Abstract
Vibrio cholerae is the causative agent of cholera, a severe and devastating diarrheal disease. V. cholerae lives naturally in various aquatic habitats during interepidemic periods. Recent studies reveal that quorum-sensing systems, which exist in many bacteria and help them monitor their population densities and regulate various cellular functions, control V. cholerae pathogenesis, biofilm formation, and protease production. In this study we surveyed quorum-sensing systems in 16 geographically diverse V. cholerae strains from epidemic-causing O1 and O139 strains as well as non-O1/non-O139 and environmental isolates and discovered an unexpectedly high rate of dysfunctional components. We also found that a functional quorum-sensing system conferred a survival advantage on bacteria in biofilms when the bacteria were exposed to seawater, though quorum sensing was less important to survival in a planktonic state under the same conditions. These findings suggest that variations in quorum-sensing systems are due to environmental selective pressures and might be beneficial to V. cholerae's fitness under certain conditions found in its natural reservoirs.
The gram-negative bacterium Vibrio cholerae is the causative agent of cholera, a severe diarrheal disease that affects millions of people and causes well over 100,000 deaths on an annual basis (8). V. cholerae communicates by monitoring its population density and alters its gene expression accordingly. This phenomenon, known as quorum sensing, is highly important for the V. cholerae infectious cycle in humans, controlling phenotypic factors critical to pathogenesis, such as virulence factors, biofilms, and protease production (19, 26, 32). It has been postulated that biofilms aid V. cholerae in the passage through the gastric barrier of the stomach, while quorum-sensing-controlled detachment from biofilms in the small intestine aids in the colonization of the intestinal epithelium and eventual infection of new hosts (32). In addition, quorum sensing and biofilms have also been implicated in the long-term survival of V. cholerae in its natural marine environment (19, 26).
Quorum sensing in V. cholerae is controlled by two central regulators, LuxO and HapR. LuxO responds to accumulated molecules in the environment called autoinducers via membrane receptors CqsS and LuxPQ. Two autoinducer molecules are important for quorum sensing in V. cholerae. These two molecules are cholera autoinducer 1 (CAI-1), a molecule of undetermined structure, and autoinducer 2 (AI-2), a furanosyl borate diester also produced by Vibrio harveyi and many other bacteria. At high cell densities and autoinducer concentrations, this system causes LuxO to turn off its repression of hapR (23, 25). HapR can then repress the expression of aphA, whose gene products are key activators of virulence regulons (22). HapR also represses the vps (Vibrio polysaccharide synthesis) operon, thus negatively regulating biofilm formation (15, 31, 32). In addition, HapR directly up-regulates the expression of hapA, which produces a secreted hemagglutinin (HA)/protease responsible for the detachment of vibrios from the intestinal epithelium (11, 18).
Although V. cholerae has over 200 known serotypes, only strains with O1 and O139 antigens are known to cause epidemic disease (26). The O1 serogroup is further divided into O1 classical and El Tor biotypes on the basis of other physiological properties. O1 classical strains were responsible for six pandemics prior to 1961, when the first El Tor strain associated with a major epidemic appeared in Indonesia, which subsequently took over as the major disease-causing O1 biotype (9). Pathogenic O139 strains are relatively new, presumably having acquired virulence genes via mobile genetic elements. They were first isolated in 1992 from an epidemic outbreak of cholera in southern Asia (10). Both O1 El Tor and O139 V. cholerae strains are causes of annual outbreaks today. Some environmental and non-O1/non-O139 strains have also been shown to cause symptoms of cholera, although it is generally accepted that these strains lack the capability to cause major epidemic disease. Recent comparative genomic microarray analysis of pathogenic non-O1/non-O139 strains indicates that these strains carry a type III secretion system that may be involved in virulence and environmental fitness (5).
In this study we investigated the V. cholerae quorum-sensing system in 16 diverse strains and its relationship to virulence factors, biofilm, and protease regulation. The examined strains belong to the epidemic-causing serotypes of O1 and O139 antigen groups, as well as non-O1/non-O139 and environmental strains lacking the CTX phage encoding cholera toxin. We found a wide variety of active and defective quorum-sensing controls in these strains, raising questions as to the importance and role of this system in infection, environmental survival, and ultimately, of the evolution of the quorum-sensing system itself.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
V. cholerae strains used in this study are listed in Table 1 and were propagated in LB containing appropriate antibiotics at 37°C, unless otherwise noted. The plasmid containing a constitutively expressed hapR was constructed by subcloning of Ptac-hapR fragment from pJZ146 (33) into pBBR1-MCS2 (20).
TABLE 1.
Quorum-sensing components and quorum-regulated phenotypes in various V. cholerae strains
| Strain (reference) | Place and yr of isolation | Biotype and serogroup | Quorum-sensing component
|
Quorum-sensing regulation
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| hapR sequence | HapR productiona | lux expressionb | Mean CT production (μg/ml/OD600) (SD)c | Biofilm formation in LB/SW (OD570)d | Mean HA/protease production (azocasein units) (SD)e | Mean protease recovery (azocasein units) (SD)f | |||
| N16961 (4) | Bangladesh, 1971 | El Tor, O1 | Frameshift | − | − | 0.86 (0.07) | 0.38/0.54 | 3.3 (1.6) | 30.1 (1.7) |
| C6706 (4) | Peru, 1991 | El Tor, O1 | +g | + | + | 2.97 (0.03) | 1.03/0.59 | 22.9 (4.3) | NA |
| HK1 (4) | Hong Kong, 1961 | El Tor, O1 | + | − | − | 2.86 (0.92) | 2.45/1.16 | 2.9 (0.3) | 27.1 (0.5) |
| NCTC8457 (4) | Saudi Arabia, 1910 | El Tor, O1 | + | + | + | 0.08 (0.08) | 1.79/0.84 | 27.6 (7.3) | NA |
| MAK757 (4) | Celebes Islands, 1937 | El Tor, O1 | + | + | − | 0.31 (0.18) | 0.20/0.18 | 1.3 (1.2) | 12.1 (0.8) |
| 857h | Bangladesh, 1996 | El Tor, O1, environmental | + | + | − | NA | 0.20/0.79 | 5.5 (3.5) | 45.1 (0.1) |
| 2740-80 (4) | Gulf Coast, 1980 | El Tor, O1, environmental | + | + | + | NA | 1.25/0.76 | 58.1 (1.9) | NA |
| O395 (4) | India, 1965 | Classical, O1 | Frameshift | − | − | 8.87 (0.97) | 0.15/0.29 | 3.0 (0.1) | 23.6 (1.3) |
| CA401 (12, 16) | India, 1953 | Classical, O1 | + | + | Constitutive | 4.46 (1.40) | 0.15/0.29 | 51.5 (4.7) | NA |
| MO10 (4) | India, 1992 | El Tor, O139 | + | + | − | 0.69 (0.26) | 0.69/0.97 | 0.7 (0.0) | 26.4 (0.3) |
| MDO14 (6, 17, 27) | India, 1992 | El Tor, O139 | + | + | + | 1.42 (0.36) | 0.16/0.58 | 48.7 (2.6) | NA |
| MDO14-T (6, 17, 27) | India, 1992 | El Tor, O139 | No hapR | − | − | 0.69 (0.26) | 0.70/1.04 | 3.8 (0.1) | 38.6 (1.9) |
| SG21 (14) | India, 1992 | El Tor, O139 | C-terminal deletion | Short | Low | 0.70 (0.11) | 0.85/0.74 | 3.3 (0.8) | 24.5 (1.2) |
| SG38-2 (14) | India, 1992 | El Tor, O139 | + | + | Constitutive | 0.06 (0.05) | 1.07/1.50 | 56.2 (0.2) | NA |
| MZO-2 (5) | Bangladesh, 2001 | Non-O1/non-O139 | + | + | + | NA | 0.94/0.83 | 24.4 (4.2) | NA |
| AM15622 (5) | Bangladesh, 2001 | Non-O1/non-O139 | + | + | + | NA | 0.27/0.52 | 52.2 (0.6) | NA |
Results summarized from the Western blot analysis of HapR protein (Fig. 2). −, not produced; +, produced.
Results summarized from strains carrying pBB1 plasmid (Fig. 1). −, not expressed; +, expressed.
Classical strains were grown in LB (pH 6.5) at 30°C. Others were grown under AKI conditions. NA (not applicable) refers to CTX− strains.
Strains were grown in LB or sea water (SW) on a glass surface, and biofilm mass was quantified using dimethyl sulfoxide. The standard deviations were ≤10%.
Cell-free supernatants from cultures grown at 37°C for 7 h were collected and measured for protease activity as described previously.
A plasmid carrying Ptac-hapR was introduced into these strains, and protease activities were then measured. NA, not applicable.
+ the hapR sequence encodes intact HapR protein.
From the strain collection of John Mekalanos.
Luminescence assay.
The pBB1 cosmid, carrying the V. harveyi lux operon (1) was introduced into V. cholerae strains by conjugation or electroporation. The resulting strains were grown in LB with appropriate antibiotics at 30°C overnight, diluted to a concentration of 1:100 in fresh LB, transferred to white opaque 96-well plates, and incubated while shaking at 30°C. Luminescence was read at 1-h intervals for 7 h, with an initial reading at time zero, using a Bio-Tek Synergy HT spectrophotometer.
Western immunoblots of HapR.
Strains were grown in LB overnight and inoculated at 1:100 into 3 ml LB broth and grown for 7 h to late log phase. Cell pellets were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and detected with affinity-purified polyclonal anti-HapR rabbit antiserum.
Detection of CT, HA/protease, and autoinducers.
GM1 ganglioside enzyme-linked immunosorbent cholera toxin (CT) assays (13) were carried out following overnight incubations of V. cholerae strains with shaking at 37°C in AKI medium (for strains other than classical biotypes) or 30°C in LB (pH 6.5) (for classical strains) (17a).
HA/protease azocasein assays were performed as described previously (2). Briefly, strains were grown in 1 ml LB overnight with shaking and at 37°C. One hundred microliters of cell-free supernatant was combined with 100 μl azocasein (MP Biomedicals) in 100 mM Tris-Cl (pH 8) and incubated at 37°C for 1 h. After incubation, 400 μl 10% trichloroacetic acid was added and mixed, followed by a 15-min centrifuge step to pellet resulting precipitate. Three hundred microliters of this supernatant was transferred to 350 μl NaOH (525 mM), and the optical density at 442 nm (OD442) was taken. Results are reported as azocasein units calculated as follows: (OD unit − background) × 100.
The AI-2 bioassay using the V. harveyi reporter strain BB170, sensitive only to AI-2, has been reported previously (29). CAI-1 production was assayed as previously published using V. cholerae bioassay strain MM920 (25). Assays were performed with an opaque 96-well plate, and luminescence was read at 4 h postinoculation with a Bio-Tek Synergy HT spectrophotometer. Results were calculated as log of the change in the level from the background level (blank LB).
Biofilm formation assays.
Strains were suspended in 1 ml LB broth from growth on LB plates to an OD600 of 0.6. One milliliter of LB broth or seawater supplemented with 0.1% casein amino acids was inoculated at 1:100 in duplicate in borosilicate glass tubes (10 × 75 mm) and incubated at room temperature for 24 h. Biofilms were stained using crystal violet, and biofilm mass was measured as previously reported (32).
Biofilm and planktonic fitness.
Biofilms were grown for 24 h in 1 ml LB inoculated 1:100 from cultures of C6706 and C6706 hapR mutant at an OD600 of 0.6. After incubation, culture broth was removed and resuspended in 1.5 ml seawater (sampled from New Jersey coastline, filtered using Whatman paper, and sterilized) for planktonic survival assay. Biofilms were washed and then submerged in 1.5 ml seawater. Test tubes were placed in 4°C. After 5-day incubation, CFU (colony formation unit) was quantified from both planktonic and biofilm samples on LB plates.
RESULTS
Induced luminescence shows quorum-sensing variability across V. cholerae biotypes and serogroups.
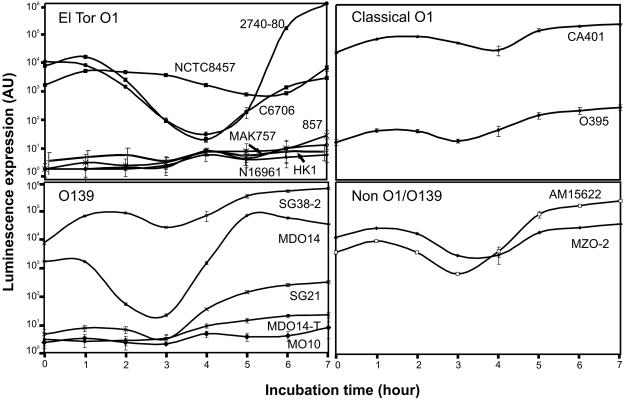
As previously shown, quorum-sensing systems in V. cholerae are able to activate the V. harveyi luxCDABE operon (23, 25). On the basis of luminescence levels, a functional quorum-sensing system generates a U-shaped curve on a plot of luminescence versus time of incubation (1). To study quorum-sensing regulation in various V. cholerae strains, the pBB1 cosmid containing the V. harveyi luxCDABE operon was introduced into 16 clinical or environmental V. cholerae isolates, and luminescence was measured as a function of cell growth. The strains were grown at 30°C and displayed similar growth rates. Using this assay, three patterns of lux expression were observed (Fig. 1 and Table 1). The first pattern of cell-density-dependent curves, indicative of the presence of functional quorum-sensing systems, was observed in six strains (C6706, NCTC8457, 2740-80, MDO14, MZO-2, and AM15622). The second pattern of low, noninduced luminescence was observed in eight strains (N16961, HK1, MAK757, 857, O395, MO10, SG21, and MDO14-T). The third pattern was observed in two strains (CA401 and SG38-2), displaying an apparent constitutive expression of luminescence. The lux expression patterns of the latter two categories suggest that these strains have defective quorum-sensing systems. There was no obvious correlation between the quorum-sensing regulation pattern and the biotype, serogroup, or year of isolation of V. cholerae.
FIG. 1.
Quorum-sensing-regulated luminescence expression in various V. cholerae strains. Overnight cultures were diluted 1:100 in LB and incubated at 30°C. Luminescence was measured at the time indicated. Lux expression curves were grouped on the basis of the strain biotypes. The values are means ± standard deviations (error bars). The results are representative of three experiments. AU, arbitrary unit.
HapR expression, hapR sequence, and HapR's effect on luminescence.
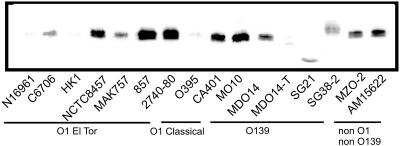
Two key proteins, LuxO and HapR, regulate quorum sensing in V. cholerae (25, 33). HapR is a functional V. harveyi LuxR homolog, which positively regulates luminescence in these organisms, while LuxO represses hapR expression at low density, and thus luminescence, through a set of small RNAs (23). To determine whether the V. cholerae strains that were defective in quorum sensing were also deficient in their expression of hapR, Western blotting using HapR antiserum was performed. Surprisingly, most strains produced HapR at or above the levels of the previously studied C6706 strain (23), with a few notable exceptions (Fig. 2 and Table 1). Strains N16961, HK1, O395, and MDO14-T did not produce any detectable HapR. The Western blot of strain SG21 showed a smaller band, indicating the production of a truncated protein. Also, strain SG38-2 produced a band representing a slightly larger protein than that of the other V. cholerae strains. All strains lacking HapR expression, N16961, HK1, O395, and MDO14-T, exhibited very low levels of luminescence induction. Conversely, strains MAK757, MO10, and 857 all expressed HapR at levels higher than that of C6706 but were still deficient in inducing luminescence. This is indicative of a nonfunctional HapR, possibly due to a mutation. Interestingly, SG21 induced low levels of luminescence in a quorum-sensing-dependent manner, indicating a HapR partially functional in luminescence induction, even though its HapR protein is smaller than the full-length HapR.
FIG. 2.
Production of quorum-sensing regulator HapR in 16 V. cholerae strains. Equal amounts of total protein (10 μg) of late-log cultures were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and detected with affinity-purified polyclonal anti-HapR rabbit antiserum (23).
We have reported that there is a naturally occurring frameshift mutation in the hapR sequence in both the sequencing strain N16961 and the widely used classical strain O395 (33). To further characterize the functionality of HapR, we sequenced hapR in each of the strains to identify any mutations (Fig. 3 and Table 1). Three strains, N16961, O395 and SG21, all had premature stop codons producing predicted peptides with 79, 68, and 119 amino acids, respectively, compared with the 204 amino acids of the peptide of strain C6706. These data are consistent with a lack of functional HapR expression (Fig. 2) and a lack of luminescence induction of pBB1 in strains N16961 and O395. Strain SG21, however, shows a response typical of quorum sensing and a distinct band on the Western blot assay. This indicates a possible conservation of function in this truncated protein. Point mutations found in strains MO10, 857, and MAK757, leading to one, two, and seven altered amino acids, respectively (Fig. 3), had no effect on HapR expression, though these strains did not induce significant levels of luminescence with the introduced lux operon (pBB1), raising the possibility that these mutations have had a deleterious effect on HapR function. Point mutations also existed in five strains (2740-80, SG38-2, NCTC8457, MZO-2, and AM15622) though these strains had normal HapR expression and positive induction of luminescence.
FIG. 3.
Alignment of HapR amino acid sequences. The well-studied HapR protein from strain C6706 was used as a reference, and all other sequences were aligned against this sequence using BLOSUM 62 scoring matrix. The amino acids that are different from those in HapR from C6706 are highlighted. Dashes indicate absence of amino acids.
We failed to PCR amplify the hapR region from strain MDO14-T. MDO14-T is a translucent variant of MDO14 (opalescent) that changes from an opalescent to translucent type at a rate of approximately 1% on LB plates. No hybridization was detected on a Southern blot, using C6706 hapR DNA fragments as probes, in MDO14-T (data not shown). This leads to the conclusion that the translucent variant of MDO14 completely lacks hapR. The mechanism of this deletion is currently under investigation.
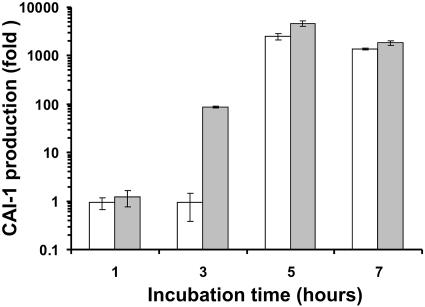
The remaining four strains (C6706, HK1, CA401 and MDO14) showed no alterations in their HapR sequences. El Tor strain HK1 has a C6706-type sequence of hapR, yet no HapR was detected on the Western blot. This might indicate a defect in another component of its quorum-sensing system, including autoinducer production. V. cholerae has been shown to produce at least two of these autoinducers, CAI-1 and AI-2, which synergistically modulate the activity of LuxO. We tested each strain's ability to produce such autoinducers by incubating V. harveyi and a V. cholerae reporter strain in each of the strain's culture supernatants, under conditions that induce luminescence activity only in the presence of specific inducers. We found all strains to have normal CAI-1 and AI-2 production (data not shown) except for HK1, which had a delayed production of CAI-1 based on incubation time (Fig. 4). This delay may be responsible for an elongated period of HapR repression due to the activity of LuxO, which is critical for the control of hapR expression. Alternatively, it is possible that other unknown quorum-sensing components are defective in HK1.
FIG. 4.
V. cholerae autoinducer CAI-1 production in strains HK1 and C6706. Overnight cultures were diluted 1:100 in fresh LB and grown at 30°C. Cell-free supernatants were withdrawn at the time indicated and assayed for CAI-1 activity using V. cholerae bioassay strain MM920 (25). CAI production is represented as the change in luminescence induction from the background level in strains HK1 (open bars) and C6706 (gray bars). The values are means ± standard deviations (error bars). The results are representative of three experiments.
Strains CA401 and SG38-2 induced luminescence of the lux operon in a constitutive manner. This, again, indicates a possible dysfunction of the LuxO repressor or other signaling components in these strains, negating the effects of cell densities on HapR function. We sequenced the luxO locus in these strains and found that they both have C6706-type LuxO amino acid sequences, indicating that other signaling steps in the quorum-sensing cascade might be defective. With the exception of SG21, all strains that produced a quorum-sensing curve in the luminescence assay had a wild-type or functional LuxO sequence as well as strong HapR expression.
Quorum-sensing-controlled phenotypes: HA/protease, CT, and biofilms.
Many of the above experiments used the nonnative Lux target as an indicator of quorum-sensing behavior. To investigate the expression of endogenous V. cholerae quorum-sensing targets, we examined protease production, cholera toxin, and biofilm formation in these strains. At high cell densities, HapR positively regulates protease production and negatively regulates virulence genes and biofilm production (15, 25, 33).
A variety of phenotypes related to quorum-sensing functions was observed. Protease production (Table 1) was deficient in all strains that either had a premature stop codon or deleterious mutations (as defined by a lack of luminescence induction) in the HapR protein. El Tor strain HK1 was also deficient in producing protease, which is consistent with its lack of HapR expression. Strains CA401 and SG38-2 produced high levels of protease even at low density (data not shown), reinforcing the notion that they have a possible quorum-sensing signaling cascade dysfunction that is locked in the regulatory state characteristic of high cell density. The remaining strains all induce protease activity in a density-dependent manner with wild-type HapR protein expression and luminescence induction (data not shown).
To see whether a constitutively expressed HapR could recover protease production, we introduced a plasmid containing Ptac-controlled hapR into the protease-deficient strains. All strains recovered normal levels of protease production using this method (Table 1), indicating that the protease-deficient phenotype results solely from a dysfunctional HapR.
HapR has also been shown to inhibit expression of virulence genes, such as the toxin-coregulated pilus pathogenicity island and cholera toxin genes, by repressing the expression of aphA, which encodes a key virulence regulator in V. cholerae (21, 28). In the in vitro induction condition, early expression of the hapR gene, such as in luxO mutants, inhibits the virulence regulon (32). We measured cholera toxin production of these isolates studied here using a CT enzyme-linked immunosorbent assay. Environmental strains 857 and 2740-80 as well as non-O1/non-O139 strains MZO-2 and AM15622, lack the genes encoding CT and subsequently produced no toxin. This is also true for O1 El Tor strain NCTC8457, which has been shown to lack the entire CTX phage, and thus produces no detectable toxin (4). The strains with a functional HapR sequence and HapR expression showed variability in their CT expression. Of these strains, only SG38-2 produced no detectable CT under AKI-inducing conditions. SG38-2 has what seems to be a LuxO deficiency that allows for a high level of HapR expression independent of cell density, and thus, can explain the very low levels of CT production in this strain. On the other hand, the classical strain CA401 produced high levels of CT, even though it also has a possible constitutive quorum-sensing system. Kovacikova et al. reported that the aphA promoter of classical strains is different from that of El Tor, and as such, HapR cannot repress aphA expression in classical strains (21). This may explain why CA401 still produced CT. The remaining strains produced a variable amount of CT whether they had an intact or mutated HapR, confirming previous in vitro results that mutations in hapR do not affect cholera toxin production (33).
Quorum sensing also negatively regulates biofilm formation in V. cholerae. Strains with HapR deletions have been shown to produce more robust biofilms than their wild-type counterparts (15, 30, 32). Biofilms are an important physiological characteristic of V. cholerae and have been implicated in many bacteria as an adaptation that provides protection against environmental stresses, such as acid shock (such as in the stomach), detergents, and antibiotics and also the predatory actions of certain protozoa (3, 24). All strains surveyed in this study produced some biofilm on a glass surface in both LB medium and seawater, but they differed greatly in the amount of biofilm produced. Strain HK1, which expressed no HapR, produced a very robust biofilm. MDO14-T, which lacks the hapR gene, produced more biofilm than MDO14, which has an intact hapR gene. NCTC8457, however, with a functional type HapR sequence and normal expression, produced a very robust biofilm, highlighting the variation of this trait in individual strains. Also, O1 classical strains produced, on average, the least biofilm mass of the serotypes and biotypes tested, also highlighting the variation of biofilm production in V. cholerae isolates.
Quorum sensing and V. cholerae environmental survival.
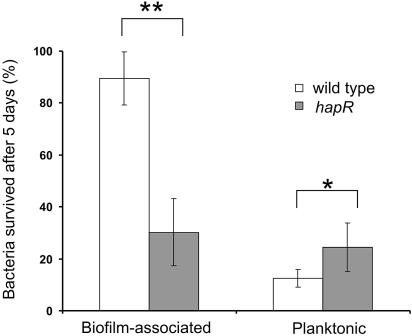
The results of the quorum-sensing survey above indicate that V. cholerae quorum-sensing systems are naturally diverse. This raises an important question regarding the selection of quorum sensing in the evolution of V. cholerae in terms of environmental survival. V. cholerae, which has the capacity to adapt to changes in salinity, temperature, and availability of nutrients, can successfully occupy one or more ecological niches in aquatic habitats as free-living bacteria or in association with phytoplankton or zooplankton (7). Because the bacterium exists in these two distinct phases, we hypothesized that the frequent mutations in the quorum-sensing systems of V. cholerae are favorable in certain aquatic environments by allowing the bacterium to more readily respond to environmental stresses. To investigate this possibility, we tested the longevity of a well-studied wild-type strain and an isogenic hapR mutant strain existing solely in biofilms versus that of planktonic cells in 4°C seawater for 5 days.
First, we investigated the importance of quorum sensing in biofilm-associated cells. Both wild-type and hapR mutant strains were grown in LB for 24 h to produce saturated biofilms. The LB was removed, and biofilms were then exposed to cold unsupplemented seawater. This procedure models a fast switch from a nutrient-rich to a nutrient-deprived environment that V. cholerae might experience in its natural reservoirs. The wild-type C6706 strain survived better than the isogenic hapR mutant under these experimental conditions (P < 0.05) (Fig. 5). This indicates a possible evolutionary role of quorum-sensing systems as an adaptation for environmental survival. This is quite surprising, considering the observed quorum-sensing dysfunctions in V. cholerae in this study. We also tested the survival rate of cells existing in a planktonic state under the same conditions. We found that the survival rate for planktonic hapR mutant cells was slightly higher than that for wild-type cells (P< 0.5). Although the statistical significance is low with these conditions and relatively small sample sizes, we believe these data suggest that a loss or alteration of quorum-sensing functions may actually confer a survival advantage for planktonic bacteria, which might explain why many quorum-sensing-deficient V. cholerae isolates exist. This possibility warrants further study.
FIG. 5.
Effect of quorum sensing on V. cholerae survival in an extreme environment. Biofilm-associated and planktonic wild-type C6706 cells and quorum-sensing-deficient mutant hapR cells were exposed to 4°C seawater. The number of CFU was determined after 5 days of incubation and represented as the percentage of the initial CFU. The values are means ± standard deviations (error bars). The results are representative of three experiments. Values that are significantly different from the wild-type value by the Student t test are indicated by asterisks (**, P < 0.05; *, P < 0.5).
DISCUSSION
Bacteria consistently sense and respond to environmental stimuli to modulate gene expression to adapt to changing environments. For example, many bacteria utilize quorum-sensing systems to monitor their population densities. In this study, we studied the V. cholerae quorum-sensing system and its regulated genes in 16 geographically diverse strains. Our data indicate three different states of quorum-sensing systems in the strains used in this study. First, six strains (C6706, NCTC8457, 2740-80, MZO-2, AM15622, and MDO14) have both functional hapR sequences and expression along with functional HapR-mediated regulation of protease production and V. harveyi luminescence. Second, eight strains (N16961, HK1, MAK757, 857, O395, MO10, MDO14-T, and SG21), have mutated HapR proteins that lack the ability to induce or induce very low levels of protease and luminescence. Third, two strains (CA401 and SG38-2) seem to have a constitutively expressed quorum-sensing system even though they have functional hapR and luxO sequences. Our subsequent data suggest that the improved stress survival of V. cholerae wild-type biofilm and hapR mutant planktonic cells provides possible explanations for the conservation and high rate of dysfunction of quorum-sensing components in V. cholerae, depending on what features are being positively selected for in the environment. We speculate that V. cholerae has the capacity to change quorum-sensing regulatory components in response to different environmental pressures. Recent reports have shown that the frequency of quorum-sensing regulator mutations under certain laboratory conditions is relatively high (15, 30), and thus, this phenomenon might be an important survival tactic of V. cholerae.
Since quorum sensing in V. cholerae negatively regulates virulence gene expression, having a dysfunctional quorum-sensing system does not reduce V. cholerae virulence factor production per se, and thus in this sense is not related to a strain's ability to cause cholera symptoms. For example, hapR mutant strains can colonize infant mice as well as wild-type strains can (25, 33). Therefore, quorum sensing is less likely to play a role in the natural selection of pathogenic strains, but it is important in the infectious cycle due to its benefits of aiding survival between outbreaks and its role in late stages of infection.
Quorum sensing may also be important for enhancing V. cholerae colonization if V. cholerae enters hosts as a part of a biofilm. When V. cholerae biofilms reach the upper intestine, quorum sensing down-regulates vps gene expression, causing V. cholerae detachment from biofilm structures and thus promoting colonization of the intestinal surface (32). Furthermore, previous research also suggests that quorum sensing down-regulates virulence genes and activates the production of extracellular proteases at the late infectious stage (33). These proteases are proposed to serve as detachases, allowing the bacteria to exit the host. Finally, although this study focused on HapR-dependent quorum-sensing systems, we cannot rule out the possible existence of other distinct quorum-sensing regulatory pathways in various V. cholerae isolates.
Acknowledgments
We are grateful to John Mekalanos for providing strains and sequence information of hapR alleles used in this study. We thank Deborah Hung and Jeffrey Weiser for helpful discussions and critically reviewing the manuscript. We also thank Bonnie Bassler for providing HapR antiserum.
This study was supported by the NIH/NIAID K22 award (AI060715) and a Penn Genomics Institute seed grant.
Editor: V. J. DiRita
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 2.Benitez, J. A., A. J. Silva, and R. A. Finkelstein. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69:6549-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 4.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziejman, M., D. Serruto, V. C. Tam, D. Sturtevant, P. Diraphat, S. M. Faruque, M. H. Rahman, J. F. Heidelberg, J. Decker, L. Li, K. T. Montgomery, G. Grills, R. Kucherlapati, and J. J. Mekalanos. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. USA 102:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehara, M., S. Shimodori, F. Kojima, Y. Ichinose, T. Hirayama, M. J. Albert, K. Supawat, Y. Honma, M. Iwanaga, and K. Amako. 1997. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol. Lett. 154:293-301. [DOI] [PubMed] [Google Scholar]
- 7.Faruque, A. S., D. Mahalanabis, A. Islam, and S. S. Hoque. 1994. Severity of cholera during concurrent infections with other enteric pathogens. J. Diarrhoeal Dis. Res. 12:214-218. [PubMed] [Google Scholar]
- 8.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., and G. B. Nair. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 46:59-66. [DOI] [PubMed] [Google Scholar]
- 10.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkelstein, R. A., and L. F. Hanne. 1982. Purification and characterization of the soluble hemagglutinin (cholera lectin) produced by Vibrio cholerae. Infect. Immun. 36:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardel, C. L., and J. J. Mekalanos. 1994. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 235:517-526. [DOI] [PubMed] [Google Scholar]
- 14.Garg, P., A. Aydanian, D. Smith, J. G. Morris, Jr., G. B. Nair, and O. C. Stine. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 16.Hamood, A. N., R. D. Sublett, and C. D. Parker. 1986. Plasmid-mediated changes in virulence of Vibrio cholerae. Infect. Immun. 52:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 17a.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 18.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 19.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 21.Kovacikova, G., W. Lin, and K. Skorupski. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 23.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 24.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102:16819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 26.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta, D. K., M. Boesman-Finkelstein, and R. A. Finkelstein. 1996. Antibody against the capsule of Vibrio cholerae O139 protects against experimental challenge. Infect. Immun. 64:343-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 29.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]