Abstract
The nucleocapsid (N) protein of hantaviruses encapsidates both viral genomic and antigenomic RNAs, although only the genomic viral RNA (vRNA) is packaged into virions. To define the domain within the Hantaan virus (HTNV) N protein that mediates these interactions, 14 N- and C-terminal deletion constructs were cloned into a bacterial expression vector, expressed, and purified to homogeneity. Each protein was examined for its ability to bind the HTNV S segment vRNA with filter binding and gel electrophoretic mobility shift assays. These studies mapped a minimal region within the HTNV N protein (amino acids 175 to 217) that bound vRNA. Sequence alignments made from several hantavirus N protein sequences showed that the region identified has a 58% identity and an 86% similarity among these amino acid sequences. Two peptides corresponding to amino acids 175 to 196 (N1) and 197 to 218 (N2) were synthesized. The RNA binding of each peptide was measured by filter binding and competition analysis. Three oligoribonucleotides were used to measure binding affinity and assess specificity. The N2 peptide contained the major RNA binding determinants, while the N1 peptide, when mixed with N2, contributed to the specificity of vRNA recognition.
Viruses within the Hantavirus genus, family Bunyaviridae, can cause one of two serious illnesses in humans, hantavirus pulmonary syndrome or hemorrhagic fever with renal syndrome (19). Sin Nombre virus and Hantaan virus (HTNV) are the prototypic hantaviruses for each disease, respectively. While researchers continue to identify additional hantaviruses throughout the world (19), we know only a few details of the molecular and biochemical functions of the viral proteins that orchestrate the virus life cycle. Hantaviruses have a negative-strand, tripartite genome that encodes an RNA-dependent RNA polymerase (L segment), the nucleocapsid (N) protein (S segment), and the G1 and G2 glycoproteins (M segment) (20, 21). We have been interested in the defining the mechanism by which N protein binds viral RNA (vRNA), because this function is essential to its role in encapsidation and ribonucleoprotein (RNP) complex formation and might have regulatory roles in the viral life cycle. The N protein encapsidates genomic and antigenomic vRNAs; however, only the genomic vRNA is packaged into the mature virion (12). Each genomic vRNA associates with the N protein, and probably the viral polymerase, to create three distinct RNPs (22). Apart from the role it plays in the formation of the RNP, the hantavirus N protein has been suggested to play a functional role in replication and transcription. In particular, the N protein may modulate the switch of virus RNA synthesis from transcription to replication, as reported for analogous proteins in other negative-strand viruses (4, 11, 18).
The N protein interacts with the three different vRNAs—vRNA, cRNA, and mRNA—during replication; although the molecular basis for the differential activities is not completely understood. It is likely that specific sequences or structures present in the vRNA molecules are involved. These structures could provide a point of nucleation for subsequent encapsidation of the entire genomic (vRNA) or antigenomic (cRNA) RNA segment, but not the mRNA. Two in vitro studies have addressed the interaction of the N protein with RNA by measurement of the binding affinity of bacterially expressed and purified N protein with vRNA and non-vRNA. In one study, a specific interaction was not observed, although binding of vRNA to filter-immobilized N protein was reported (9). In the second study, the HTNV N protein demonstrated a preference for its S segment vRNA as compared to its binding with an RNA encoding the open reading frame (ORF) of the S segment (23). Furthermore, a strong preference was noted for the S segment vRNA as compared to nonspecific RNA (23). Additional experiments found the 5′ end of the S segment vRNA to be necessary and sufficient for the binding reaction (24). Recently, preferential binding of the Bunyamwera virus (BUNV) N protein to the 5′ end of the S segment vRNA has also been reported (17). BUNV is also a member of the Bunyaviridae family, genus Bunyavirus, and has a replication strategy similar to that of the hantaviruses. Both the HTNV and BUNV N proteins have similar dissociation constants (53 and 80 nM, respectively) for their S segment vRNA substrates in the presence of magnesium ions (17, 23). These values are comparable to the dissociation constant reported for the human immunodeficiency virus type 1 (HIV-1) nucleocapsid (NC)-SL2 complex, 110 ± 50 nM (3), which has been proposed to be a major determinant in genome recognition.
A specific RNA binding domain (RBD), one that shows preference for viral genomic RNAs, has not been defined for any hantavirus N protein. For HTNV and Puumala virus, a nonspecific RNA binding activity was reported to reside in the carboxyl terminus of their respective N proteins (9). To map the domain of the N protein that specifically interacts with viral nucleic acids, we prepared several deletion constructs encoding the HTNV N protein truncated at either the N terminus, the C terminus, or both. The constructs were expressed in Escherichia coli, and the protein products were purified by Ni2+-nitrilotriacetic acid (NTA) chromatography. The HTNV N protein and its derivatives were assayed for their binding affinity for vRNA to map the site in the N protein that specifically interacted with vRNA.
MATERIALS AND METHODS
Oligonucleotides and enzymes.
Oligonucleotide PCR primers were synthesized by Integrated DNA Technologies (Coralville, Iowa). Restriction enzymes and Vent polymerase were purchased from New England Biolabs, Beverly, Mass. T4 DNA ligase and kinase were purchased from Gibco-BRL (Grand Island, N.Y.). All chemicals were purchased from Sigma (St. Louis, Mo.).
Construction of HTNV N protein truncations NΔ90, NΔ209, NΔ300, CΔ130, and CΔ255.
PCR was used to amplify and construct deletions of the HTNV N protein ORF from pHTNV-N (23). Primers were designed to reamplify the entire plasmid yet delete the region of interest in the HTNV S segment ORF. Specific details are provided in the following paragraph. In general, PCR products were purified with QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.). The nucleotide sequences of all clones were confirmed by bidirectional sequencing with the LiCOR (Lincoln, Nebr.) model 4200 IR2 automated sequencer. Each clone was constructed with a hexahistidine tag at the C terminus, which has been previously shown to not interfere with the RNA binding properties of the N protein (9, 23).
Deletions were created in the N terminus by using a universal reverse primer (pET23b-R primer) and different forward primers that incorporated an NdeI site (shown in italics below) to facilitate removal of sequences in the 5′ end of the S segment ORF. The PCR products were recircularized with T4 DNA ligase, transformed into E. coli DH-5α cells (Life Technologies), and screened with NdeI. The pET23b-R primer (GACGTCATATGTATATCTCCTTCTTAAAGTTAAAC-3′) was used as the reverse primer for cloning NΔ90, NΔ209, and NΔ300 and contained sequence from 238 to 268 of the pET21b and pET23b plasmids (underlined). The forward primers were as follows: NΔ90 primer (5′-GACGTCATATGGAGAGGTCAATGCTCAGTTAT-3′), containing nucleotides 274 to 294 of the HTNV S segment ORF (underlined); NΔ209-5′ primer (5′-GACGTCATATGGGGCTCTACCCTGCACAG-3′), containing nucleotides 610 to 628 of the HTNV S segment ORF (underlined); and the NΔ300 primer (5′-GACGTCATATGGAGTCACCATCATCAATATGG-3′), containing nucleotides 897 to 918 of the HTNV S segment ORF (underlined).
Deletions were created in the C terminus by using a universal forward primer (pET23b-F primer) and different reverse primers which incorporated a XhoI site (shown in italics in each primer below) to facilitate removal of sequences in the 3′ end of the S segment ORF. The PCR products were recircularized with T4 DNA ligase, transformed into E. coli DH-5α cells, and screened with XhoI. pET 23b-F primer, 5′-ATCCGCTCGAGCACCACCACCACCACCAC-3′, containing a hexahistidine tag from the pET21b or pET23b plasmid(underlined), was used as a forward primer for cloning CΔ130 and CΔ255. The following primers were synthesized with an XhoI site at their 5′ end (italics): CΔ130-3′ primer (5′-ATCCGCTCGAGAATATCTTCAATCATGCTACAGCCAGC-3′), containing nucleotides 897 to 870 of the HTNV S segment ORF (underlined); and CΔ255-3′ (5′-GTACTGCTCGAGTGGTTTCCGGATACCGTTAACATC-3′), containing nucleotides 522 to 598 of the HTNV S segment ORF (underlined).
Construction of HTNV N proteins with N- and C-terminal deletions.
Vent polymerase (New England Biolabs) was used to PCR amplify each fragment. PCR products were digested with NdeI and XhoI and ligated with T4 DNA ligase directly into pET21b, which was also restriction digested with NdeI and XhoI. The nucleotide sequences of the clones were confirmed by bidirectional sequencing of the constructs and analysis with a LiCOR 4200 IR2 automated sequencer. In the following expression vector constructions, the numbers following NP refer to the HTNV N protein amino acid sequence. The 5′ primer used for NP137-214 was 5′-GGGACGTCATATGGCTCTGTATATGTTGACAACAAGGGG-3′, which contained nucleotides 409 to 435 of the HTNV S segment ORF (underlined) and an NdeI site at the 5′ end. The 3′ primer for NP137-214 was 5′-GTACTGCTCGAGCTGCCGTGCCTTAATCTGTGCAGG-3′, which contained nucleotides 642 to 618 of the HTNV S segment ORF (underlined) and an XhoI site at the 5′ end. The 5′ primer used for NP208-262 was 5′-CGACGTTCATATGGCACAGATTAAGGCACGGCAGATG-3′ and contained nucleotides 624 to 648 of the HTNV S segment ORF (underlined) and an NdeI site at the 5′ end. The 3′ primer for NP208-262 was 5′-GTACTGCTCGAGGTTTGTTGCAGGACCACCAAGGAG-3′ and contained nucleotides 786 to 762 of the HTNV S segment ORF (underlined) and an XhoI site at the 5′ end. The forward primer for NP constructs NP175-217, NP175-232, NP175-249, NP175-270, and NP175-300 was 5′-GTACTGGCTAGCAAACATCTTTACGTGTCCTTG-3′ and contained nucleotides 524 to 543 of the HTNV S segment ORF (underlined). The forward primer has an NheI site in the 5′ end that cloned the gene adjacent to a start codon in pET21B. The reverse primers for each of these constructs contained an XhoI site at the 5′ end and were as follows: NP175-300 (5′-GTACTGCTCGAGCTCAATATCTTCAATCATGC-3′), NP175-270 (5′-GTACTGCTCGAGTTGCCGCTGCCGTAAG-3′) NP175-249 (5′-GTACTGCTCGAGATCTGGAAGAAGCTTGC), NP175-232 (5′-GTACTGCTCGAGCCAGTCCTTTGCTAATG-3′) and NP175-217 (5′-GTACTGCTCGAGACTGATCATCTGCCGTG-3′). The sequence representing the HTNV S segment ORF is underlined.
Expression and purification of truncated wild-type protein and deletions of the HTNV N protein.
NΔ209/pET23b, NΔ300/pET23b, NP137-214/pET21b, NP208-262/pET21b, CΔ319/pRSET, NP213-316/pRSET, NP175-217/pET21b, NP175-232/pET21b, NP175-249/pET21b, NP175-270/pET21b, and NP175-300/pET21b were transformed into BL21(DE3) cells, and NΔ90/pET 21b, CΔ130/pET 21b, and CΔ255/pET 21b were transformed into HMS174(DE3) cells. CΔ319/pRSET and NP213-316/pRSET were gifts from Alexander Dekonenko, U.S. Army Medical Research Institute for Infectious Diseases (USAMRIID), Ft. Detrick, Frederick, Md. The wild-type HTNV N protein/pET21b was expressed in HMS174(DE3). The N protein and its derivatives were expressed with the T7 polymerase expression system and purified as hexahistidine-tagged fusion proteins from the insoluble fraction as described previously (23). The final preparation was dialyzed against 0.5 M NaCl, 20 mM HEPES (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 20% (vol/vol) glycerol. Protein concentrations were measured by the Bradford method (5) by the Bio-Rad Micro-Assay.
In vitro transcription and labeling of RNA substrates.
pGEM1/HTNVS, which contains the HTNV S segment cDNA cloned into the PstI site, was used for in vitro transcription of genomic vRNA as described previously (24). pGEM7Zf was used for in vitro transcription of a nonviral 67-nucleotide RNA used as a control RNA. Briefly, the plasmids were purified with the QIAquick miniprep kit (Qiagen) according to the manufacturer's protocol. pGEM1/HTNVS was digested with XbaI, and pGEM7Zf was digested with SmaI. Radiolabeled HTNV S segment RNA transcripts were produced from the linear DNAs with a MaxiScript Sp6 RNA transcription kit (Ambion, Austin, Tex.). The RNeasy kit (Qiagen) was used to purify transcripts. Purified RNA was stored at −80°C for up to 2 weeks.
Filter binding assay.
Purified HTNV N proteins and N protein deletions were serially diluted (22.7, 7.57, 2.52, 0.84, and 0.28 μM) in a final volume of 20 μl of binding buffer (40 mM HEPES [pH 7.4], 100 mM NaCl, 5% glycerol). The samples were incubated at 37°C for 5 min, 1 ng of [α-32P]UTP-labeled RNA was then added to each reaction mixture, and the reaction mixtures were incubated for an additional 15 min at 37°C. Binding reactions were slot blotted (Bio-Rad) onto nitrocellulose filters as previously described (23). Signals were quantitated with a Molecular Dynamics PhosphorImager and analyzed by ImageQuaNT version 4.2 software (Molecular Dynamics).
GEMSA.
For the gel electrophoresis mobility shift assay (GEMSA), 1 ng of 32P-radiolabeled vRNA S segment, prepared as described above, was incubated with 22.7 μM purified N protein or N protein deletion in binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 1 mM MgCl2, 0.2 mM DTT, 5% glycerol, 20 U of RNase inhibitor [Ambion]) in a final reaction volume of 20 μl and incubated at 37°C for 20 min. Two microliters of sample buffer (80% glycerol and 0.2% bromphenol blue) was added to each reaction mixture, the reaction mixtures were loaded onto 1% agarose gel, separated by electrophoresis in 0.5× Tris-borate-EDTA (TBE) buffer at 120 V (constant voltage) for 1.5 h, and visualized by autoradiography.
Competition experiments.
Competition experiments were analyzed with the filter binding assay. A constant concentration of the HTNV N peptides (3.5 × 10−7 M) was incubated with 1 ng of [γ-32P]ATP-labeled RNA (0.05 nM) for 10 min at 37°C. RNAs were synthesized and polyacrylamide gel electrophoresis (PAGE) purified by Integrated DNA Technologies. The RNAs used were HTNV 5′-end vRNA(1-39) (5′-UAGUAGUAUGCUCCCUAAAAAGACAAUCAAGGAGCAAUC-3′) HTNV 3′-end vRNA(1661-1696) (5′-CGUUGUUCUAGUAGCUCUUUAGGGAGUCUACUACUA-3′), and random RNA (5′-ACCAACAAAGAUGAGUGUUACAGCUCUUGC-3′). Various concentra-tions (0.05 to 500 nM) of unlabeled RNA were added to the binding reaction mixtures, and a further 10-min incubation followed. The reaction mixtures were slot blotted onto nitrocellulose filters as described previously (24). Analyses of the competition assays from the filter binding experiments were performed by a nonlinear fit of the data by using the Origin program (MicroCal).
RESULTS
Design and construction of HTNV N protein deletions.
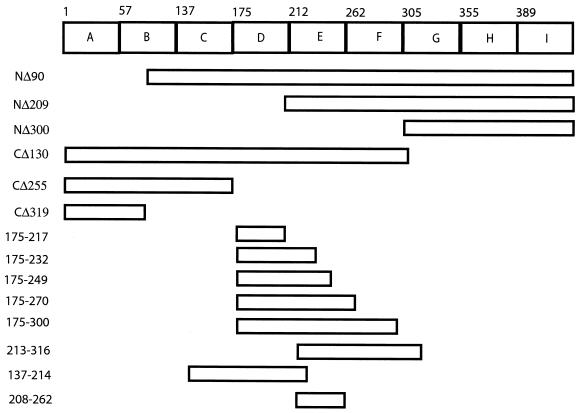
The BLOCKS database was used to identify highly conserved regions in the hantavirus N protein family (10). This database prepares multiply aligned, ungapped amino acid sequences for a protein family. By using the amino acid sequences of several hantavirus N proteins, the Block Maker program revealed nine distinct, conserved regions, or blocks, which were designated A to I (Fig. 1). To map the RNA binding region of HTNV N protein, 14 truncated constructs were prepared to remove the conserved blocks in the N-terminal, C-terminal, or both regions of the N protein. HTNV N protein derivatives included NΔ90, NΔ209, and NΔ300, which had N-terminal deletions of 90, 209, and 300 amino acids (aa), respectively (Fig. 1). NΔ90 lies just inside block B, and represents one of several deletions constructs (data not shown) made in this region. C-terminally deleted HTNV N proteins included CΔ130, CΔ255, and CΔ319, which removed 130, 255, and 319 aa from the C terminus. NP137-214, NP208-262, NP213-316, NP175-217, NP175-232, NP175-249, NP175-270, and NP175-300 had both N-terminal and C-terminal deletions. The nomenclature of the proteins with N- and C-terminal deletions provides the HTNV N protein amino acids expressed in the numbers that follow NP.
FIG. 1.
Schematic diagram of the blocks predicted for the hantavirus N protein and the locations of deletions constructed from the HTNV N protein. Bioinformatic analysis of hantaviral N amino acid sequences by the BLOCKS database revealed nine distinct conserved regions (blocks), which are designated PF00846A to -I. The alignment of nucleocapsid sequences used to generate the BLOCKS map are based on the following virus sequences (with accession numbers given in parentheses): Hantaan (Swiss Prot P05133), Seoul (Swiss Prot ID P17881), Prospect Hill (Swiss Prot ID P22047), and Puumala (Swiss Prot ID P41268) viruses. NΔ90 contained aa 91 to 429, NΔ209 contained aa 210 to 429, NΔ300 contained aa 301 to 429, CΔ130 contained aa 1 to 299, CΔ255 contained aa 1 to 174, and CΔ319 contained aa 1 to 110. The remaining N protein deletion constructs were named based on the amino acids of the HTNV N protein that were contained in the clone. For example, NP175-217 contains aa 175 through 217.
Expression and purification of truncated HTNV N protein deletions.
Several different host strains, HMS174(DE3), Novablue(DE3), BL21(DE3), BLR, and BL21(DE3)pLysS, were examined to see if the level of HTNV N protein expression could be improved. None of the hosts tested showed a substantial increase in expression levels when compared to the other strains, except for HMS174(DE3), which showed a slightly higher level for the wild-type N protein (data not shown). Therefore, we chose to express HTNV N protein and deletion constructs in either BL21(DE3) cells or HMS174(DE3) cells. In these two hosts, all constructs showed similar levels of expression, except NΔ90, which expressed poorly, and NP208-262, which had a slightly lower level of expression (data not shown). Additional deletions were created—NΔ105, for example—but none of these showed a stably expressed protein product (data not shown). Thus, creation of stable expression constructs with deletions in the N terminus from aa 90 to 105 proved to be difficult.
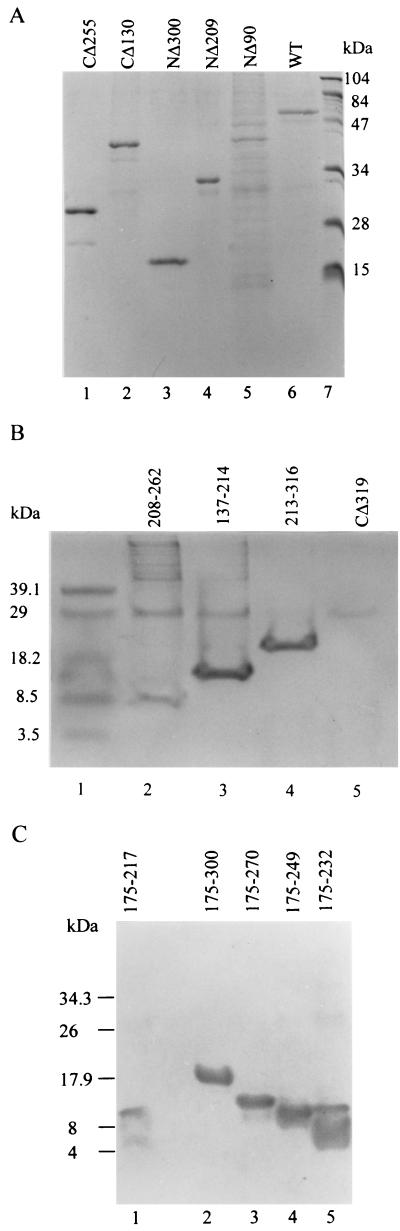
Each of the truncated HTNV N proteins was purified by Ni2+-NTA chromatography and examined for its level of purity by sodium dodecyl sulfate (SDS)-PAGE or Tricine SDS-PAGE (Fig. 2). The column fraction eluates from the N protein truncations—CΔ255 (Fig. 2A, lane 1), CΔ130 (Fig. 2A, lane 2), NΔ300 (Fig. 2A, lane 3), and NΔ209 (Fig. 2A, lane 4) NP213-316 (Fig. 2B, lane 4), and CΔ319 (Fig. 2B, lane 5)—showed a high level of purity (greater than 95%). Protein products—NP208-262 (Fig. 2B, lane 2) and NP137-214 (Fig. 2B, lane 3)—were slightly less homogenous in the preparations shown. The expression product NΔ90 (Fig. 2A, lane 5) was difficult to purify, and its low yield and purity made it unsuitable for subsequent functional studies. The HTNV N proteins with truncation in the N and C terminus—NP175-217, NP175-300, NP175-270, NP175-249, and NP175-232 (Fig. 2C, lanes 1-5)—were easily purified to greater than 95% homogeneity. Western blot analysis of the purified proteins showed them to be stable throughout the purification procedures as noted by an absence of proteolysis (data not shown).
FIG.2.
SDS-PAGE analysis of the full-length (wild-type [WT]) HTNV N protein and truncated HTNV N protein purified by nickel affinity chromatography. (A) Purified proteins shown are shown as follows: lane 1, CΔ255; lane 2, CΔ130; lane 3, NΔ300; lane 4, NΔ209; lane 5, NΔ90; and lane 6, wild-type N protein. (B) Purified proteins are shown as follows: lane 2, NP208-262; lane 3, NP137-214; lane 4, NP213-316; and lane 5, CΔ319. Molecular mass markers (M) are shown in lanes 1 and 7 in panels A and B, respectively. (C) Purified proteins are shown as follows: lane 1, NP175-217; lane 2, NP175-300; lane 3, NP175-270; lane 4, NP175-249; and lane 5, NP175-232. In panels A and B, the symbols CΔ and NΔ along with the number refer to the amino acids truncated in the N or C terminus, respectively. In panels B and C, the protein designation refers to the region of the HTNV N protein amino acid expressed by the construct.
Filter binding analysis of the binding affinity of the HTNV N protein and deletion derivatives to HTNV S segment RNA.
The binding affinity of purified wild-type N protein and deletions of the N protein to in vitro-transcribed radiolabeled HTNV S segment vRNA was measured by filter binding. The percentage of vRNA bound was measured for several concentrations of each protein. At the highest concentrations of protein assayed, both wild-type and CΔ130 bound 100% of the input RNA (Fig. 3). NΔ209, NP208-262, and NP137-214 bound ∼50, 64, and 73% of the vRNA, respectively. CΔ255 and NP213-316 bound less than 5% of the vRNA, and NΔ300 and CΔ319 did not bind the vRNA. These data demonstrated that the entire RBD was contained in CΔ130 (aa 1 to 299), while CΔ255 (aa 1 to 174), which had less than 5% binding activity, probably contained few amino acids, if any of the RBD. Therefore, the RBD was inferred to be within aa 174 to 299. Five additional deletions of the HTNV N protein were made in order to resolve the boundaries of the RBD within aa 174 to 299. Purified protein expression products from each of the five constructs, NP175-300, NP175-270, NP175-249, NP175-232, and NP175-217, were examined for their binding affinity for S-segment vRNA in a filter binding assay (Table 1). At the highest concentration, all of the deletion products showed approximately 70% or greater binding compared to the wild-type N protein. NP175-249 and NP175-232, showed the greatest amount of binding (85 and 79%, respectively). We consistently observed higher filter binding of protein derivatives that contained only aa 217 to 249 than those that were larger (aa 175 to 300) and smaller (aa 175 to 217). The reason for the decrease in binding with constructs that contain aa 217 to 249 were unclear. It is possible that these fragments contain domains that have additional functions such as protein-protein interactions that compete with the RNA binding function (2). Together, the filter binding data defined a minimal RBD between amino acid residues 175 and 217 that may extend into aa 217 to 249.
FIG. 3.
Filter binding analysis of the full-length HTNV N protein and truncated N proteins. N proteins were incubated with the S segment vRNA and examined for their binding affinity with a filter-binding assay as described in Materials and Methods. Shown are the mean and standard error of a minimum of two assays done in duplicate with the fraction of substrate retained on the membrane. An additional control was included in the analysis: the wild-type N protein with a nonviral 67-nucleotide RNA transcribed from pGEM7Zf, shown as the wild-type control (WT-ctr). The x axis shows the N protein concentration, and the y axis shows the amount of vRNA bound (percent).
TABLE 1.
Percentage of S segment vRNA bound to HTNV N protein and HTNV N protein derivatives as measured by filter binding
| N protein (μM) | % of vRNA bound to N protein derivative:
|
|||||
|---|---|---|---|---|---|---|
| Wild type | NP175- 217 | NP175- 232 | NP175- 249 | NP175- 270 | NP175- 300 | |
| 22 | 100 | 70 | 79 | 85 | 73 | 67 |
| 7.6 | 47 | 7.8 | 20 | 19 | 15 | 15 |
| 2.5 | 5 | 3 | 4 | 5 | 2 | 3 |
| 0.8 | 1 | 1 | 2 | 1 | 0.5 | 0.8 |
| 0.3 | 0.6 | 0.7 | 0.9 | 0.4 | 0.5 | 0.7 |
GEMSA analysis of complexes formed with the HTNV N protein and deletion derivatives with HTNV S segment RNA.
The interaction between wild-type or the truncated HTNV N proteins with the vRNA was also investigated with a GEMSA. HTNV N protein and each of the deleted proteins were incubated with the in vitro-transcribed radiolabeled vRNA S segment, and the complexes were resolved by separation on agarose gels (Fig. 4). Substantial levels of protein-RNA complexes were observed with reactions containing HTNV N protein wild type (Fig. 4A, lane 2), CΔ130 (Fig. 4A, lane 5), NP208-263 (Fig. 4A, lane 9), and NP137-214 (Fig. 4A, lane 10). A small amount of protein-vRNA complex was observed in the reaction mixtures containing the N protein derivatives NΔ209 (Fig. 4A, lane 3) and NP213-316 (Fig. 4A, lane 8). To further map the RBD within the region 175 to 300, five additional HTNV N protein deletions were made, purified, and analyzed by GEMSA. The following constructs provided a deletion analysis of the C-terminal boundary of the RBD (in order of decreasing size): NP175-300, NP175-270, NP175-249, NP175-232, and NP175-217. Each of the deletion products showed a similar level of binding in the GEMSA (Fig. 4B). The levels of complex formation were similar to those observed in the filter binding assay (Fig. 4 and Table 1). Two of the proteins analyzed in these studies, NP137-214 and NP175-232, consistently showed an additional higher order of complex formation in the GEMSA. NP175-232 also showed the highest amount of binding by filter binding analysis. These results suggest that this region of the protein when expressed without adjacent regions has the ability to form higher-order protein complexes or aggregates. At this time, it is unclear as to whether or how this is biologically relevant to what is known regarding hantavirus N protein oligomerization (2).
FIG. 4.
GEMSA of HTNV N protein, the wild type (WT), and deletions. (A) Binding affinities were examined by GEMSA with in vitro-transcribed full-length HTNV S segment vRNA and the expressed wild-type N protein (lane 2), NΔ209 (lane 3), NΔ300 (lane 4), CΔ130 (lane 5), CΔ255 (lane 6), NPΔ319 (lane 7), NP213-316 (lane 8), NP137-214 (lane 9), and NP208-262 (lane 10). A no-protein control is presented in lane 1. (B) Binding affinities were examined by GEMSA with in vitro-transcribed full-length HTNV S segment vRNA and the expressed wild-type N protein (lane 2), NP175-217 (lane 3), NP175-232 (lane 4), NP175-249 (lane 5), NP175-270 (lane 6), NP175-300 (lane 7), CΔ130 (lane 8), and CΔ255 (lane 9). A no-protein control is presented in lane 1. In panels A and B, a 22.7 μM concentration of wild-type N protein and each deletion mutant was incubated with radiolabeled vRNA S segment. The reaction mixtures were loaded onto a 1% agarose gel after an incubation period of 20 min, and the protein-RNA complexes were separated from free RNA by gel electrophoresis as described in Materials and Methods.
Competition analysis of the minimal RBD of HTNV N protein.
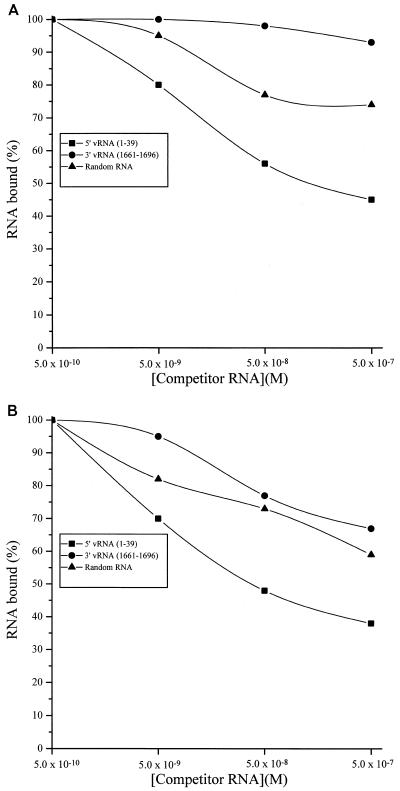
To corroborate the location of the functional RBD of the HTNV N protein, two synthetic peptides, N1 and N2, were made based on the HTNV N protein amino acid sequence 175 to 218 (N1, KHLYVSLPNAQSSMKAEEIT; and N2, PGRYRTAVCGLYPAQIKARQMIS). We were unable to produce the entire region as one peptide due to a breakdown somewhere near the two consecutive serines (Sigma-Genosys, personal communication). We investigated the ability of unlabeled oligoribonucleotides to compete with complexes made with 32P-labeled HTNV S segment oligoribonucleotides and each peptide. A constant concentration of two N protein oligopeptides with amino acid sequences corresponding to Lys-175 through Thr-195 (N1) and Pro-196 through Ser-218 (N2) was incubated with [γ-32P]ATP-labeled HTNV vRNA(1-39) (0.05 nM) and increasing concentrations (0.05 to 500 nM) of unlabeled competitor RNA (Fig. 5A). The level of each complex was measured by filter binding. The vRNA(1-39) representing the 5′ end of the HTNV S segment vRNA showed the greatest ability to compete compared to the 3′-end vRNA(1661-1696) or random RNA (Fig. 5A). Competition of complexes of the combined peptides (N1 and N2) with 32P-labeled 5′-end vRNA(1-39) by the unlabeled 5′-end vRNA(1-39) was approximately 55% at 500 nM.
FIG. 5.
Competition experiments. (A) Two N protein oligopeptides (N1 and N2) were bound to 1 ng of [γ-32P]ATP-labeled 5′-end vRNA(1-39) in the presence of variable concentrations (0.5 to 500 nM) of 5′-end vRNA(1-39), 3′-end vRNA(1661-1696), and a random RNA. (B) Competition experiments with N2 oligopeptide and the three synthetic oligoribonucleotides. Analyses of competition assays were performed by fitting a binding curve to the empirical filter binding data by using the Origin program (Microcal).
The two peptides were also analyzed individually in the competition assay. N2 had good binding activity (Fig. 5B), while N1 showed no RNA binding activity (data not shown). In the competition analysis of the N2-vRNA complex, the 5′-end vRNA(1-39) at a concentration of 500 nM reduced binding by approximately 68% (Fig. 5B). The 3′-end vRNA(1661-1696) and the random RNAs were not effective competitors of the N2-vRNA complex; although at the highest levels of competitor RNA, we observed a 30% reduction in the binding. When compared to the competition data with both peptides, these data suggest that N2 provides the major determinant of the binding affinity of the RBD. However, N2 alone has reduced specificity compared to N1 and N2 combined. The specificity of this activity was enhanced substantially by the addition of N1. The neurogranin peptide (AAKIQASFRGHMARKK) was used as a control in these filter-binding assays. Neurogranin peptide has a similar molecular weight and is composed of a similar number of hydrophobic amino acids as compared to peptides N1 and N2. Neurogranin peptide did not bind to any of the oligoribonucleotides (data not shown). In summary, these data suggest that the region of the N protein corresponding to amino acids Pro-196 through Ser-218 (N2) as well as determinants in peptide N1(175-195) are required for a functional RBD.
DISCUSSION
The hantavirus N protein plays an important role in the formation of the RNP complex of each genomic segment of vRNA and assembly of the RNPs into the virion. The N protein may also play a vital role in viral replication and transcription. During these various activities, the N protein must interact differentially with the vRNAs. There has been only one report that has addressed the location of the hantavirus RBD in the N protein. In that study, a nonspecific RBD was localized to the carboxyl-terminal amino acids of the HTNV and Puumala virus N proteins (9). Motifs common to other RNA binding proteins (25) have not been identified in hantaviral N proteins.
To identify the RBD of the HTNV N protein that specifically interacts with the vRNAs, we prepared and examined expression products from a series of several N- and C-terminal deletion constructs. Filter and GEMSA binding studies showed a minimal RBD from aa 175 to 217 (Fig. 6 and Table 2). Additional binding determinants may also extend to aa 232. We hypothesize that the RBD does not extend past aa 232. In addition to the evidence provided by deletion mapping, the amino acids that follow 232 are highly nonconserved (aa 233 to 309). It is generally held that nonconserved regions are less likely to contain functional domains. Exploration of the region identified showed it to have a 58% identity and an 86% similarity among the six hantavirus N protein sequences (Fig. 6). Studies with other RNA viruses have also reported that the nucleocapsid proteins bind vRNA through a localized, conserved domain. For example, a conserved region in the C terminus of the rabies virus N protein, aa 298 to 352, binds directly to the vRNA (14). In the influenza virus nucleoprotein, a highly conserved region among A-, B- and C- type viruses was mapped (1, 13). Similarly, a central, conserved region of the mouse hepatitis virus N protein, aa 169 to 308, comprises the RBD (15, 16). As with the region we mapped in the HTNV N protein, none of the canonical consensus RNA binding motifs has been identified in these proteins. Furthermore, at present, no apparent similarity has been noted among these RBDs. Identification of common features of RBDs among the hantaviral nucleocapsid proteins may require elucidation of their three-dimensional structures.
FIG. 6.
Alignment of the minimal RBD of the HTNV N protein with the same region in other hantavirus N proteins. Top, Topogravof virus; PUU, Puumala virus; ISLA, Isla Vista virus; PH, Prospect Hill virus; SN, Sin Nombre virus CC107; and HTN, Hantaan virus.
TABLE 2.
Summary of RNA binding levels for HTNV N protein and derivatives with the S segment vRNA
| Protein | Deletiona | aa | Filter binding (%) | Gel shift resultb |
|---|---|---|---|---|
| Wild type | None | 1-429 | 100 | ++++ |
| NΔ209 | N | 210-429 | 50 | ++ |
| NΔ300 | N | 301-429 | <5 | − |
| CΔ130 | C | 1-299 | 100 | ++++ |
| CΔ255 | C | 1-174 | <5 | − |
| CΔ319 | C | 1-119 | <5 | − |
| NP213-316 | N and C | 213-316 | <5 | + |
| NP137-214 | N and C | 137-214 | 73 | +++ |
| NP208-262 | N and C | 208-262 | 64 | ++ |
| NP175-300 | N and C | 175-300 | 67 | ++++ |
| NP175-270 | N and C | 175-270 | 73 | ++++ |
| NP175-249 | N and C | 175-249 | 85 | ++++ |
| NP175-232 | N and C | 175-232 | 79 | ++++ |
| NP175-217 | N and C | 175-217 | 70 | ++++ |
Deletion created in the N and/or C terminus of the HTVN N protein.
Symbols refer to approximate level compared to (++++): the wild type +++, ∼75 to 100%; ++, ∼50 to 75%; +, <25%; −, no binding.
To confirm the contribution of amino acids or the effect of the absence of amino acids in binding the RNA, we investigated the ability of unlabeled synthetic oligoribonucleotides to compete with 32P-labeled HTNV 5′-end vRNA in a filter-binding assay. Neither 3′-end vRNA nor random RNA was an effective competitor of the N1/N2-vRNA or N2-vRNA complexes. These data suggest that the N2 provides major determinants of the binding affinity noted by the RBD, but the specificity of this activity was enhanced substantially by the presence of N1. In a previous study by Gott et al., the C-terminal region of the HTNV and PUUV N proteins were shown to have a nonspecific RNA binding ability (9). The central region (aa 120 through 334) of the HTNV protein was not examined. It is difficult to compare our results with these, because Gott et al. used a different method to measure the RNA binding interaction. However, it is plausible that additional regions of the N protein contribute to the interaction with the vRNAs. In other studies that have attempted to map the RBDs of other viral nucleoproteins, different groups have mapped distinct regions. For example, in addition to the influenza virus work discussed above (1, 13), a recent mutational analysis of the influenza virus N protein found that several amino acids scattered throughout the protein were crucial for viral RNA binding (8). Studies of the HIV-1 nucleocapsid protein have also mapped a distinct region involved in the interaction with the virus genome (3) as well as the involvement of basic amino acids throughout the nucleocapsid protein (6). In the case of HIV-1, it has been proposed that the nonspecific interactions of the basic residues drive the assembly of the Gag polymerization and virion assembly though their interaction with RNA. The regions of the HTNV N protein reported herein and the previous report may also reflect the specific and nonspecific interactions of the protein with RNA.
Amino acids that are known to interact with nucleic acids include Lys, Arg, Asp, Glu, Thr, Ser, and Tyr (7). Fifteen of the 43 aa of the HTNV N protein RBD (Fig. 6) contain these residues: Lys and Arg comprise 11.6% of the amino acids, while Asp and Glu comprise an additional 4.7%, and Thr, Ser, and Tyr represent 18.6% of the amino acids in this region. The full-length HTNV N protein has a similar level of the basic residues, 11% Lys and Arg, but twice the amount of acidic residues: 10.6% are Glu/Asp. The full-length protein also has slightly fewer Thr, Tyr, and Ser residues: 15.9%. Future work will define the importance of each of these three groups in mediating the N protein's interaction with vRNA and non-vRNA.
Acknowledgments
We thank Alexander Dekonenko, Virology Division, USAMRIID, for gifts of HTNV N protein expression plasmids.
This work was supported by NIH grant 1RO3AI41114-01 to C.B.J. N.V. was supported by NIH Biomedical Research Training for Honors Undergraduates grant GMO7667-23.
REFERENCES
- 1.Albo, C., A. Valencia, and A. Portela. 1995. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J. Virol. 69:3799-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarasinghe, G. K., R. N. De-Guzman, R. B. Tuner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to step-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 4.Beaton, A. R., and R. M. Krug. 1986. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl. Acad. Sci. USA 83:6282-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duane, M. 1993. Molecular biophysics, p. 432-455. Oxford University Press, New York, N.Y.
- 8.Elton, D., L. Medcalf, K. Bishop, D. Harrison, and P. Digard. 1999. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 73:7357-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gott, P., R. Stohwasser, P. Schnitzler, G. Darai, and E. K. F. Bautz. 1993. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology 194:332-337. [DOI] [PubMed] [Google Scholar]
- 10.Henikoff, S., and J. G. Henikoff. 1994. Protein family classification based on searching a database of blocks. Genomics 19:97-107. [DOI] [PubMed] [Google Scholar]
- 11.Honda, A., K. Ueda, K. Nagata, and A. Ishihama. 1988. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J. Biochem. 104:1021-1026. [DOI] [PubMed] [Google Scholar]
- 12.Jin, H., and R. M. Elliot. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, M., T. Toyoda, D. M. Adyshev, Y. Azuma, and A. Ishihama. 1994. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J. Virol. 68:8433-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouznetzoff, A., M. Buckle, and N. Tordo. 1998. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J. Gen. Virol. 79:1005-1013. [DOI] [PubMed] [Google Scholar]
- 15.Masters, P. S. 1992. Localization of an RNA binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 125:141-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson, G. W., and S. A. Stohlman. 1993. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J. Gen. Virol. 74:1975-1979. [DOI] [PubMed] [Google Scholar]
- 17.Osborne, J. C., and R. M. Elliot. 2000. RNA binding properties of Bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946-9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patton, J. T., N. L. Davis, and G. W. Wertz. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J. Virol. 49:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmaljohn, C. S. 1996. Bunyaviridae: the viruses and their replication, p. 1447-1472. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippencott-Raven, Philadelphia, Pa. [Google Scholar]
- 21.Schmaljohn, C. S. 1996. Molecular biology of hantaviruses, p. 337. In R. M. Elliot (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 22.Schmaljohn, C. S., S. E. Hasty, S. A. Harrison, and J. M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005-1012. [DOI] [PubMed] [Google Scholar]
- 23.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan N protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 24.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siomi, H., and G. Dreyfuss. 1997. RNA binding proteins as regulator of gene expression. Curr. Opin. Gen. Dev. 7:345-353. [DOI] [PubMed] [Google Scholar]