and also elicited more inflammation in the brain than those in NP1-MC cells (Fig. 3h).
FIG. 3.
Inflammatory responses in NP1-SM- and NP1-MC-infected mice. An extensive inflammatory response was detected in the lungs of NP1-SM (i.t.)-infected mice (a), whereas NP1-MC (i.t.)-infected mice exhibited a minimal inflammatory response and developed large cryptococcomas (b). A high magnification (HP) (×400) demonstrated more infiltration of mononuclear cells in the lung tissue of NP1-SM (c)- than of NP1-MC (d)-infected mice. In contrast, both NP1-SM- and NP1-MC-infected (i.v.) mice exhibited cryptococcomas (e and f) in the brain, although the cryptococcomas in the NP1-SM (e)-injected mice were smaller. Upon higher magnification (×200), it also appeared that NP1-SM elicited more inflammation in the brain (g) compared to the cryptococcomas in NP1-MC-infected mice (f and h). H&E, hematoxylin and eosin.
Phenotypic switching is important for pathogenesis.
Visual inspection of colonies derived from lung tissue of BALB/c and SCID mice infected with either NP1-SM or NP1-MC in the i.t. and i.v. models demonstrated the existence of both phenotypes in the lungs, but in most cases, only the smooth phenotype was recovered from the brains (Fig. 4a and b; Table 2). In SCID mice infected i.t. with NP1-SM, pulmonary clearance was prolonged and the pulmonary fungal burden was higher than that in infected BALB/c mice. Hence, in these mice there was more opportunity for switching to the mucoid colony type, and a higher percentage of MC colonies were recovered (Table 2). Thus, we concluded that phenotypic switching occurs reversibly in vivo between the smooth and mucoid colony phenotypes in both immunocompetent and immunocompromised mice.
FIG. 4.
Both NP1-SM and NP1-MC undergo phenotypic switching. (a) For NP1-MC-infected mice, both SM (white arrow) and MC (black arrows) colonies are detected in lung homogenates. (b) In contrast, only SM colonies are detected in the brain homogenates of NP1-MC-infected mice.
TABLE 2.
Colony phenotypes in lung and brain homogenates
| Mouse group | Colony phenotype (%)
|
|
|---|---|---|
| Lungs | Brains | |
| NP1-SM i.t. infected SCID | MC (40-90) | All SM |
| NP1-MC i.t. infected SCID | MC | SM |
| NP1-SM i.t. infected BALB/c | MC (2-20) | NDa |
| NP1-MC i.t. infected BALB/c | MC | All SM |
| NP1-SM i.v. infected BALB/c | SM + MC (few) | SM |
| NP1-MC i.v. infected BALB/c | MC (>90) | SM |
ND, not done.
Mechanism of selection pressure.
We then investigated what kind of host-pathogen interactions determined whether the NP1-MC or -SM colony phenotype dominated the pathogen population.
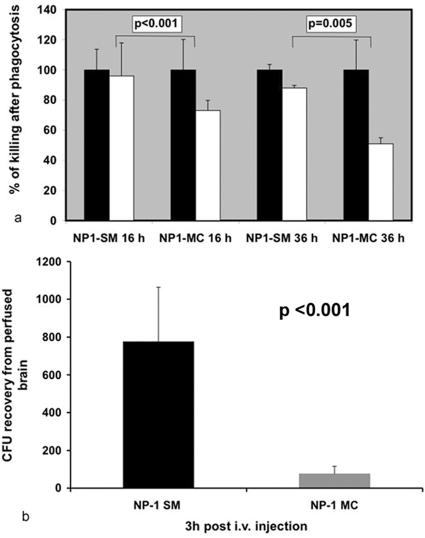
Macrophages are the primary phagocytic cells in pulmonary cryptococcosis. Because NP1-MC dominated pulmonary infections, we performed phagocytosis and killing assays to test if these parameters were affected by the differences in the polysaccharide capsule. Most yeast cells were phagocytosed by macrophages after 2 h. Antibody-mediated phagocytosis by macrophages was comparable for NP1-SM and NP1-MC cells (PI of NP1-SM, 14.5 ± 3.6; PI of NP1-MC, 15 ± 8.3 [P = 0.86]). In contrast, intracellular survival was significantly better for NP1-MC cells than for NP1-SM cells 16 and 36 h after infection of macrophage cell layers (Fig. 5a). Hence, NP1-MC was more resistant to intracellular killing by macrophages. Consistent with these findings, we found yeast cells with large capsules in macrophages.
FIG. 5.
Selection pressures for NP1-SM and NP1-MC differ. (a) Cocultivation in a macrophage cell line demonstrated enhanced intracellular survival of the NP1-MC variant after 16 and 36 h compared to that of NP1-SM. Black bars denote yeast cells grown in the presence of macrophages that are not intracellular (no antibody added). White bars reflect the percentages of phagocytosed yeast cells killed after specific times. (b) Studies investigating transmigration across the blood-brain barrier demonstrated that significantly (P < 0.001; t test) more NP1-SM than NP1-MC colonies could be recovered from brains of i.v. injected mice.
As described above, the smooth colony phenotype predominated in the brains of mice. Hence, we tested the hypothesis that NP1-SM cells can better cross the blood-brain barrier. For these experiments, mice were injected with NP1-SM and -MC cells, and CFU were determined 3 h after infection in perfused brain homogenates. These experiments yielded significantly (P < 0.001) larger numbers of colonies from NP1-SM-injected mice than from NP1-MC-injected mice. In a repeat experiment, the same difference was detected (data not shown). The capsule sizes of injected NP1-SM and -MC cells were 0.63 μm ± 0.4 μm and 1.5 μm ± 0.25 μm, respectively (Table 1). At the time of death, the capsule sizes of NP1-MC cells in the brains and lungs of infected mice did not differ (6.8 μm ± 3.8 μm versus 6.0 μm ± 1.6 μm), although the colony morphology of the brain CFU was smooth and that of the lung CFU was mucoid. We concluded that NP1-SM cells are better able to cross the blood-brain barrier, and it appears that the capsule size of NP1-MC cells is subsequently upregulated again in the brain without a reversion of colony morphology. Consistent with this finding, we also noted smaller polysaccharide capsules in the brains of NP1-SM-infected mice (Fig. 3e).
DISCUSSION
We previously reported the phenomenon of phenotypic switching in several serotype A and serotype D strains of C. neoformans (17). We extend our observation now to C. neoformans var. gattii. Both the smooth and mucoid colony variants of the C. neoformans var. gattii strain NP1 were isolated directly from a patient's CSF sample, suggesting that phenotypic switching occurred during infection. A more detailed analysis of this phenotypic switching system demonstrated that this strain is different from previously described strains because it switches in vivo reversibly between the two phenotypes. The local microenvironment in the host determined whether the NP1-MC or -SM colony phenotype dominated the pathogen population. In that regard, phenotypic switching of this strain plays a crucial role in pathogenesis, as it facilitates dissemination to the CNS.
Phenotypic switching enables pathogens to undergo rapid microevolution and to adapt to different microenvironments (18, 27, 37, 38). For human pathogens, the host represents a microenvironment of particular interest, and phenotypic switching in this setting may affect the host-pathogen relationship with consequences that can translate into changes in virulence (4). An example of how phenotypic switching can alter the host-pathogen relationship is provided by Trypanosoma cruzi, where phenotypic switching generates antigenically different variants that can escape the antibody response (31). Interestingly, in encapsulated microbes, phenotypic switching often controls the expression of the capsule, which is pivotal in determining whether the microorganism becomes an invasive pathogen (35, 44). Encapsulated bacteria that cross the blood-brain barrier often have to change their cellular phenotype. For Streptococcus pneumoniae, transparent colonies contain less capsular polysaccharide and more cell wall phosphorylcholine than opaque colony variants and are associated with invasive disease (35).
For fungi, phenotypic switching was first described for Candida albicans over 20 years ago (39, 40) and has been extensively studied. In more recent years, phenotypic switching has also been demonstrated with other fungi, including C. neoformans (15-17). In all of the examined C. neoformans strains, phenotypic switching was associated with changes in the polysaccharide capsule, which constitutes a major virulence factor (3).
The NP1 switching system is reminiscent of the previously described RC-2 system, which is a serotype D strain that switches in vivo between smooth and mucoid colonies (16). Presumably, the parent strain in the serotype D strain is smooth and the mucoid colonies represent switch variants, whereas for the NP1 serotype B strain, the parent strain is more likely the mucoid variant. In both strains, the mucoid polysaccharide is very viscous, consistent with the shiny colony surface; however, detailed NMR analysis of the GXM structures of the NP1 variants could not elicit a difference in the repeat structure or in the degree of acetylation. Hence, the biochemical basis of the mucoid polysaccharide remains to be resolved. It is conceivable that phenotypic switching to the mucoid phenotype may provide the ubiquitous fungus C. neoformans var. gattii with a survival advantage in an immunocompetent host, where a fierce innate immune response with potent phagocytic cells would not permit colonization and infection.
C. neoformans is a facultative intracellular pathogen that employs similar survival strategies in mammalian and unicellular hosts, such as amoebae (11, 25, 43). Hence, changes in the polysaccharide capsule can either affect phagocytosis or prevent the rapid destruction of intracellular yeast cells by potent inflammatory cells such as host alveolar macrophages. This biological advantage of better intracellular survival may constitute a selection pressure that promotes the selection of NP1-MC variants in the lung after i.t. infection. Survival studies, fungal burdens, and histological examinations are consistent with this conclusion. In contrast to the MC variant of RC-2, NP1-MC did not elicit a damage-driven immune response but rather appeared to downregulate the immune response in the lung, as there was much less recruitment of inflammatory cells. Eventually, an accumulation of large cryptococcomas was observed in the lung, which is a finding commonly described for patients infected with C. neoformans var. gattii (42).
Our results also further our understanding of C. neoformans as a major cause of fungal meningitis in humans. The mechanisms underlying transversal from the circulation, across the blood-brain barrier, and into the subarachnoid space have been recently elucidated in two elegant studies (5, 6). Both studies concluded that crossing of the blood-brain barrier occurred early after inoculation at the level of the cortical capillaries. In addition, the second study established that the expression of antigenic epitopes in the polysaccharide capsule was modified to facilitate crossing of the blood-brain barrier (6). Other murine infection studies measuring in vivo capsule size in brain and lung tissue have also demonstrated a decreased cell size and capsule size of yeast cells in the brain compared to those of yeast cells recovered from the lungs (36). In NP1, the differences in cell and capsule size most likely contribute to the difference in migration. In addition, capsule size was more upregulated in vivo in NP1-MC cells than in NP1-SM cells. In this regard, we found more capsule induction in NP1-MC yeast cells in vivo. At the time of death, capsule sizes in the brains and lungs did not differ in NP1-MC-infected mice. The capsule size of yeast cells in the brains of NP1-MC-infected mice was larger than that in NP1-SM-infected mice. Interestingly, the colony morphology in NP1-MC-infected mice was smooth in brain homogenates, despite the large capsule. Hence, capsule size alone does not determine colony morphology.
In summary, phenotypic switching occurs in C. neoformans var. gattii (serotype B). Similar to the case with serotype A and D strains, the phenotypic switch is associated with changes in the polysaccharide capsule and changes in virulence. In contrast to the serotype A and D strains, the serotype B strain switches reversibly. We hypothesize that this occurs due to in vivo selection pressure for both variants. For NP1-MC, enhanced intracellular survival in macrophages selects the mucoid variant in the lungs, whereas better transmigration across the blood-brain barrier selects for NP1-SM in the CNS. Although smooth colonies are rarely observed with C. neoformans var. gattii strains, we hypothesize that this switch in colony morphology may represent a “locked in” phenotype that may normally be transient and thus not detected. Hence, this strain may serve as a tool to identify genes that are regulated during in vivo infection and affect capsule induction and transmigration across the blood-brain barrier.
Acknowledgments
We thank J. Nosanchuk for critical reading.
This work was supported by grant R0-1 AI 59681 to B.C.F. and by a grant from the AIDS International Training and Research Program of the Albert Einstein College of Medicine (program director, Vinayak Prasad [NID D43-TW01403]) to U.B. and N.J. Furthermore, N.J. is thankful to ICMR (80/500/ECD-1/2003) for a senior research fellowship.
Editor: J. L. Flynn
REFERENCES
- 1.Brandt, M. E., M. A. Pfaller, R. A. Hajjeh, E. A. Graviss, J. Rees, E. D. Spitzer, R. W. Pinner, and L. W. Mayer. 1996. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated cryptococcosis. Cryptococcal Disease Active Surveillance Group. J. Infect. Dis. 174:812-820. [DOI] [PubMed] [Google Scholar]
- 2.Brandt, M. E., M. A. Pfaller, R. A. Hajjeh, R. J. Hamill, P. G. Pappas, A. L. Reingold, D. Rimland, and D. W. Warnock. 2001. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates in the United States: 1992 to 1994 and 1996 to 1998. Antimicrob. Agents Chemother. 45:3065-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulmer, G. S., and M. D. Sans. 1968. Cryptococcus neoformans. III. Inhibition of phagocytosis. J. Bacteriol. 95:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall, A., and L. A. Pirofski. 2003. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. C., M. F. Stins, M. J. McCaffery, G. F. Miller, D. R. Pare, T. Dam, M. Paul-Satyaseela, K. S. Kim, and K. J. Kwon-Chung. 2004. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 72:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlier, C., F. Chretien, M. Baudrimont, E. Mordelet, O. Lortholary, and F. Dromer. 2005. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 166:421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, and K. Byth. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 8.Cherniak, R., L. Morris, B. Anderson, and S. Meyer. 1991. Facilitated isolation, purification, and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect. Immun. 59:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherniak, R., H. Valafar, L. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmesser, M., Y. Kress, P. Novikoff, and A. Casadevall. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries, B. C., E. Cook, X. Wang, and A. Casadevall. 2005. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob. Agents Chemother. 49:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fries, B. C., D. L. Goldman, and A. Casadevall. 2002. Phenotypic switching in Cryptococcus neoformans. Microbes Infect. 4:1345-1352. [DOI] [PubMed] [Google Scholar]
- 15.Fries, B. C., D. L. Goldman, R. Cherniak, R. Ju, and A. Casadevall. 1999. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect. Immun. 67:6076-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fries, B. C., C. P. Taborda, E. Serfass, and A. Casadevall. 2001. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 108:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman, D., B. Fries, S. Franzot, L. Montella, and A. Casadevall. 1998. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc. Natl. Acad. Sci. USA 95:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt, S., R. Hilse, J. van Putten, R. Gerady-Schahn, A. Unkmair, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang, L. M., J. A. Maguire, P. Doyle, M. Fyfe, and D. L. Roscoe. 2004. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997-2002): epidemiology, microbiology and histopathology. J. Med. Microbiol. 53:935-940. [DOI] [PubMed] [Google Scholar]
- 20.Jain, N., B. L. Wickes, S. M. Keller, J. Fu, A. Casadevall, P. Jain, M. A. Ragan, U. Banerjee, and B. Fries. 2005. The molecular epidemiology of clinical Cryptococcus neoformans strains from India. J. Clin. Microbiol. 43:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller, S. M. 2004. The association of mating type with virulence in serotype A Cryptococcus neoformans strains. Ph.D. dissertation. The University of Texas Health Science Center at San Antonio, San Antonio, Tex.
- 22.Khanal, B., S. Sharma, and M. Deb. 2002. Cryptococcal meningitis in a non-AIDS patient. J. Nepal Med. Assoc. 41:325. [Google Scholar]
- 23.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. Macdougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon-Chung, K., T. Boekhout, J. Fell, and M. Diaz. 2002. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51:804-806. [Google Scholar]
- 25.Levitz, S. M., S. H. Nong, K. F. Seetoo, T. S. Harrison, R. A. Speizer, and E. R. Simons. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litvintseva, A. P., R. Thakur, L. B. Reller, and T. G. Mitchell. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J. Infect. Dis. 192:888-892. [DOI] [PubMed] [Google Scholar]
- 27.Lysnynansky, I., R. Rosengarten, and D. Yogev. 1996. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangement. J. Bacteriol. 178:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, T., and J. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee, S., M. Feldmesser, and A. Casadevall. 1996. J774 murine macrophage-like cell interactions with Cryptococcus neoformans in the presence and absence of opsonins. J. Infect. Dis. 173:1222-1231. [DOI] [PubMed] [Google Scholar]
- 31.Myler, P., J. Allison, N. Agabian, and K. Stuart. 1984. Antigenic variation in African trypanosomes by gene replacement or activation of alternate telomeres. Cell 39:203-211. [DOI] [PubMed] [Google Scholar]
- 32.Nussbaum, G., W. Cleare, A. Casadevall, M. D. Scharff, and P. Valadon. 1997. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J. Exp. Med. 185:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perfect, J. R., and A. Casadevall. 2002. Cryptococcosis. Infect. Dis. Clin. N. Am. 16:837-874, v-vi. [DOI] [PubMed] [Google Scholar]
- 34.Pfaller, M. A., J. Zhang, S. A. Messer, M. E. Brandt, R. A. Hajjeh, C. J. Jessup, M. Tumberland, E. K. Mbidde, and M. A. Ghannoum. 1999. In vitro activities of voriconazole, fluconazole, and itraconazole against 566 clinical isolates of Cryptococcus neoformans from the United States and Africa. Antimicrob. Agents Chemother. 43:169-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera, J., M. Feldmesser, M. Cammer, and A. Casadevall. 1998. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 66:5027-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan, T. G., and B. J. Hinnebush. 1998. Bloodstream- versus tick-associated variants of a relapsing fever bacterium. Science 280:1938-1940. [DOI] [PubMed] [Google Scholar]
- 38.Silverman, M., J. Zieg, M. Hilmen, and M. Simon. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. USA 76:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slutsky, B., J. Buffo, and D. R. Soll. 1985. High-frequency switching of colony morphology in Candida albicans. Science 230:666-669. [DOI] [PubMed] [Google Scholar]
- 40.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorrell, T. C. 2001. Cryptococcus neoformans variety gattii. Med. Mycol. 39:155-168. [PubMed] [Google Scholar]
- 42.Speed, B., and D. Dunt. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28-34. [DOI] [PubMed] [Google Scholar]
- 43.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swartley, L., A. Marfin, S. Edupuganti, L. Liu, P. Cieslak, B. Perkins, and J. Wenger. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taborda, C. P., and A. Casadevall. 2002. CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity 16:791-802. [DOI] [PubMed] [Google Scholar]
- 46.Trilles, L., B. Fernandez-Torres, S. Lazera Mdos, B. Wanke, and J. Guarro. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]