Abstract
To characterize the roles of Porphyromonas gingivalis and its components in disease processes, we investigated the cytokine profiles induced by live P. gingivalis, its lipopolysaccharide (LPS), and its major fimbrial protein, fimbrillin (FimA). A cytokine antibody array revealed that human monocyte-derived macrophages were induced to produce chemokines (e.g., monocyte chemoattractant protein 1, macrophage inflammatory protein 1β [MIP-1β], and MIP-3α) as early as 1 h after exposure to P. gingivalis, with production declining after 24 h of exposure. As expected, an extensive repertoire of inflammatory mediators increased subsequent to infection, most predominantly tumor necrosis factor alpha (TNF-α), interleukin 1β (IL-1β), IL-6, IL-10, and granulocyte-macrophage colony-stimulating factor. The induction of cytokines by P. gingivalis was not triggered simply by bacterial cell surface components, since purified P. gingivalis LPS and FimA induced similar patterns of cytokines, while the pattern of cytokines induced by live P. gingivalis was significantly different, indicating that the host defense system senses live bacteria differently than it does the cell surface components LPS and FimA. To further understand the mechanisms by which live P. gingivalis and its components exert their effects, we used a high-throughput immunoblot screening approach (Becton-Dickinson PowerBlot) to analyze intracellular proteins involved in P. gingivalis infection in human macrophages. Exposure of human macrophages to either live P. gingivalis, its LPS, or its FimA protein led to the up-regulation of 12, 8, and 10 proteins and the down-regulation of 15, 8, and 17 proteins, respectively. The expression of proteins involved in gene transcription (e.g., monocyte enhancer factor 2D [MEF2D], signal transducer and activator of transcription 1 [STAT1], STAT3, STAT6, and IL enhancer binding factors [ILF3]), of protein kinases (e.g., mitogen-activated protein kinase 3 [MAPK3], MAP3K8, double-stranded RNA-activated protein kinase [PRKR], and MAP2K4), and of proteins involved in immune responses (e.g., TNF super family member 6 [TNFSF6] and interferon-induced protein with tetratricopeptide repeat 4 [IFIT4]), apoptosis (e.g., genes associated with retinoid interferon-induced mortality 19 [GRIM19]), and other fundamental cellular processes (e.g., clathrin heavy-chain polypeptide, culreticulin, and Ras-associated protein RAB27A) was found to be modulated differentially by P. gingivalis, LPS, and FimA. These differential changes are interpreted as preferential signal pathway activation in host immune/inflammatory responses to P. gingivalis infection.
Periodontitis is a chronic infectious disease to which genetic, microbial, immunological, and environmental factors combine to influence disease risk and progression. Both bacterial virulence factors and host responses contribute to the connective tissue destruction and alveolar bone resorption characteristic of this disease (8, 13, 23). Porphyromonas gingivalis is a predominant periodontal pathogen which expresses a number of virulence factors involved in the pathogenesis of periodontitis. Among them, fimbriae play a critical role in mediating the bacterial interaction with host tissues, promoting bacterial adhesion to and invasion of the targeted sites (31). Fimbriae and LPS of this bacterium have been implicated in both the initiation and progression of disease. Previous studies have demonstrated that protein-free P. gingivalis LPS retains immunostimulatory activity in TLR4-deficient C3H/HeJ mice (37). Furthermore, it has been shown that, unlike enterobacterial LPS, P. gingivalis LPS and fimbriae use mostly TLR2 to induce innate immune responses in both human and mouse macrophages (32, 52).
In most cases of chronic inflammation triggered by an infection, a mononuclear cell infiltrate is typically present, and the prominent cell type found in mononuclear infiltrates is the monocyte. The mononuclear phagocyte plays an important role in regulation of the inflammatory host response, in part through its ability to secrete mediators, particularly cytokines, in response to microorganisms and their products. It has been demonstrated that monocytes constitute a substantial proportion of the cells recovered from the gingival tissues of patients with periodontitis (45). In addition, the numbers of monocytes in the inflammatory tissues of patients with periodontitis have been found to be higher than those from normal tissue (42). Overall, monocytes play a central role in orchestrating the response to gram-negative bacteria.
Understanding the molecular basis of the host response to bacterial infections is critical for preventing infection and also for minimizing the tissue damage resulting from an overly aggressive host response. The cellular and molecular events during the interaction of individual pathogenic components with host monocytes/macrophages have been characterized to some extent. Although LPS is generally considered to be a bacterial component that alerts the host to infection, P. gingivalis LPS is not as potent an activator of human monocytes as is Escherichia coli LPS, as measured by its relative activation of inflammation (1, 38). P. gingivalis LPS may selectively modify the host response as a means of facilitating colonization. Indeed, there have been reports suggesting a differential regulation of particular signaling pathways, such as the phosphatidylinositol-3-kinase-Akt pathway and the p38 MAPK pathway, in P. gingivalis LPS-induced production of proinflammatory and anti-inflammatory cytokines in human monocytes (5, 26). In addition, studies have indicated that P. gingivalis fimbriae activate human peripheral blood monocytes utilizing specific cellular receptors (32) and phosphorylated proteins (29), inhibit caspase-3-mediated apoptosis of monocytic cells (33), and induce monocyte adhesion to the endothelial cell surface and infiltration of monocytes into periodontal tissues of adult individuals with periodontitis (12). However, very little is known about the effect of live P. gingivalis bacteria relative to LPS and fimbriae on the human peripheral blood monocyte response.
Our previous study demonstrated qualitative and quantitative differences between the responses of monocytes to live P. gingivalis and to its fimbriae or LPS (52), supporting our hypothesis that unique signaling mechanisms are induced by live P. gingivalis. On this basis it is reasonable to suggest that a comprehensive understanding of host-bacterium interactions is currently incomplete, because most studies have examined the response to bacterial components rather than to live bacteria. To improve our understanding of the mechanisms by which human monocytes interact with live P. gingivalis, we attempted in the present study to identify proteins that are differentially modulated after treatment with live P. gingivalis relative to its purified LPS or fimbrial components. For this purpose, we employed the Becton-Dickinson PowerBlot Western Array screening system. This is a high-throughput Western blotting method which uses carefully formulated mixtures of subsets of ∼1,000 monoclonal antibodies to evaluate the differences in levels of cellular signaling proteins between total cell extracts from different cells or tissue. Treatments with live P. gingivalis, its LPS, or its FimA protein resulted in changes in the levels of several proteins, including transcription factors, signaling mediators, DNA synthesis and repair proteins, and proteins related to immune responses.
The P. gingivalis-, P. gingivalis LPS-, and P. gingivalis FimA-modulated proteins identified include some that have not been previously associated with the immune response. Our results provide additional support for the contention that P. gingivalis and its purified components modulate specific genes and proteins associated with the innate immune response, cell growth, and apoptosis.
MATERIALS AND METHODS
Abbreviations.
The following terms used herein are abbreviated as indicated: lipopolysaccharides (LPS), Toll-like receptor 4 (TLR-4), mitogen-activated protein kinase (MAPK), sodium dodecyl sulfate (SDS), tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), enzyme-linked immunosorbent assay (ELISA), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1β (MIP-1β), fibroblast growth factor 4 (FGF-4), insulin-like growth factor binding protein 2 (IFGBP-2), pulmonary and activated-regulated chemokine (PARC), granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-related oncogene (GRO), tissue inhibitor of metalloproteinase 2 (TIMP-2), epithelial neutrophil-activating peptide 78 (ENA-78), neutrophil activating protein 2 (NAP-2), macrophage-derived chemokine (MDC), interferon-induced protein 10 (IP-10), regulated upon activation normal T-cell expressed and secreted (RANTES), oncostatin M (OSM), transforming growth factor β2 (TGF-β2), IL enhancer-binding factor 2 (LIF3), signal transducer and activator of transcription factor 1 (STAT1), monocyte enhancer factor 2D (MEF2D), TNF superfamily member 6 (TNFSF6), culreticulin (CALR), double-stranded RNA-activated protein kinase (PRKR), clathrin heavy-chain polypeptide (CLTC), Ras-associated protein (RAB27A), and cell division cycle 42 (CDC42).
Bacterial strain and its components.
P. gingivalis 381 (ATCC) was cultured in brain-heart infusion broth (Gibco) enriched with hemin (5 μg/ml) and menadione (1 μg/ml) in an anaerobic atmosphere (52). LPS from P. gingivalis 381 was extracted with phenol-water and purified by cesium chloride isopycnic density gradient ultracentrifugation followed by repurification, as we have previously described (52). The purity of LPS preparations was confirmed by amino acid analysis and by SDS-polyacrylamide gel electrophoresis with silver staining. FimA from P. gingivalis 381 was purified by size exclusion chromatography of sonicated extracts of P. gingivalis 381 (52). Fractions that were negative of any contaminating substances on silver-stained SDS-polyacrylamide gels were pooled, dialyzed, and lyophilized. The identity of the 41-kDa FimA protein was confirmed by N-terminal amino acid sequencing. The total FimA protein was quantified with the NanoOrange protein quantification kit (Molecular Probes, Eugene, OR).
Macrophage culture.
Freshly elutriated human monocytes (>95% pure) were purchased from Advanced Biotechnologies (Columbia, MD). Monocytes were plated for 5 days to reach a density of 2 × 107 cells per 10 ml of Dulbecco modified Eagle medium (Invitrogen) with 20% fetal calf serum (Invitrogen), 10% human serum type AB (Nabi, Boca Raton, FL), and 50 μg/ml gentamicin (Invitrogen) in 10-cm-diameter tissue culture plates at 37°C in a humidified atmosphere containing 5% CO2 to enable differentiation into macrophages. On days 5 and 7, half of the medium was removed and replaced with medium lacking fetal calf serum. Medium on the cultured macrophages was changed to 5 ml of Dulbecco modified Eagle medium with 1% human serum on day 9, 1 h before experiments were begun.
Infection or treatment of macrophages with P. gingivalis or P. gingivalis components.
Adherent macrophages were infected with live P. gingivalis bacteria at a ratio of 25:1 (bacteria to macrophages) at 37°C. Frozen stocks of P. gingivalis 381 were thawed, cultured, and diluted in medium to a concentration of 5 × 108 bacteria per 50 μl, to give multiplicities of infection (MOIs) of 25:1, and added to cultures of macrophages. Dilutions were plated on brain-heart infusion agar plates for anaerobic culturing, and colonies were counted to confirm the accuracy of dilution and viability of bacteria. In additional cultures, 10 μg/ml of purified LPS or FimA from P. gingivalis was added to medium containing 1% human serum and cells were incubated at 37°C. After incubation with P. gingivalis or its components, cells and supernatants were harvested at the indicated times.
Cytokine antibody array.
The supernatants from P. gingivalis-infected and LPS- or FimA-stimulated macrophages derived from freshly elutriated human monocytes were analyzed with cytokine antibody arrays using RayBio Human Cytokine Antibody Array V (RayBiotech, Inc., Norcross, GA) according to the manufacturer's instructions. Briefly, cytokine array membranes were blocked in 2 ml of 1× blocking buffer for 30 min and then incubated with 1 ml of samples at room temperature for 1 to 2 h. Samples then were decanted from each container, and the membranes were washed three times with 2 ml of 1× wash buffer I, followed by two washes with 2 ml of 1× wash buffer II at room temperature with shaking. Membranes then were incubated in 1:250-diluted biotin-conjugated primary antibodies at room temperature for 1 to 2 h and washed as described above, before incubation in 1:1,000-diluted horseradish peroxidase-conjugated streptavidin for 30 to 60 min. Membranes were then washed thoroughly and exposed to a peroxidase substrate (detection buffers C and D; RayBiotech, Inc., Norcross, GA) for 5 min in the dark before imaging. Membranes were exposed to X-ray film (Kodak X-Omat AR) within 30 min ofaddition of the substrate. Signal intensities were quantified with Bio-Rad VersaDoc Imaging System 3000 and analyzed with Quantity One software (Bio-Rad). Biotin-conjugated immunoglobulin G served as a positive control at six spots, where it was used to identify membrane orientation and to normalize the results from different membranes that were being compared. For each spot, the net optical density level was determined by subtracting the background optical level from the total raw optical density level.
Sample preparation for PowerBlot and Western blot analysis.
Cells were grown in 10-cm-diameter tissue culture plates and treated with live P. gingivalis, P. gingivalis LPS, or FimA for 24 h. Whole-cell lysates were prepared as follows: after removal of the medium by aspiration, cells were rinsed with phosphate-buffered saline three times. Boiling lysis buffer (10 mM Tris [pH 7.4], 1.0 mM sodium orthovanadate, and 1.0% SDS) was added to the plates to denature cellular proteins rapidly. The cell lysates were collected into a 50-ml conical tube and heated briefly in boiling water. The lysates were then sonicated for 10 s to shear genomic DNA. The protein concentration was determined with the Bio-Rad protein assay kit, and the samples were frozen at −80°C and sent on dry ice to BD Biosciences/Pharmingen (San Diego, CA) for PowerBlot analysis.
PowerBlot analysis.
The levels of proteins were determined by PowerBlot analysis by BD Biosciences/Pharmingen (San Diego, CA). Briefly, samples containing 500 μg of protein in 500 μl of sample buffer were loaded in one large well on top of a 7.5 to 15% gradient SDS-polyacrylamide slab gel. Electrophoresis was conducted overnight at a constant current. The proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad). After transfer, the membranes were incubated in blocking buffer containing 5% nonfat dried milk. The membrane was clamped with a grid that isolates 40 lanes (chambers) oriented from the top to the bottom of the membrane. Mixtures containing four to five mouse monoclonal antibodies that recognize proteins of distinct molecular weights and antigenicities were added to each lane and allowed to hybridize for 1 h. A total of 995 antibodies were applied to seven templates per sample for this analysis. The blot was removed from the manifold, washed, and hybridized with goat anti-mouse antibody conjugated with horseradish peroxidase. The blots were developed with a chemiluminescence system (SuperSignal West Pico; Pierce, Rockford, IL). Each of the samples was analyzed on triplicate blots.
After exposure to X-ray films, the developed films were scanned, and the “trace” of each band was processed for densitometric analysis. Briefly, the trace includes a measure of both band intensity and band area. Standard average is the average trace for all bands on a blot. The individual band intensity was expressed as a percentage of the corresponding trace for average standards and for standards designated “percent lane average” to normalize for different exposures and protein loadings between gels. The percent lane average for bands of samples treated by P. gingivalis and its components (percent lane average for treated bands) was expressed as a percentage of the percent lane average for the corresponding control bands. The “percentage of control” was determined to express increases or decreases in protein expression. Changes are expressed as fold increase or decrease between the control percent lane average and the treated percent lane average. A summary file was provided, listing all protein expression changes detected in order of confidence, 1 through 5, with 5 being the highest level of confidence. The confidence level was based on fold change, reproducibility, and signal intensity (a good quality signal easily passes a visual inspection; it is hard for a low signal to pass a visual inspection). Levels are defined as follows: level 5, changes of >2-fold in triplicate from good quality signals; level 4, changes of 1.5- to 1.99-fold in triplicate from good quality signals; level 3, changes of 1.25- to 1.49-fold in triplicate from good quality signals; level 2, changes of >2-fold in triplicate from low signals; level 1, changes of >2-fold in duplicate from good quailty signals. In this article, we present only changes from levels 2 and higher.
Western blot analysis.
Cell lysates prepared as described above or with cold lysis buffer (25 mM HEPES [pH 7.7], 400 mM NaCl, 1.5 mM MgCl2, 2 mM EDTA, 0.5% Triton X-100, 3 mM dithiothreitol, 20 mM β-glycerophosphate, 1mM sodium orthovanadate, and 25 mM para-nitrophenylphosphate and protease inhibitor mixture [Roche]) were analyzed by conventional Western blotting. Protein (50 μg) was electrophoresed through an SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad) by electroblotting. The membranes were incubated in blocking buffer (5% nonfat dried milk, 10 mM Tris [pH 7.5], 100 mM NaCl, and 0.1% Tween 20). Immunoblotting for protein expression was performed with mouse monoclonal antibodies (BD Biosciences/Transduction Laboratories) against eight proteins selected from the proteins identified by the PowerBlot analysis. We used horseradish peroxidase-conjugated goat anti-mouse as a secondary antibody. The blots were developed with an enhanced chemiluminescence system (Amersham). Western blotting was repeated at least two times.
Cytokine ELISA.
The supernatants from live P. gingivalis-, LPS-, or FimA-stimulated human macrophages were analyzed for TNF-α, IL-6, and IL-10 by ELISA (R&D Systems, Minneapolis, MN).
Statistics.
Statistical analysis was performed by the two-tailed Student t test for comparison between two groups. A P value of <0.05 was considered statistically significant. The correlation between two groups was quantified by use of the linear correlation coefficient.
RESULTS
Cytokine expression in human monocytes exposed to live P. gingivalis.
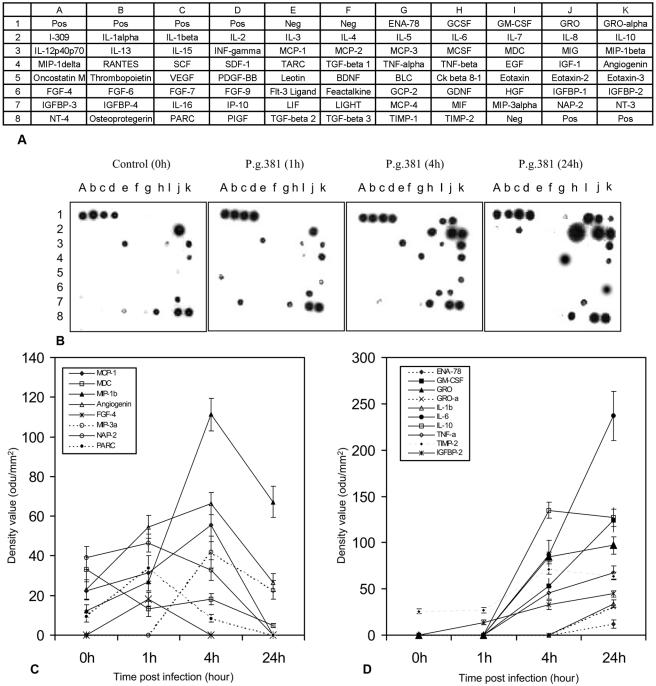
To assess the impact of P. gingivalis on macrophage phenotype and function, freshly elutriated human monocytes were cultured as adherent monocyte-derived macrophages and exposed to live P. gingivalis. The cell culture supernatants were subjected to cytokine antibody array analysis (Fig. 1). In these isolated primary macrophage cultures, live P. gingivalis induced MCP-1, MIP-1β, angiogenin, FGF-4, IGFBP-2, and PARC as early as 1 h after exposure (Fig. 1C). Then, GM-CSF, GRO, IL-6, IL-10, TNF-α, MIP-3α, and TIMP-2 were detected at 4 h, while ENA-78, GRO-α, and IL-1β were detected at 24 h (Fig. 1D). In addition, among the earliest-induced cytokines, FGF-4 and PARC levels decreased or were not detected at 4 h, while MCP-1, MIP-1β, MIP-3α, and angiogenin levels decreased or were not detected at 24 h (Fig. 1C). NAP-2 and MDC were constitutively expressed in adherent monocyte-derived macrophages, but they were completely inhibited or degraded by live P. gingivalis after 24 h (Fig. 1B and C).
FIG. 1.
Cytokines induced by P. gingivalis in freshly elutriated human peripheral blood monocytes. Monocytes were cultured to enable differentiation into macrophages and then were exposed to live P. gingivalis bacteria at an MOI of 25:1 for 1 h, 4 h, or 24 h. Untreated cell cultures served as controls (0 h). Cell culture supernatants were subjected to a cytokine antibody array. (A) The layout shows the locations of each antibody in the array membrane. (B) The cytokine array image shows the results of one of two independent experiments obtained by exposure of membranes to X-ray film. (C and D) Average net optical intensities (mean ± standard deviation; n = 3) in optical density units (odu) for each cytokine spot induced by P. gingivalis at the indicated time points. P.g., P. gingivalis.
Differential effects of P. gingivalis and its purified components on the induction of cytokines in human monocytes.
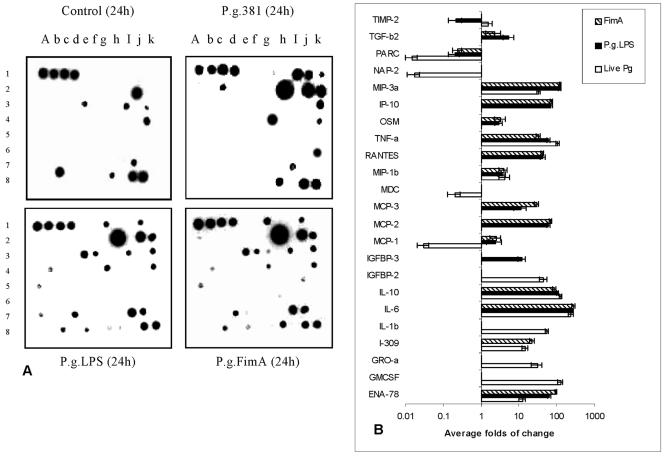
Following the initial infection, P. gingivalis can release its cell surface components such as LPS and FimA, which contribute to the progression of periodontitis. To establish whether P. gingivalis stimulated a profile of cytokine expression different from its LPS, compared to its FimA protein, in human monocytes, cultured monocyte-derived macrophages were treated with intact microorganisms, purified LPS, or purified FimA for 24 h and cytokine induction detected by cytokine array was compared with that of parallel unstimulated cell populations (Fig. 2). In these macrophage cultures, untreated controls were found constitutively to express IL-8, PARC, MCP-1, MDC, MIP-1β, angiogenin, NAP-2, and TIMP-2 (Fig. 2A), while all of the other cytokines in the array were undetectable. Expression levels of IL-6, IL-10, TNF-α, MIP-3α, MIP-1β, and ENA-78 in macrophages exposed to live P. gingivalis, purified LPS, and FimA were similar at 24 h, with IL-6, IL-10, TNF-α, and MIP-3α being the dominant cytokines, whether triggered by P. gingivalis or by its components. A constitutively expressed cytokine, PARC, was not detectable either with P. gingivalis or with its two purified components after 24 h of exposure. However, expression of IL-1β, GM-CSF, GRO-α, and IGFBP-2 was induced only by live P. gingivalis, whereas, inversely, expression of IP-10, RANTES, MCP-1, MCP-2, MCP-3, OSM, and TGF-β2 was induced only by its purified components LPS and FimA (Fig. 2B and C). MCP-1 was induced by P. gingivalis after a 4-h exposure but was down-regulated after 24 h of exposure (Fig. 1). Both P. gingivalis and its FimA protein induced I-309/CCL-1, while LPS induced IGFBP-3. TIMP-2 was up-regulated by live P. gingivalis but down-regulated by the purified LPS.
FIG. 2.
Differential cytokine profiles of freshly elutriated human peripheral blood monocytes infected with P. gingivalis or exposed to its purified LPS or FimA protein. Freshly elutriated human monocytes were cultured to enable differentiation into macrophages and then exposed to live P. gingivalis bacteria at an MOI of 25:1; parallel cell cultures were exposed to 10 μg/ml of either P. gingivalis LPS or P. gingivalis FimA for 24 h. Untreated cell cultures were used as controls. Cell culture supernatants were subjected to a cytokine antibody array. (A) Cytokine array image showing the results of one of two independent experiments that found similar patterns of expression; the spot orientation is as presented in Fig. 1A. (B) Cytokines whose average changes in level (mean ± standard deviation; n = 3) were greater than twofold in P. gingivalis-treated, LPS-treated, or FimA-treated cells relative to control cells are shown. Cytokines were quantified by net optical intensities with Bio-Rad VersaDoc Imaging System 3000 and analyzed by Quantity One software. If the cytokine spot was undetectable, its optical intensity was designated 1. P.g., P. gingivalis.
In order to check the reliability of the relative differences observed between cultures treated with P. gingivalis and with its components, we quantified the levels of TNF-α, IL-6, and IL-10, three cytokines that showed large fold differences in levels among cultures. The actual TNF-α, IL-6, and IL-10 levels in the cell supernatants used in the antibody arrays were determined by a conventional ELISA, as described in Materials and Methods. The actual TNF-α, IL-6, and IL-10 levels (in pg/ml) showed the same patterns seen in the arrays (Table 1). The levels of TNF-α, IL-6, and IL-10 detected by ELISA correlated well with the net optical intensity differences seen in the antibody array experiments. The linear correlation coefficients (r values) for TNF-α, IL-6, and IL-10 were 0.9911, 0.9936, and 0.9962, respectively.
TABLE 1.
Comparison of cytokine levels obtained by cytokine antibody array analysis and by ELISA
| Cell culture treatment | Level (mean ± SD) ofa:
|
|||||
|---|---|---|---|---|---|---|
| TNF-α
|
IL-6
|
IL-10
|
||||
| Cytokine array (odu/mm2) | ELISA (pg/ml) | Cytokine array (odu/mm2) | ELISA (pg/ml) | Cytokine array (odu/mm2) | ELISA (pg/ml) | |
| None | — | 34 ± 12 | — | 54 ± 21 | — | 28 ± 14 |
| LPS | 79.6 ± 8.2 | 1,493 ± 42 | 253.3 ± 32.5 | 4,244 ± 166 | 102.3 ± 8.6 | 1,992 ± 104 |
| FimA | 59.6 ± 8.6 | 996 ± 84 | 236.4 ± 48.1 | 3,445 ± 82 | 84.8 ± 14.2 | 1,532 ± 98 |
| Live P.g. | 102.4 ± 42.8 | 1,943 ± 238 | 272.3 ± 26.4 | 4,435 ± 296 | 126.9 ± 17.1 | 2,231 ± 312 |
The cytokine spot intensities in array membranes were quantified by optical density unit (odu) with Bio-Rad VersaDoc Imaging System 3000 and analyzed with Quantity One software. —, no spot detected in the array membrane.
Identification of differentially expressed proteins induced by P. gingivalis or its purified components as revealed by PowerBlot analysis.
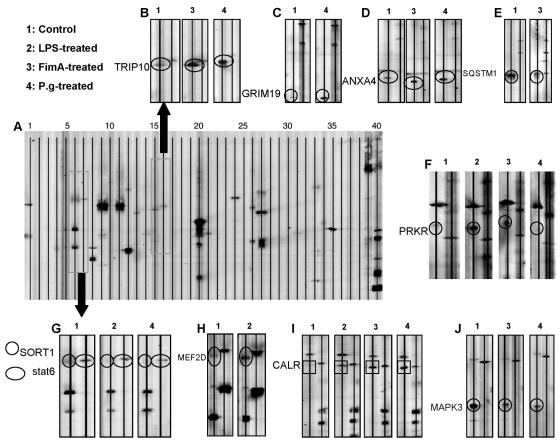
Extracts of human monocyte-derived macrophages grown for 24 h in control medium or in media supplemented with live P. gingivalis, LPS, or FimA were subjected to PowerBlot analysis. A representative blot of proteins extracted from untreated cells is shown in Fig. 3A. Enlarged parts of this control blot compared with corresponding regions from blots derived from P. gingivalis-treated or cell surface component-treated cells demonstrating changes (increases or decreases) in the levels of several proteins (circled or enclosed within rectangles) are presented in Fig. 3B through J. The analysis of the PowerBlot antibody array data indicated that P. gingivalis and its purified components induced both increases and decreases in the levels of specific proteins. Thus, treatment of the human macrophages with live P. gingivalis, its LPS, or its FimA protein led to up-regulation of 12, 8, and 10 proteins and down-regulation of 15, 8, and 17 proteins, respectively (Table 2). Included in this table are proteins involved in cell signaling, cell adhesion, cell growth, gene transcription, DNA repair, apoptosis, and other fundamental cellular processes. The changes induced in protein levels by P. gingivalis and its purified LPS and FimA could be categorized into several types: (i) changes that were similar for the three stimuli, (ii) changes that were induced by two of the three stimuli, (iii) changes that were induced by only one of the stimuli, and (iv) changes that were modulated uniquely by each stimulus.
FIG. 3.
PowerBlot analysis of total cell protein from untreated cells and from cells treated with P. gingivalis or its purified components. Freshly elutriated human monocytes were grown in multiple 10-cm-diameter dishes in medium and were exposed to either live P. gingivalis bacteria at an MOI of 25:1 or to 10 μg/ml of either P. gingivalis LPS or P. gingivalis FimA for 24 h. Untreated cell cultures were used as controls. After 24 h, the cells were harvested and the total protein fraction was extracted as described in Materials and Methods. Samples of the proteins were subjected to slab gel electrophoresis and PowerBlot analysis, as described in Materials and Methods. (A) A blot of proteins extracted from a culture of untreated (control) cells developed by the chemiluminescence method. The lines and numbers above the blot represent the 40 lanes used for hybridization with a mixture of four or five antibodies per lane. (B through J) Segments of blots containing two lanes each from blots of different protein extracts from untreated cultures (lanes 1), LPS-treated cultures (lanes 2), FimA-treated cultures (lanes 3), or live P. gingivalis-treated cultures (lanes 4). The proteins showing differential levels are circled or included within a rectangle on the blots. P.g., P. gingivalis.
TABLE 2.
Modulated proteins as determined by PowerBlot analysis
| Protein name | Description | Locus Link ID | Hugo ID | Change in level bya:
|
||
|---|---|---|---|---|---|---|
| LPS | FimA | P. gingivalis | ||||
| Cell growth and maintenance | ||||||
| GNPNAT1 | Glucosamine-phosphate N-acetyltransferase 1 | 64841 | 19980 | 7.42 + 2 | ||
| CAPZA2 | Capping protein (actin filament) muscle Z-line, alpha 2 | 12343 | 1490 | 2.16 + 5 | ||
| RBBP4 | Retinoblastoma binding protein 4 | 5928 | 9887 | 1.82 + 4 | ||
| CLTC | Clathrin, heavy polypeptide (Hc) | 54241 | 2092 | 2.15 − 5 | 1.77 − 4 | 3.39 − 5 |
| CDC42 | GTP-binding protein G25K, brain | 998 | 1736 | 7.59 + 2 | ||
| ENAH | Enabled homolog (Drosophila) | 13800 | 18271 | 9.32 − 2 | ||
| BCCIP | BRCA2 and CDKN1A interacting protein | 56647 | 978 | 6.68 − 2 | ||
| GRIM19 | Cell death-regulatory protein GRIM19 | 67184 | 2.44 + 5 | |||
| CUL2 | Cullin 2 | 8453 | 2552 | 6.82 + 2 | 8.74 + 2 | |
| UBE2L3 | Ubiquitin-conjugating enzyme E2, 18-kDa UBCH7 | 7332 | 12488 | 2.39 + 2 | ||
| PPP3CA | Phosphoprotein phosphatase 3-alpha catalytic chain | 3330 | 9314 | 4.17 + 2 | ||
| TUBA1 | Tubulin, alpha 1 (testis specific) | 64158 | 12407 | 3.22 − 5 | 2.49 − 1 | |
| KIF3A | Kinesin family member 3A | 11127 | 6319 | 6.95 − 2 | 3.03 − 2 | |
| GGA2 | Golgi associated, ARF binding protein 2 | 74105 | 16064 | 3.67 − 5 | 7.78 − 5 | |
| Protein kinase activity | ||||||
| AURKC | Aurora kinase C | 6795 | 11391 | 3.86 + 2 | ||
| MAPK3 | Mitogen-activated protein kinase 3 | 50689 | 6877 | 2.31 − 2 | 4.20 − 5 | |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 116596 | 6860 | 5.39 − 2 | 1.92 − 2 | |
| PRKR | dsRNA-activated protein kinase | 5610 | 9437 | 3.7 + 2 | 5.59 − 5 | 3.04 − 2 |
| CDC2 | Protein kinase cdc2 | 983 | 1722 | 3.60 − 2 | ||
| DCAMKL1 | Doublecortin and CaM kinase-like 1 | 83825 | 2700 | 2.78 − 2 | ||
| DMPK | Dystrophia myotonica-protein kinase | 1760 | 2933 | 4.66 + 2 | ||
| MAP2K4 | Mitogen-activated protein kinase kinase 4 | 6416 | 6844 | 2.31 − 2 | 2.55 − 5 | |
| Transcription factor activity | ||||||
| HSF4 | Heat shock transcription factor 4 | 3299 | 5227 | 3.29 + 2 | ||
| MEF2D | MADS box transcription enhancer factor 2, polypeptide D | 17261 | 6997 | 2.62 + 5 | ||
| STAT3 | Signal transducer and activator of transcription 3 | 104593 | 11364 | 2.06 − 2 | 2.88 + 5 | |
| SRF | Transcription factor SRF | 6722 | 11291 | 5.58 − 5 | 15.04 − 5 | |
| STAT6 | Signal transducer and activator of transcription 6 | 6778 | 11368 | 3.46 − 5 | ||
| STAT1 | Interferon-dependent positive-acting transcription factor | 6772 | 11362 | 3.35 − 5 | 16.63 − 5 | |
| ILF3 | Interleukin enhancer binding factor 3 | 6038 | 3807 | 2.43 + 2 | ||
| Translation regulator | ||||||
| SKP1A | S-phase kinase-associated protein 1A | 6500 | 10899 | 5.64 + 5 | ||
| Protein transport | ||||||
| CENTB2 | Centaurin, beta 2 | 16469 | 23527 | 4.22 − 5 | ||
| SORT1 | Sortilin 1/Neurotensin receiptor 3 | 6272 | 11186 | 3.04 − 5 | 10.58 − 5 | |
| Immune response | ||||||
| TNFSF6 | Fas antigen ligand | 356 | 11936 | 2.22 + 2 | ||
| BDKRB2 | Bradykinin receptor B2 | 624 | 1030 | 3.54 − 5 | ||
| BECN1 | Beclin 1 (coiled-coil, myosin-like BCL2 interacting protein) | 8678 | 1034 | 12.48 + 5 | ||
| IFIT4 | Interferon-induced protein with tetratricopeptide repeats 4 | 3437 | 5411 | Div/0 + 5 | ||
| Structural constituent of cytoskeleton | ||||||
| KATNB1 | Katanin p80 (WD repeat containing) subunit B 1 | 6217 | 26828 | 5.71 + 2 | 12.37 + 2 | |
| SQSTM1 | Sequestosome 1 | 8878 | 11280 | 2.87 − 5 | ||
| Adaptor | ||||||
| TDE1 | Tumor differentially expressed 1 | 98830 | 11699 | 5.10 − 2 | ||
| CAV1 | Caveolin | 857 | 1527 | 3.36 − 2 | ||
| Calcium signaling | ||||||
| ANXA4 | Annexin A4 | 307 | 542 | 1.65 + 4 | 2.05 + 5 | |
| ITGAV | Integrin, alpha V | 3685 | 9909 | 2.21 + 2 | ||
| CALR | Calreticulin precursor | 12317 | 2867 | 2.98 + 5 | 5.08 + 5 | 6.15 + 5 |
| Other | ||||||
| LMNA | Lamin A/C | 4000 | 6636 | 15.57 + 2 | ||
| RABGEF1 | RAB guanine nucleotide exchange factor 1 | 56715 | 17676 | 3.01 − 2 | ||
| CENTA1 | Centaurin, alpha 1 | 231821 | 16486 | Div/0 − 5 | ||
| RAB27A | RAB27A, member RAS oncogene family | 9766 | 2255 | 2.29 − 2 | 2.51 − 2 | 2.31 − 2 |
| RAP2A | RAP2A, member of RAS oncogene family | 9861 | 4212 | 8.84 − 5 | ||
| PTPN1 | Protein tyrosine phosphatase, non-receptor type 1 | 5770 | 9642 | 3.05 + 5 | ||
Changes of greater than twofold relative to controls in cells treated with P. gingivalis or its components that were reproduced at least twice by PowerBlot analysis are indicated as follows: blank, no change; +, increase; −, decrease; numbers on the left side of each plus or minus sign, average fold change; numbers on the right, confidence level of change; Div/0, the protein is present in the treated sample but absent in the control, and fold change is unmeasurable.
Validation of PowerBlot results by conventional Western blotting.
To confirm the PowerBlot data, we selected eight proteins (ILF3, STAT1, MAPK3, MEF2D, TNFSF6, CALR, STAT6, and PRKR) with various levels of differential expression and representing different confidence levels of changes detected by PowerBlot for analysis by conventional Western blotting (Fig. 4). With one exception (LPS-induced MAPK3), the conventional Western blotting analysis confirmed the differential expression of these eight proteins, which are listed in Table 3. The amounts of MEF2D, CALR, and PRKR induced by LPS relative to controls increased 2.03-, 2.54-, and 3.64-fold, respectively (Fig. 4), concordant with their 2.62-, 2.98-, and 3.7-fold differential expressions observed in the PowerBlot analysis results (Table 2). The amounts of ILF3, TNFSF6, CALR, PRKR, STAT1, and MAPK3 induced by FimA relative to controls increased 1.53-, 1.68-, 5.62-, 1.87-, 0.64-, and 0.37-fold, respectively (Fig. 4), in good agreement with their 1.43-, 2.22-, 2.98-, and 5.59-fold increases and 3.35- and 2.31-fold decreases, respectively, observed in the PowerBlot analysis results (Table 2). The relative ratios of STAT1, MAPK3, STAT6, PRKR, and CALR to controls induced by P. gingivalis were 0.08, 0.43, 0.32, 0.66, and 4.21, respectively (Fig. 4); these are comparable with the 16.63-, 4.20-, and 3.46-fold decrease and the 6.15-fold increase in differential expression observed for these proteins, respectively, in the PowerBlot analysis. MAPK3 induced by LPS was the only exception that was observed: its levels decreased according to the Western blot analysis but were unchanged according to the PowerBlot analysis. P. gingivalis, its purified LPS, and FimA all increased CALR expression in human monocytes in the PowerBlot analysis. This was also confirmed by two-dimensional gel electrophoresis followed by mass spectrometry (data not shown).
FIG. 4.
Conventional Western blotting used for validation of PowerBlot data. Freshly elutriated human monocytes were cultured and treated with P. gingivalis or with its purified components, and their total protein fractions were extracted. Samples containing ∼50 μg protein were subjected to Western blotting as described in Materials and Methods. Each blot was rehybridized with antiactin antibodies to compare loadings in different lanes. The results of chemiluminescence analysis of the blots are presented. The numbers above the lanes indicate average ratios relative to controls, normalized (with β-actin) using the Bio-Rad image analysis program. Asterisks indicate changes that were statistically significant, as determined by the Student t test (P < 0.05; n = 3). P.g., P. gingivalis.
TABLE 3.
Proteins analyzed by conventional Western blotting to validate their differential levels in cells treated with P. gingivalis or its components and in untreated cells
| Protein | Description | Locus Link ID | Hugo ID | Change in level bya:
|
||
|---|---|---|---|---|---|---|
| LPS | FimA | P. gingivalis | ||||
| ILF3 | Interleukin enhancer binding factor 3 | 6038 | 3807 | NC | + | NC |
| STAT1 | Interferon-dependent positive-acting transcription factor | 6772 | 11362 | NC | − | − |
| MAPK3 | Mitogen-activated protein kinase 3 | 50689 | 6877 | −/NC | − | − |
| MEF2D | MADS box transcription enhancer factor 2, polypeptide D | 17261 | 6997 | + | NC | NC |
| TNFSF6 | Fas antigen ligand | 356 | 11936 | NC | + | NC |
| CALR | Calreticulin precursor | 12317 | 2867 | + | + | + |
| STAT6 | Signal transducer and activator of transcription 6 | 6778 | 11368 | NC | NC | − |
| PRKR | dsRNA-activated protein kinase | 5610 | 9437 | + | + | − |
Changes in protein levels in cells treated with P. gingivalis or its components relative to controls are indicated as follows: NC, no change detected by PowerBlot or Western blot analysis; +, increase in protein levels detected by both PowerBlot and Western blot analyses; −, decrease in protein levels detected by both PowerBlot and Western blot analyses; −/NC, decrease in protein levels detected by Western blotting but no change detected by PowerBlot analysis.
DISCUSSION
The innate immune response to P. gingivalis is initiated when the microorganisms interact with cell surface pattern receptors, including TLRs, to signal and/or become internalized. Recognition occurs largely through the bacterial cell surface constituents, such as LPS and fimbriae, and those cell surface constituents bind to TLR2 and also may bind to the glycosylphosphatidylinositol-linked protein CD14 (9, 10, 48) to induce inflammatory responses. It is thought that many of the effects of bacteria on host immune and inflammatory responses are mediated by changes in gene transcription. Indeed, changes in gene expression have been identified in cells treated with P. gingivalis and its purified LPS or fimbriae using differential display (35) and cDNA array (15) techniques, both of which are focused on mRNA analysis. Changes at the mRNA level may not correspond to changes in protein levels or to changes in activity, especially when those depend on posttranslational modifications and/or protein stability. Therefore, it is important to investigate the effects of P. gingivalis and its components on protein levels. Thus far, few reports have addressed the effects of P. gingivalis and its components on the modulation of proteins in human peripheral monocytes using large-scale proteomics. We designed this study to compare the effects of P. gingivalis and its components to see whether they cause distinct biological effects in human peripheral monocyte-derived macrophages. This article provides the first description of host proteins by P. gingivalis and its components LPS and FimA.
Using cytokine antibody arrays, we first investigated cytokines, the major inflammatory mediators induced by live P. gingivalis. At the initial interaction with macrophages, P. gingivalis induced chemokines (e.g., MCP-1, MIP-1β, and PARC) involved in macrophage and neutrophil recruitment and in the antigen-specific T helper cell type 1 immune response (14, 47). However, this immune response may be suppressed after 24 h of exposure to P. gingivalis, since those chemokines, along with DMC/CCL22 and NAP-2, were dramatically reduced or disappeared completely after that time (Fig. 1 and 2). Following the initial response, expression of an extensive set of cytokines (including TNF-α, IL-6, and IL-1β) and other mediators transiently escalated subsequent to infection. Simultaneously, increases in IL-10 and TGF-β may have contributed to the dampening of the initial activation response (3, 4). TNF-α and IL-1β, proinflammatory cytokines that activate multiple signal transduction pathways to influence both immune and nonimmune cell functions, are known to inhibit macrophage apoptosis (24). This agrees with previous observations that infection with P. gingivalis appears to protect macrophages from apoptosis (30, 33, 36, 50), thereby maintaining the pool of infected and infectible targets.
The immune and inflammatory responses to live bacteria may not be triggered simply by bacterial cell surface components, although these are among the best-characterized pattern recognition receptor ligands. Our cytokine antibody array data show that LPS and FimA induce similar patterns of cytokine production by human monocyte-derived macrophages; however, the patterns of cytokines produced by human macrophages stimulated by LPS or FimA differed substantially from that observed when the cells were challenged with live P. gingivalis (Fig. 2). It seems that the host defense system senses live bacteria differently than individual bacterial constituents, and thus it launches a different immune response. Our data are consistent with previous studies demonstrating that both P. gingivalis LPS and FimA signal through a common pattern recognition receptor, TLR2, whereas live P. gingivalis signals through both TLR2 and TLR4 in the induction of cytokine production by mouse peritoneal macrophages (9, 52).
In order to identify intracellular signaling pathways activated during P. gingivalis infection, we identified intracellular proteins modulated in human macrophages after interaction with live P. gingivalis and its by-products, LPS and FimA, by using the PowerBlot array screening system. The analysis demonstrates differences in the levels of several proteins after macrophage exposure to P. gingivalis, its LPS, or its FimA protein (Table 2). These proteins included those associated with immune response, both Ser/Thr and Tyr protein kinases, heat shock response proteins, transcription factors, DNA repair enzymes, and proteins engaged in vesicular transport, apoptosis, cell cycle, and cell adhesion. Additional studies are required to determine whether some or any of the changes in the levels of proteins are due to transcriptional regulation via ligand-specific receptors and whether they are the cause or consequence of the biological effects. It is noteworthy that we have analyzed changes that occurred after 24 h of cell treatment, and so some of the changes may be earlier than changes in cell cycle or cell viability. Here we have elected to highlight several of the modulated proteins that may be related to the mechanisms underpinning the effects of P. gingivalis and its constituents on infection, inflammation, and immune response.
We observed that similar changes in CLTC, CALR, RAB27A, and MAPK3 levels were induced by P. gingivalis, its LPS, and its FimA protein, suggesting a common mechanism of action. CLTC, a clathrin heavy-chain protein, was down-regulated after exposure to all three treatments, which may result in attenuated macrophage chemotaxis and phagocytosis (18, 46). A Ca2+ binding protein, CALR, which was reported to modulate the host immune response by binding to complement C1q, thereby participating in the inflammatory processes associated with vascular or atherosclerotic lesions, autoimmune diseases, or infections (7), was up-regulated by these three stimuli. The GTP-binding protein RAB27A, which appears to be a key effector of cytotoxic granule exocytosis, a pathway essential for immune homeostasis (28), was also down-regulated under all three conditions. P. gingivalis, its LPS, and its FimA protein all repressed the basal expression of MAPK3 (ERK1) in macrophages; suppression of MAPK3 was reported to facilitate an adaptive regulation in the process of cell proliferation under cell growth arrest conditions (11). All of these functions may favor its establishment and propagation within the host after P. gingivalis infection or enhance the host's chronic inflammatory reactions.
Purified LPS from P. gingivalis specifically induced MEF2D, IFIT4, and CDC42. MEF2D is a member of the myocyte enhancer factor 2 family of transcription factors and has been shown to increase CD14 expression in monocytes/macrophages (34); up-regulation of the MEF2D protein by P. gingivalis LPS may enhance macrophage responses to LPS by possibly increasing CD14 expression. That LPS up-regulates the interferon-inducible antiviral protein IFIT4 was also found by other investigators (16); however, its role in biological processes and molecular function here is still unknown and needs further investigation. The GTPase CDC42 is a GTP-binding protein that is involved in macrophage phagocytosis via the calcium-promoted Ras inactivator (51). However, P. gingivalis LPS may not induce phagocytosis and oxidative burst by activating CDC42, because we did not detect the up-regulation of the calcium-promoted Ras inactivator in macrophages treated with P. gingivalis LPS.
FimA has been shown to up-regulate TNFSF6 (CD95L) and ILF3. A type II transmembrane protein of the TNF family of death factors, TNFSF6 has shown to be associated with the establishment of immune privilege and tumor survival (39). Interaction between TNFSF6 and its receptor can regulate key events in cellular activation, proliferation, differentiation, cell death, and survival of immune cells and other tissues (22). ILF3 is a transcription factor required for T-cell expression of IL-2 (25). ILF3 binds to a sequence in the IL-2 enhancer known as antigen receptor response element 2. In addition, ILF3 can bind RNA and is an essential component for encapsidation and protein priming of hepatitis B viral polymerase (17). That FimA induced TNFSF6 and ILF3 expression may indicate that it plays an important role in affecting the host immune system.
Live P. gingivalis dramatically down-regulated expression of the transcription factor STAT6. This is an intriguing finding, in light of other data that STAT6 deficiency confers enhanced immunity against tumors (44) as well as against parasite infection (40, 43). STAT6-deficient mice generate excess nitric oxide in their macrophages compared to the macrophages from wild-type mice, which produce arginase instead of nitric oxide (44). Nitric oxide-producing macrophages have a strong ability to induce antigen-specific CD4+ T-cell proliferation in response to unrelated antigens (41). It seems that during the acute infection stage (24 h after infection), P. gingivalis turns on nitric oxide production by macrophages as a host defense mechanism, accomplished by down-regulating the expression of STAT6.
The level of STAT1 was decreased in macrophages treated with either FimA or live P. gingivalis. STAT1 plays an essential role in the caspase-independent cell death of activated macrophages through the p38 MAPK/STAT1/reactive oxygen species pathway (20). Decreased STAT1 levels were reported to decrease superoxide production in macrophages (20). Down-regulation of STAT1 by P. gingivalis or by its FimA protein may be one of the several survival mechanisms used by P. gingivalis to evade host defenses.
STAT3 is a transcription factor mediating the anti-inflammatory properties of IL-10 (49). It has been reported that mice with a STAT3 deficiency display excessive local and systemic inflammation and fail to facilitate bacterial clearance (27). We noticed that P. gingivalis LPS down-regulated and FimA up-regulated STAT3 expression in macrophages; this may indicates that LPS plays a negative role while FimA plays a positive role in stimulation of anti-inflammatory activity and bacterial clearance during P. gingivalis infection. In addition, the STAT3 level in macrophages was not changed by whole bacterial treatment compared to that of untreated macrophages; it seems that the whole bacteria balanced the role of LPS and FimA on inducing STAT3 expression in macrophages.
PRKR, an interferon-induced, double-stranded RNA (dsRNA)-activated protein kinase (p68 kinase), acts as a tumor suppressor by actively participating in apoptosis (21). PRKR is also essential for antiviral activity by inhibiting both viral and cellular protein synthesis (19). Interestingly, it was increased only by LPS and decreased by both live P. gingivalis and FimA. The abilities of live P. gingivalis and FimA to decrease PRKR levels may be important for the antiapoptotic activities observed by other investigators (50). However, the ability of P. gingivalis LPS to increase PRKR levels may not related to apoptogenic activity, because P. gingivalis LPS also possesses antiapoptotic properties (30).
Another interesting observation of this study was the down-regulation of TPL-2 (MAP3K8) by live P. gingivalis and its LPS. P. gingivalis LPS is a TLR2 agonist; it was reported to inhibit a TLR4 agonist, E. coli LPS-induced TNF-α production, with IκB kinase activity being unperturbed (6). This TLR “heterotolerance” induced by P. gingivalis LPS may come about through down-regulation of TPL-2, via an IκB kinase-independent pathway, because TPL-2 is essential for TLR4 activation of the ERK MAPK cascade in E. coli LPS-stimulated macrophages and subsequent up-regulation of genes involved in innate immune responses (2). The down-regulation of TPL-2 may also be associated with the down-regulation of MAPK3 detected in this study, which could weaken the innate host immune responses to bacterial infection.
In summary, the large-scale screening of proteins modulated after treatment of human monocyte-derived macrophages with live P. gingivalis, its LPS, and its FimA protein has enabled us to identify proteins that are implicated in the effects of P. gingivalis and its components on the innate immune responses of human monocytes and macrophages. P. gingivalis and its components appear to stimulate different protein expression patterns in macrophages that may reflect intrinsic functional differences in these stimuli. This differential modulation of proteins in host cells may well coordinate with their specific roles in triggering host inflammatory/immune responses during P. gingivalis infection.
Acknowledgments
This work was supported by grant RO1 DE 15989 from NIDCR.
Editor: D. L. Burns
REFERENCES
- 1.Agarwal, S., N. P. Piesco, L. P. Johns, and A. E. Riccelli. 1995. Differential expression of IL-1 beta, TNF-alpha, IL-6, and IL-8 in human monocytes in response to lipopolysaccharides from different microbes. J. Dent. Res. 74:1057-1065. [DOI] [PubMed] [Google Scholar]
- 2.Beinke, S., M. J. Robinson, M. Hugunin, and S. C. Ley. 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 24:9658-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaillon, J. M. 2001. Pro- versus anti-inflammatory cytokines: myth or reality. Cell. Mol. Biol. (Noisy-le-Grand) 47:695-702. [PubMed] [Google Scholar]
- 4.Chen, W., and S. M. Wahl. 1999. Manipulation of TGF-beta to control autoimmune and chronic inflammatory diseases. Microbes Infect. 1:1367-1380. [DOI] [PubMed] [Google Scholar]
- 5.Darveau, R. P., S. Arbabi, I. Garcia, B. Bainbridge, and R. V. Maier. 2002. Porphyromonas gingivalis lipopolysaccharide is both agonist and antagonist for p38 mitogen-activated protein kinase activation. Infect. Immun. 70:1867-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrovolskaia, M. A., A. E. Medvedev, K. E. Thomas, N. Cuesta, V. Toshchakov, T. Ren, M. J. Cody, S. M. Michalek, N. R. Rice, and S. N. Vogel. 2003. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-kappa B signaling pathway components. J. Immunol. 170:508-519. [DOI] [PubMed] [Google Scholar]
- 7.Ghebrehiwet, B., and E. I. Peerschke. 2004. cC1q-R (calreticulin) and gC1q-R/p33: ubiquitously expressed multi-ligand binding cellular proteins involved in inflammation and infection. Mol. Immunol. 41:173-183. [DOI] [PubMed] [Google Scholar]
- 8.Graves, D. T., H. Al-Mashat, and R. Liu. 2004. Evidence that diabetes mellitus aggravates periodontal diseases and modifies the response to an oral pathogen in animal models. Compend. Contin. Educ. Dent. 25:38-45. [PubMed] [Google Scholar]
- 9.Hajishengallis, G., M. Martin, H. T. Sojar, A. Sharma, R. E. Schifferle, E. DeNardin, M. W. Russell, and R. J. Genco. 2002. Dependence of bacterial protein adhesins on toll-like receptors for proinflammatory cytokine induction. Clin. Diagn. Lab. Immunol. 9:403-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harokopakis, E., and G. Hajishengallis. 2005. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur. J. Immunol. 35:1201-1210. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, R., F. Garcia, I. Encio, and C. De Miguel. 2004. Promoter analysis of the human p44 mitogen-activated protein kinase gene (MAPK3): transcriptional repression under nonproliferating conditions. Genomics 84:222-226. [DOI] [PubMed] [Google Scholar]
- 12.Hirose, K., E. Isogai, and I. Ueda. 2000. Porphyromonas gingivalis fimbriae induce adhesion of monocytic cell line U937 to endothelial cells. Microbiol. Immunol. 44:17-22. [DOI] [PubMed] [Google Scholar]
- 13.Honig, J., C. Rordorf-Adam, C. Siegmund, W. Wiedemann, and F. Erard. 1989. Increased interleukin-1 beta (IL-1 beta) concentration in gingival tissue from periodontitis patients. J. Periodontal Res. 24:362-367. [DOI] [PubMed] [Google Scholar]
- 14.Huang, D. R., J. Wang, P. Kivisakk, B. J. Rollins, and R. M. Ransohoff. 2001. Absence of monocyte chemoattractant protein 1 in mice leads to decreased local macrophage recruitment and antigen-specific T helper cell type 1 immune response in experimental autoimmune encephalomyelitis. J. Exp. Med. 193:713-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, G. T., H. B. Zhang, H. N. Dang, and S. K. Haake. 2004. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 37:303-312. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, K., M. Kurita-Taniguchi, M. Aoki, T. Kimura, Y. Kashiwazaki, M. Matsumoto, and T. Seya. 2005. Gene-inducing program of human dendritic cells in response to BCG cell-wall skeleton (CWS), which reflects adjuvancy required for tumor immunotherapy. Immunol. Lett. 98:280-290. [DOI] [PubMed] [Google Scholar]
- 17.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janciauskiene, S., and S. Lindgren. 1999. Human monocyte activation by cleaved form of alpha-1-antitrypsin involvement of the phagocytic pathway. Eur. J. Biochem. 265:875-882. [DOI] [PubMed] [Google Scholar]
- 19.Katze, M. G., J. Tomita, T. Black, R. M. Krug, B. Safer, and A. Hovanessian. 1988. Influenza virus regulates protein synthesis during infection by repressing autophosphorylation and activity of the cellular 68,000-Mr protein kinase. J. Virol. 62:3710-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, H. S., and M. S. Lee. 2005. Essential role of STAT1 in caspase-independent cell death of activated macrophages through the p38 mitogen-activated protein kinase/STAT1/reactive oxygen species pathway. Mol. Cell. Biol. 25:6821-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. B., and M. Esteban. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104:487-501. [DOI] [PubMed] [Google Scholar]
- 23.Makris, G. P., and J. L. Saffar. 1986. Destruction and repair of periodontal tissues during burst episodes of activity in hamster periodontitis. J. Biol. Buccale 14:101-113. [PubMed] [Google Scholar]
- 24.Mangan, D. F., G. R. Welch, and S. M. Wahl. 1991. Lipopolysaccharide, tumor necrosis factor-alpha, and IL-1 beta prevent programmed cell death (apoptosis) in human peripheral blood monocytes. J. Immunol. 146:1541-1546. [PubMed] [Google Scholar]
- 25.Marcoulatos, P., E. Avgerinos, D. V. Tsantzalos, and N. C. Vamvakopoulos. 1998. Mapping interleukin enhancer binding factor 3 gene (ILF3) to human chromosome 19 (19q11-qter and 19p11-p13.1) by polymerase chain reaction amplification of human-rodent somatic cell hybrid DNA templates. J. Interferon Cytokine Res. 18:351-355. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M., R. E. Schifferle, N. Cuesta, S. N. Vogel, J. Katz, and S. M. Michalek. 2003. Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J. Immunol. 171:717-725. [DOI] [PubMed] [Google Scholar]
- 27.Matsukawa, A., K. Takeda, S. Kudo, T. Maeda, M. Kagayama, and S. Akira. 2003. Aberrant inflammation and lethality to septic peritonitis in mice lacking STAT3 in macrophages and neutrophils. J. Immunol. 171:6198-6205. [DOI] [PubMed] [Google Scholar]
- 28.Menasche, G., E. Pastural, J. Feldmann, S. Certain, F. Ersoy, S. Dupuis, N. Wulffraat, D. Bianchi, A. Fischer, F. Le Deist, and G. de Saint Basile. 2000. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 25:173-176. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, Y., S. Hanazawa, A. Watanabe, K. Naganuma, H. Iwasaka, K. Kawakami, and S. Kitano. 1994. Porphyromonas gingivalis fimbriae induce a 68-kilodalton phosphorylated protein in macrophages. Infect. Immun. 62:5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, D. A., and J. M. Wilton. 2003. Lipopolysaccharide from the periodontal pathogen Porphyromonas gingivalis prevents apoptosis of HL60-derived neutrophils in vitro. Infect. Immun. 71:7232-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa, I., A. Amano, H. Inaba, S. Kawai, and S. Hamada. 2005. Inhibitory effects of Porphyromonas gingivalis fimbriae on interactions between extracellular matrix proteins and cellular integrins. Microbes Infect. 7:157-163. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa, T., Y. Asai, M. Hashimoto, and H. Uchida. 2002. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 32:2543-2550. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki, K., and S. Hanazawa. 2001. Porphyromonas gingivalis fimbriae inhibit caspase-3-mediated apoptosis of monocytic THP-1 cells under growth factor deprivation via extracellular signal-regulated kinase-dependent expression of p21 Cip/WAF1. Infect. Immun. 69:4944-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, S. Y., H. M. Shin, and T. H. Han. 2002. Synergistic interaction of MEF2D and Sp1 in activation of the CD14 promoter. Mol. Immunol. 39:25-30. [DOI] [PubMed] [Google Scholar]
- 35.Park, Y., O. Yilmaz, I. Y. Jung, and R. J. Lamont. 2004. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect. Immun. 72:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preshaw, P. M., R. E. Schifferle, and J. D. Walters. 1999. Porphyromonas gingivalis lipopolysaccharide delays human polymorphonuclear leukocyte apoptosis in vitro. J. Periodontal Res. 34:197-202. [DOI] [PubMed] [Google Scholar]
- 37.Pulendran, B., P. Kumar, C. W. Cutler, M. Mohamadzadeh, T. Van Dyke, and J. Banchereau. 2001. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J. Immunol. 167:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reife, R. A., R. A. Shapiro, B. A. Bamber, K. K. Berry, G. E. Mick, and R. P. Darveau. 1995. Porphyromonas gingivalis lipopolysaccharide is poorly recognized by molecular components of innate host defense in a mouse model of early inflammation. Infect. Immun. 63:4686-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restifo, N. P. 2000. Not so Fas: re-evaluating the mechanisms of immune privilege and tumor escape. Nat. Med. 6:493-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Sosa, M., J. R. David, R. Bojalil, A. R. Satoskar, and L. I. Terrazas. 2002. Cutting edge: susceptibility to the larval stage of the helminth parasite Taenia crassiceps is mediated by Th2 response induced via STAT6 signaling. J. Immunol. 168:3135-3139. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Sosa, M., A. R. Satoskar, R. Calderon, L. Gomez-Garcia, R. Saavedra, R. Bojalil, and L. I. Terrazas. 2002. Chronic helminth infection induces alternatively activated macrophages expressing high levels of CCR5 with low interleukin-12 production and Th2-biasing ability. Infect. Immun. 70:3656-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saglie, F. R., J. Pertuiset, M. T. Rezende, M. Nestor, A. Marfany, and J. Cheng. 1988. In situ correlative immuno-identification of mononuclear infiltrates and invasive bacteria in diseased gingiva. J. Periodontol. 59:688-696. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki, Y., T. Yoshimoto, H. Maruyama, T. Tegoshi, N. Ohta, N. Arizono, and K. Nakanishi. 2005. IL-18 with IL-2 protects against Strongyloides venezuelensis infection by activating mucosal mast cell-dependent type 2 innate immunity. J. Exp. Med. 202:607-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha, P., V. K. Clements, S. Miller, and S. Ostrand-Rosenberg. 2005. Tumor immunity: a balancing act between T cell activation, macrophage activation and tumor-induced immune suppression. Cancer Immunol. Immunother. 54:1137-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoufi, E. D., M. A. Taubman, J. L. Ebersole, D. J. Smith, and P. P. Stashenko. 1987. Phenotypic analyses of mononuclear cells recovered from healthy and diseased human periodontal tissues. J. Clin. Immunol. 7:235-245. [DOI] [PubMed] [Google Scholar]
- 46.Terashima, Y., N. Onai, M. Murai, M. Enomoto, V. Poonpiriya, T. Hamada, K. Motomura, M. Suwa, T. Ezaki, T. Haga, S. Kanegasaki, and K. Matsushima. 2005. Pivotal function for cytoplasmic protein FROUNT in CCR2-mediated monocyte chemotaxis. Nat. Immunol. 6:827-835. [DOI] [PubMed] [Google Scholar]
- 47.Vitiello, P. F., M. G. Shainheit, E. M. Allison, E. P. Adler, and R. A. Kurt. 2004. Impact of tumor-derived CCL2 on T cell effector function. Immunol. Lett. 91:239-245. [DOI] [PubMed] [Google Scholar]
- 48.Vogel, S., M. J. Hirschfeld, and P. Y. Perera. 2001. Signal integration in lipopolysaccharide (LPS)-stimulated murine macrophages. J. Endotoxin Res. 7:237-241. [PubMed] [Google Scholar]
- 49.Williams, L., L. Bradley, A. Smith, and B. Foxwell. 2004. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 172:567-576. [DOI] [PubMed] [Google Scholar]
- 50.Yilmaz, O., T. Jungas, P. Verbeke, and D. M. Ojcius. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 72:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J., J. Guo, I. Dzhagalov, and Y. W. He. 2005. An essential function for the calcium-promoted Ras inactivator in Fcgamma receptor-mediated phagocytosis. Nat. Immunol. 6:911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Q., T. Desta, M. Fenton, D. T. Graves, and S. Amar. 2005. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect. Immun. 73:935-943. [DOI] [PMC free article] [PubMed] [Google Scholar]