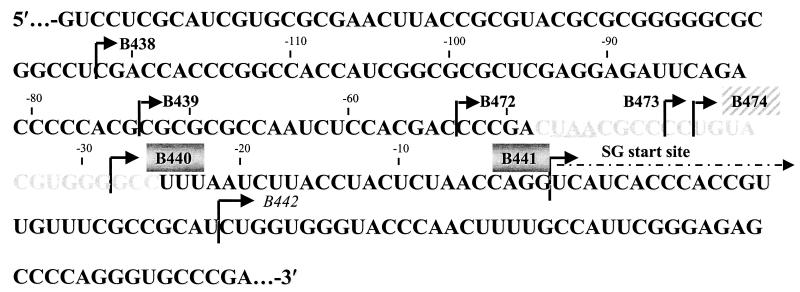Abstract
Rubella virus (RUB), the sole member of the Rubivirus genus in the Togaviridae family of positive-strand RNA viruses, synthesizes a single subgenomic (SG) RNA containing sequences from the 3′ end of the genomic RNA including the open reading frame (ORF) that encodes the virion proteins. The synthesis of SG RNA is initiated internally on a negative-strand, genome-length template at a site known as the SG promoter (SGP). Mapping the RUB SGP was initiated by using an infectious cDNA vector, dsRobo402/GFP, in which the region containing the SGP was duplicated (K. V. Pugachev, W.-P. Tzeng, and T. K. Frey, J. Virol. 74:10811-10815, 2000). In dsRobo402/GFP, the 5′-proximal nonstructural protein ORF (NS-ORF) is followed by the first SGP (SGP-1), the green fluorescent protein (GFP) gene, the second SGP (SGP-2), and the structural protein ORF. The duplicated SGP, SGP-2, contained nucleotides (nt) −175 to +76 relative to the SG start site, including the 3′ 127 nt of the NS-ORF and 47 nt between the NS-ORF and the SG start site. 5′ Deletions of SGP-2 to nt −40 (9 nt beyond the 3′ end of the NS-ORF) resulted in a wild-type (wt) phenotype in terms of virus replication and RNA synthesis. Deletions beyond this point impaired viability; however, the analysis was complicated by homologous recombination between SGP-1 and SGP-2 that resulted in deletion of the GFP gene and resurrection of viable virus with one SGP. Since the NS-ORF region was not necessary for SGP activity, subsequent mapping was done by using both replicon vectors, RUBrep/GFP and RUBrep/CAT, in which the SP-ORF is replaced with the reporter GFP and chloramphenical acetyltransferase genes, respectively, and the wt infectious clone, Robo402. In the replicon vectors, 5′ deletions to nt −26 resulted in the synthesis of SG RNA. In the infectious clone, deletions through nt −28 gave rise to viable virus. A series of short internal deletions confirmed that the region between nt −28 and the SG start site was essential for viability and showed that the repeated UCA triplet at the 5′ end of SG RNA was also required. Thus, the minimal SGP maps from nt −26 through the SG start site and appears to extend to at least nt +6, although a larger region is required for the generation of virus with a wt phenotype. Interestingly, while the positioning of the RUB SGP immediately adjacent the SG start site is thus similar to that of members of the genus Alphavirus, the other genus in the Togaviridae family, it does not include a region of nucleotide sequence homology with the alphavirus SGP that is located between nt −48 and nt −23 with respect to the SG start site in the RUB genome.
Positive-strand RNA viruses can be subdivided into viruses that utilize solely the genomic RNA as an mRNA, such as members of the Picornaviridae and Flaviviridae families, and viruses that synthesize one or more subgenomic (SG) mRNAs, such as members of the Togaviridae and Coronaviridae families. Among this latter group, two discrete mechanisms for synthesizing SG mRNAs have been characterized. In the first, documented for the Alphavirus genus of the Togaviridae (21) and plant viruses, such as brome mosaic virus (3, 11), the transcription of a single SG RNA consisting of sequences from the 3′ end of the genomic RNA is initiated at a single internal promoter site on a negative-strand, genome-length RNA template. In the second, best documented for the Coronaviridae, a nested 3′-terminal set of SG mRNAs is synthesized by a discontinuous transcription mechanism, resulting in each RNA containing a short leader from the 5′ end of the genome connected with sequences from the 3′ end of the genome; the mechanism by which this process is accomplished is not completely understood (9, 18, 19, 23).
Rubella virus (RUB) is the sole member of the Rubivirus genus in the Togaviridae family of animal viruses (for a review, see reference 4). The genome of RUB is 9,762 nucleotides (nt) long exclusive of the 3′ poly(A) tract, and the 5′ end of the SG RNA has been mapped to nt 6436 of the genomic RNA (14). The genomic RNA contains two long, nonoverlapping open reading frames (ORFs) that are separated by a 123-nt untranslated sequence known as the junction untranslated region (UTR). The 5′-proximal ORF (nonstructural protein ORF [NS-ORF]), which encodes nonstructural proteins involved in RNA replication, is translated from the genomic RNA, while the 3′-proximal ORF (structural protein ORF [SP-ORF]), which encodes virion structural proteins, is translated from the SG mRNA.
In one of the alphaviruses, Sindbis virus (SIN), the minimal promoter responsible for SG mRNA synthesis (SG promoter [SGP]) has been mapped to between nt −19 and +5 with respect to the SG RNA start site (10). Among alphaviruses, SGP sequences are highly conserved. Interestingly, there is a region within the junction region of the RUB genome which shares nucleotide sequence homology with the conserved alphavirus SGP sequence (4). However, this region of nucleotide sequence homology is 23 nt upstream from the SG start site (nt −46 to −23) rather than immediately adjacent to it. Thus, if this region is functional in SG RNA synthesis in RUB, the RUB SGP is either larger than the alphavirus SGP or there is a spacer between the SGP and the SG start site.
The goal of this study was to begin mapping the RUB SGP. Mapping of the alphavirus SGP was done by using a vector based on defective interfering RNAs with the viral RNA-dependent RNA polymerase (RDRP) supplied in trans by standard helper virus (10). No equivalent vector has been developed for RUB; however, both an infectious expression vector containing two SGPs, dsRobo402, and a replicon vector containing foreign sequences in place of the SP-ORF, RUBrep, have been constructed based on the RUB infectious clone Robo402 (15, 22). The duplicated SGP in the dsRobo402 vector consists of the 249-nt region of the genome from nt −175 to +76 relative to the SG start site, including the 3′ 127 nt of the NS-ORF followed by the entire junction region and ending at the initiating AUG of the SP-ORF. This duplication is much larger than that in the corresponding alphavirus SGP, but it was thought prudent to duplicate a region large enough to ensure capture of the SGP. In dsRobo402, the native SGP (SGP-1) is followed at the end of the junction region by a multiple cloning site (MCS) into which foreign ORFs can be introduced. Downstream from the MCS is the duplicated SGP (SGP-2), followed by the SP-ORF and the 3′ end of the genome. dsRobo402 constructs expressing reporter genes such as the green fluorescent protein (GFP) gene synthesize two SG RNAs with equal efficiencies, showing that the SGP is contained within the duplicated region. The advantage of using this construct for mapping the SGP is that deletions within SGP-2 can be made without disturbing the function of the 3′ end of the NS-ORF. While deletion mapping of the SGP-2 with this vector quickly determined that the 3′ end of the NS-ORF was not part of the SGP, the use of this vector was plagued by recombination between the SGPs resulting in deletion of the reporter gene and resurrection of virus with a single SGP. Therefore, mapping of the SGP was continued by using both RUBrep and Robo402 constructs modified to contain a unique restriction site at the 5′ end of the junction region for easy insertion of SGP mutations.
MATERIALS AND METHODS
Plasmids and recombinant DNA manipulations.
Recombinant DNA manipulations were carried out essentially as described by Sambrook et al. (17) with minor modifications. Escherichia coli MC1061 was used as the bacterial host. Restriction enzymes and T4 DNA ligase were obtained from New England BioLabs (Beverly, Mass.) or Roche Molecular Biochemicals (Indianapolis, Ind.) and used essentially as recommended by the manufacturers.
The infectious clone (Robo402), the double-SG vector (dsRobo402/GFP), and the replicon vector (RUBrep) were described previously (13, 15, 22). All site-directed mutations were made by PCR with mutagenic oligonucleotide primers. Standard PCR mixtures contained 400 ng of each primer, 20 ng of linearized plasmid template, 200 μM each deoxynucleoside triphosphate, and 5 U of Ex Taq DNA polymerase (PanVera/TakaRa, Madison, Wis.) in 1× buffer (PanVera TaKaRa) in a total volume of 50 μl. The protocol was 20 s at 90°C, 30 s at 50°C, and 2 min at 70°C for 35 cycles followed by one cycle of 10 min at 72°C.
dsRobo402 contains a unique NsiI site at the beginning of SGP-2 which facilitates the introduction of mutations. To create a Robo402 construct with an NsiI site immediately following the NS-ORF termination codon (at the beginning of the junction UTR) (NRobo402), a three-round, asymmetric PCR strategy was used (2). In the first round, mutagenic oligonucleotide 534 (Table 1), which contained the 3′-terminal 24 nt of the NS-ORF, an NsiI site, and the 5′-terminal 25 nt of the junction region, was used to prime asymmetric amplification on a PstI-linearized Robo402 template; this step resulted in a single-stranded DNA (ssDNA) extending from nt 6378 to 8228 of the genome. In the second round, oligonucleotide 1 (the sequence is complementary to nt 7328 to 7345 of the RUB genome) was used to prime asymmetric PCR amplification on the first-round ssDNA amplification product; this step created an ssDNA complementary to nt 6378 through 7345 of the genome, including the inserted NsiI site. In the third round, the second-round ssDNA product and oligonucleotide 106 (nt 5321 to 5340 of the genome) were used to prime PCR on a PstI-linearized Robo402 template. The double-stranded DNA amplification product, extending from nt 5321 to 7345 of the genome, was restricted with BglII (unique site at nt 5355) and AscI (unique site at nt 7318) and used to replace the corresponding fragment in Robo402.
TABLE 1.
Oligonucleotides used in this study
| Purpose | Oligo- nucleotide | Primer (5′ → 3′)a | Position (nt in RUB genome)b | Polarityc |
|---|---|---|---|---|
| Construction of NRobo402, NRUBrep/ GFP, and NRUBrep/CAT | 534 | CGCCAATCTCCACGACGCCGACTAATGCATCGCCCCTGTAC GTGGGGCCTTTAAT (NsiI) | 6367-6391; 6392-6416 | + |
| 1 | GAAGCGGATGCGCCAAGG | 7328-7345 | − | |
| 106 | AGCTCACCGACCGCTACGC | 5321-5340 | + | |
| 788 | ATGCTCTAGAATGGAGAAAAAAATCACTGG (XbaI) | CAT (1-20) | + | |
| 789 | TATAAGGCCTATGCATCTAGTCGAGGCACCAAT (StuI and NsiI) | CAT (677-693) | − | |
| 919 | ACTAATGCATCGCCCCTGTACGTGGGG (NsiI) | 6392-6408 | + | |
| 1033 | GTACTCTAGATTCGGGCACCCTGGGGCTC (XbaI) | 6494-6512 | − | |
| Creation of 5′-terminal SGP and SGP-2 deletions | 438 | GCATATGCATCGACCACCCGGCCAC (NsiI) | 6313-6327 | + |
| 439 | GCATATGCATCGCGCGCCAATCTCC (NsiI) | 6363-6377 | + | |
| 440 | GCATATGCATGCCTTTAATCTTACC (NsiI) | 6408-6422 | + | |
| 441 | GCATATGCATTCATCACCCACCGTT (NsiI) | 6436-6450 | + | |
| 442 | GCATATGCATCTGGTGGGTACCCAA (NsiI) | 6463-6477 | + | |
| 472 | GCATATGCATGCCGACTAACGCCCC (NsiI) | 6383-6497 | + | |
| 473 | GCATATGCATCCTGTACGTGGGGCC (NsiI) | 6396-6410 | + | |
| 474 | GCATATGCATTGTACGTGGGGCCTT (NsiI) | 6398-6412 | + | |
| 802 | GCATATGCATTACGTGGGGCCTTTA (NsiI) | 6400-6414 | + | |
| 803 | GCATATGCATCGTGGGGCCTTTAAT (NsiI) | 6402-6416 | + | |
| 803B | GCATATGCATTGGGGCCTTTAATCT (NsiI) | 6404-6418 | + | |
| 804 | GCATATGCATGGGCCTTTAATCTTA (NsiI) | 6406-6420 | + | |
| 916 | TTAAATGCATTCTTACCTACTCTAAC (NsiI) | 6416-6431 | + | |
| 917 | CTACATGCATTCTAACCAGGTCATC (NsiI) | 6426-6440 | + | |
| 977 | GCATATGCATCCTTTAATCTTACCT (NsiI) | 6409-6423 | + | |
| 978 | GCATATGCATCTTTAATCTTACCTA (NsiI) | 6410-6424 | + | |
| 979 | GCATATGCATTTTAATCTTACCTAC (NsiI) | 6411-6425 | + | |
| 980 | GCATATGCATTTAATCTTACCTACT (NsiI) | 6412-6426 | + | |
| 981 | GCATATGCATTAATCTTACCTACTC (NsiI) | 6413-6427 | + | |
| 982 | GCATATGCATAATCTTACCTACTCT (NsiI) | 6414-6428 | + | |
| 983 | GCATATGCATATCTTACCTACTCTA (NsiI) | 6415-6429 | + | |
| 1 | GAAGCGGATGCGCCAAGG | 7328-7345 | − | |
| Creation of internal SGP and SGP-2 deletions | 527 | TACCTACTCTAACCAGGΔTCACCCACCGTTGTTTC | 6419-6435Δ6439-6455 | + |
| 528 | CTTACCTACTCTAACCΔTCATCACCCACCGTTG | 6417-6432Δ6436-6451 | + | |
| 529 | CGTGGGGCCTTTAATCTTACCTACTΔTCATCACCCACCGTT GTTTCGCCGC | 6402-6426Δ6436-6460 | + | |
| 530 | CCTGTACGTGGGGCCTTTAATCTTAΔTCATCACCCACCGTT GTTTCGCCGC | 6396-6420Δ6436-6460 | + | |
| 535 | GGCCTTTAATCTTACCTΔCTAACCAGGTCATCACC | 6407-6423Δ6427-6443 | + | |
| 536 | ACGTGGGGCCTTTAATCΔCCTACTCTAACCAGGTC | 6401-6417Δ6421-6437 | + | |
| 537 | CCCTGTACGTGGGGCCTΔATCTTACCTACTCTAAC | 6395-6411Δ6415-6430 | + | |
| 813 | GACTAATGCATCGCΔTGTACGTGGGGC (NsiI) | 6392-6394Δ6398-6409 | + | |
| 814 | GACTAATGCATCGCCCΔTACGTGGGGCCTTT (NsiI) | 6392-6396Δ6400-6413 | + | |
| 815 | GACTAATGCATCGCCCCTGΔGTGGGGCCTTT (NsiI) | 6392-6399Δ6403-6413 | + | |
| 816 | GACTAATGCATCGCCCCTGTACGTGGGGΔTTAATC (NsiI) | 6392-6408Δ6412-6417 | + | |
| 918 | ACTAATGCATCGCCCCTGTACGTGCCTTTAATC (NsiI) | 6392-6405Δ6409-6417 | + | |
| 1 | GAAGCGGATGCGCCAAGG | 7328-7345 | − | |
| 546 | GCATATGCATGTCCTCGCTATCG (NsiI) | 6263-6275 | + | |
| 919 | ACTAATGCATCGCCCCTGTACGTGGGG (NsiI) | 6392-6408 | + | |
| RT-PCR and sequencing of the SGP region | 197 | CACGAAGCTTGTGCCTCGAGGGCCTTC (HindIII) | 6550-6566 | − |
| 177 | CCGGAATTCGGTGCTTTGCCGCCGTT (EcoRI) | 6124-6140 | + |
Restriction sites in oligonucleotides used for cloning the resulting PCR amplicon are underlined, and the enzymes are shown in parentheses. In oligonucleotides used to create internal deletions, the deletion site is marked with Δ.
The nucleotides in the RUB genome to which the sequence following the restriction site corresponds are shown (oligonucleotide 534 was used to insert an NsiI site; thus, there is an RUB sequence on either side of the NsiI site). In oligonucleotides used to create internal deletions, the nucleotides on either side of the deletion are shown.
+, genome polarity; −, complementary to genome polarity.
To create NRUBrep, we used a three-fragment ligation strategy with the NsiI-XbaI-digested PCR fragment amplified from the NRobo402 template and oligonucleotides 919 (NsiI site followed by the 5′ 17 nt of the junction UTR) and 1033 (XbaI site followed by a sequence complementary to the 3′ 19 nt of the junction region), the XbaI-EcoRI fragment from RUBrep/GFP (containing the GFP gene and a 3′ cis-acting element through the EcoRI linearization site), and the NsiI-EcoRI fragment from NRobo402 (containing the plasmid backbone and the 5′ end of NRobo402 through the NsiI site at the end of the NS-ORF). To construct NRUBrep/CAT, the GFP gene in NRUBrep/GFP was replaced with the chloramphenical acetyltransferase (CAT) gene from SINrep/CAT (obtained from Charles M. Rice) amplified by PCR by using oligonucleotides 788 (XbaI site followed by the 5′ 20 nt of the CAT gene) and 789 (StuI and NsiI sites followed by a sequence complementary to the 3′ 18 nt of the CAT gene), restricting the amplification product with XbaI and StuI, and using it to replace the XbaI-StuI fragment in NRURrep/GFP.
To construct deletion mutations extending from the 5′ terminus of the SGP (B438, B439, B472, N/B473, N/B474, N802, N803, N803B, N804, N/B440, N977, N978, N979, N980, N981, N982, N983, N916, N917, N/B441, and N/B442), mutagenic oligonucleotides were designed which contained an NsiI restriction site and 15 nt of the SGP downstream from the deletion (the sequences of these oligonucleotides are given in Table 1; the number of the oligonucleotide used in each mutation corresponds to the number by which the resulting mutated construct is designated). Following PCR amplification on a BglII-linearized Robo402 template with a mutagenic oligonucleotide and oligonucleotide 1 as primers, the amplification product was restricted with NsiI and AscI and used to replace the corresponding restriction fragment in dsRobo402/GFP or NRobo402. The mutations were transferred to NRUBrep/GFP by PCR amplification by using the mutated NRobo402 construct as a template, the corresponding mutagenic oligonucleotide, and oligonucleotide 1033; restricting the amplification product with NsiI and XbaI; and using it to replace the NsiI-XbaI fragment in NRUBrep/GFP (this construct has an XbaI site at the 3′ end of the junction UTR, immediately preceding the GFP gene). NRUBrep/GFP constructs with SGP mutations were converted to NRUBrep/CAT constructs by replacing the GFP gene with the CAT gene as described above.
To construct additional internal deletion mutations within the SGP (N527, N528, N529, N530, N535, N536, and N537), the three-round, asymmetric PCR amplification strategy (2) was used. In the first round, an ssDNA was synthesized by asymmetric PCR by using as a template PstI-linearized Robo402 and as a primer a mutagenic oligonucleotide containing the desired deletion with 15 nt of the SGP sequence on either side of the deletion (the sequences of these oligonucleotides are given in Table 1; the number of the oligonucleotide used in each mutation corresponds to the number by which the resulting mutated construct is designated). The first-round ssDNA product (extending from the junction UTR [the exact nucleotide depended on the oligonucleotide primer used] through nt 8228) was used as a template for the second round of asymmetric PCR primed with oligonucleotide 1, creating an ssDNA complementary to the junction UTR through nt 7345, including the SGP deletion. In the third round of PCR, the second-round ssDNA product and oligonucleotide 546 (nt 6263 to 6275) were used to prime PCR on a PstI-linearized Robo402 template, creating a double-stranded DNA extending from nt 6263 to 7345. The amplification product was restricted with NsiI and AscI and used to replace the corresponding restriction fragment in dsRobo402/GFP. The mutations were transferred to NRobo402 by PCR amplification by using the mutated dsRobo402/GFP construct as a template and oligonucleotides 919 and 1, restricting the amplification product with NsiI and AscI, and using it to replace the NsiI-AscI fragment in NRobo402. To create internal deletions N813, N814, N815, N816, and N918 in NRobo402, mutagenic oligonucleotides were designed which contained an NsiI restriction site, the 5′ nt in the junction UTR to the deletion, and 15 nt downstream from the deletion. Following PCR amplification on a BglII-linearized Robo402 template with the mutagenic oligonucleotide and oligonucleotide 1 as primers, the amplification product was restricted with NsiI and AscI and used to replace the corresponding restriction fragment in NRobo402.
With all constructs, the mutation was verified with both restriction mapping and direct sequencing of the region that was mutagenized. For mutated virus constructs (dsRobo402 and NRobo402), at least three clones of each construct with the confirmed mutation were used in separate in vitro transcriptions and transfections to ascertain the resulting phenotype. For mutated replicon constructs (NRUBrep), one clone of each construct with the confirmed mutation was used for in vitro transcription and subsequent transfection.
In vitro transcription.
Robo402, dsRobo402/GFP, NRobo402, NRUBrep, and mutagenized constructs from these plasmids were linearized with EcoRI followed by phenol-chloroform extraction and ethanol precipitation. For synthesis of 5′-capped RNA transcripts, 1 μg of linearized plasmid was transcribed at 37°C in a 25-μl reaction mixture containing reaction buffer (40 mM Tris-HCl [pH 7.5], 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol); 1 mM each ATP, GTP, CTP, and UTP; 2 mM cap analog [m7G(5′)ppp(5′)G] (New England BioLabs); 1 U of RNasin (Roche Molecular Biochemicals)/μl; and 25 U of SP6 DNA-dependent RNA polymerase (Epicentre, Madison, Wis.)/μl. The transcripts were analyzed by electrophoresis of aliquots of the reaction mixtures in 1% agarose gels in the presence of ethidium bromide. Precise yields of the transcripts were determined spectrophotometrically. Typical yields were 6 to 7 μg of RNA in a 25-μl reaction mixture containing 1 μg of linearized plasmid. The transcription reaction mixtures were used directly for cell transfection without DNase treatment or phenol-chloroform extraction.
Transfection of RNA and harvesting of virus.
For transfection with in vitro transcripts, ∼80% confluent Vero cells (maintained in Dulbecco's modified Eagle's medium [D-MEM] containing 5% fetal bovine serum [FBS]) in a 60-mm2 plate were washed two times with 4 ml of phosphate-buffered saline and one time with Opti-MEM I (Gibco/BRL). Subsequently, a mixture of a 12-μl aliquot from an RNA transcription reaction and an equal volume of Lipofectamine-2000 (Gibco/BRL) was added. Following 4 h of incubation, the transfection mixture was removed, and either 4 ml of D-MEM containing 2% FBS was added or the cells were overlaid with plaque assay agar. For cells maintained in liquid medium, GFP expression was monitored by direct examination with a Zeiss Axioplan microscope with epifluorescence capability. The culture fluid was collected following the appearance of cytopathic effects (CPE), divided into aliquots, and stored at −80°C. Subsequent passaging was done by inoculating fresh 60-mm2 plates of Vero cells with 0.1 ml of transfected or infected culture fluid. For cells overlaid with plaque assay agar, plaques to be picked were located microscopically, picked, and eluted in 1 ml of D-MEM. To visualize plaques, 6 to 7 days after transfection or infection, the agar overlay was removed and the cells were stained with 0.1% crystal violet in 10% formalin-0.1 M sodium phosphate buffer (pH 7.0).
RNA analysis and sequence confirmation.
Extraction of RNA, either total intracellular RNA or virion RNA from the medium, was done by using TRI-Reagent (Molecular Research Center, Cincinnati, Ohio) in accordance with the manufacturer's protocol. The RNA was resuspended in diethylpyrocarbonate-treated water, and concentrations were determined spectrophotometrically (optical density at 260 nm [OD260] of 0.022 = 1 μg/ml).
Reverse transcription (RT)-PCR amplification and sequencing were used to confirm the presence of intended mutations in viable virus recovered following transfection with transcripts from mutated constructs and to examine potential genomic rearrangements produced by homologous recombination between the two SGPs in dsRobo402/GFP constructs. One-tenth milliliter of medium from transfection plates maintained with liquid medium or one-fifth of the agar plug fluid from a plaque pick was used to infect a 35-mm2 plate of cells. RNA was extracted following the appearance of CPE. One-fifth of the extracted RNA was used as a template in an RT reaction (22) with oligonucleotide 197 as the primer. The single-stranded cDNA product was then used as the template for PCR amplification with oligonucleotides 197 and 177 (Table 1). Automated sequencing was performed by using an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, Calif.) with oligonucleotide 177 as the primer and an ABI373 sequencer (Perkin-Elmer Corporation, Foster City, Calif.). Additionally, RT-PCR was performed with virion RNA extracted from the medium from plates transfected with transcripts from dsRobo402/GFP constructs; the sizes of the amplification products were examined by electrophoresis in 1% agarose gels and visualization by ethidium bromide staining.
For analysis of RNA synthesis in cells transfected or infected with dsRobo402/GFP constructs, after the initial appearance of CPE, the culture fluid was removed, the cells were washed once with 1 ml of phosphate-free D-MEM (Gibco/BRL), and 1 ml of phosphate-free D-MEM containing 1% dialyzed FBS (Gibco/BRL) and 1 μg of actinomycin D/ml was added. Following 20 min of incubation at 35°C, 20 μCi of 32P-orthophosphate (ICN, Costa Mesa, Calif.) was added. After 8 h of radiolabeling, the cells were washed once with phosphate-buffered saline, and total intracellular RNA was extracted. Equivalent amounts of RNA were denatured with NorthernMax-Gly Sample Loading Dye (Ambion, Houston, Tex.) at 55°C for 30 min and electrophoresed in 1% agarose gels made in NorthernMax-Gly Gel Prep/Running Buffer (Ambion). After electrophoresis, the gels were vacuum dried at 60°C, and 32P-labeled RNA was detected by autoradiography with Kodak (Rochester, N.Y.) X-ray film. Exposures were made at room temperature. To examine RNA synthesis by NRUBrep/GTP constructs, total intracellular RNA was extracted 4 days posttransfection and subjected to Northern analysis with 32P-labeled, nick-translated pGEM/GFP as a probe (22). Densitometry of the genomic and SG RNA bands was done by Bio-Rad GS710 Calibrated Imagine Densitometry. Since the genomic bands turned out to be of equal intensity, the relative intensity of SG RNA bands is reported without normalization to the intensity of genomic RNA bands. To analyze CAT expression by NRUBrep/CAT constructs, at 4 days posttransfection, cytoplasmic lysates were made and assayed for CAT activity as previously described (15).
RESULTS
Mapping the SGP in Robo402/GFP.
The dsRobo402/GFP vector used to map the RUB SGP is shown in Fig. 1. The native SGP (SGP-1) directed the expression of GFP, while the duplicated SGP (SGP-2) directed the expression of the SP-ORF. SGP-2 was manipulated, since this could be done without affecting the function of the NS-ORF. Since SGP-2 directs the expression of the SP-ORF, mutations were screened for viability.
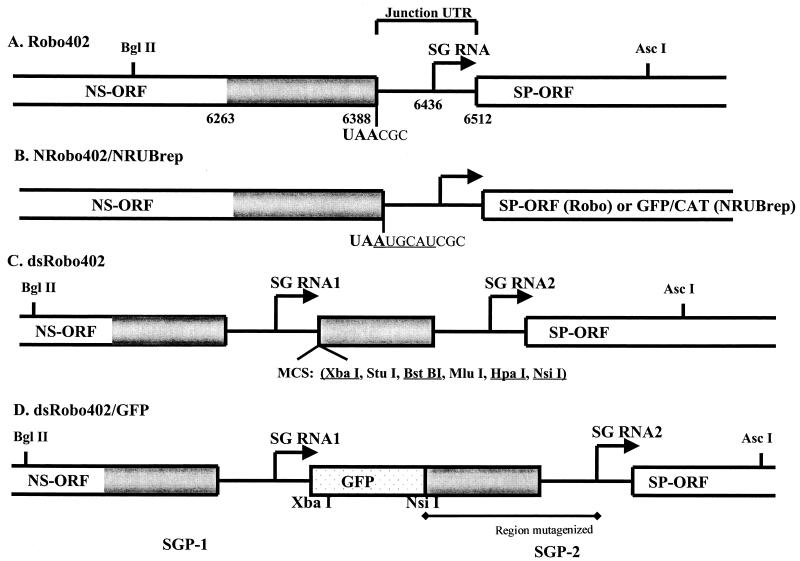
FIG. 1.
Schematic diagram of the genomic regions containing the SGPs in the RUB infectious cDNA constructs used in this study. (A) Robo402 is an infectious cDNA clone containing a full-genome-length cDNA. The 3′-terminal region of the NS-ORF and the 5′-terminal region of the SP-ORF are shown as boxes, the junction UTR is shown as a line, and the SG start site is shown as an arrow. The genomic coordinates of the 3′ end of the NS-ORF, the SG start site, and the 5′ end of the SP-ORF, as well as the BglII and AscI sites used in cloning, are given; the sequence at the 3′ end of the NS-ORF (i.e., the termination codon) and the 5′ end of the junction UTR is also shown. The region of the genome duplicated to produce a second SGP (SGP-2) for the construction of the dsRobo402 vector extends from nt 6263 in the NS-ORF through nt 6512 at the beginning of the SP-ORF; the region of the NS-ORF included in SGP-2 is shaded. (B) NRobo402 is a Robo402 derivative in which an NsiI site was introduced immediately following the NS-ORF termination codon in Robo402; the modified sequence at the NS-ORF-junction UTR boundary is shown (the NsiI site is underlined). (C) dsRobo402 is a Robo402 derivative in which an MCS (the unique sites are underlined) and SGP-2 were introduced into Robo402 to create a double-SG vector. (D) GFP was introduced into dsRobo402 between the XbaI and NsiI sites in the MCS to generate dsRobo402/GFP. In this construct, the native SGP (SGP-1) directs the expression of GFP, while the duplicated SGP (SGP-2) directs the expression of SP-ORF. The region of SGP-2 in which deletion mutations were created is underlined.
First, a series of deletions were made from the 5′ end of SGP-2. Initially, five deletions, B438, B439, B440, B441, and B442, were created which removed 50, 100, 145, 173, and 200 nt, respectively (Fig. 2). With respect to the SG start site, B438 deleted to nt −122, B439 deleted to nt −72, B440 deleted to nt −27, B441 coincided with the SG start site, and B442 deleted through nt +27 (downstream from the SG start site); the last deletion was designed as a negative control. Cells transfected with transcripts from these constructs were either maintained in medium and examined daily for both CPE and GFP expression or overlaid with plaque assay agar and stained for plaques 7 days posttransfection. Three distinct patterns were observed, as summarized in Table 2. In cells transfected with B438 and B439 transcripts, GFP expression could be detected as early as day 3 and was widespread among cells in the culture, CPE appeared on day 5, and numerous plaques were formed; this pattern was similar to that observed following transfection with dsRobo402/GFP transcripts. With B440 and B441 transcripts, the appearance of CPE was delayed and was less pronounced, GFP expression was not observed, and only a few plaques were present by day 7 posttransfection. CPE, expression of GFP, and plaques were not detected following transfection with B442 transcripts. Because of dramatic difference in phenotypes between B439 and B440 (which deleted 100 and 145 nt of SGP-2, respectively), three additional deletions were constructed—B472, B473, and B474, which deleted 120, 133, and 135 nt from the 5′ terminus of SGP-2 (to nt −53, −40, and −38 from the SG start site, respectively). As shown in Table 2, phenotypically, B472 and B473 resembled B438 and B439 (rapid appearance of CPE, widespread GFP expression, and numerous plaques), while B474 expressed an intermediate phenotype (delayed appearance of CPE, delayed GFP expression restricted to individual cells and small foci of cells, and few plaques).
FIG. 2.
5′ deletion mapping of SGP-2 in dsRobo402/GFP. The complete sequence of SGP-2 is shown. The UAA termination of the NS-ORF is underlined, the region that shares homology with the SGP of alphaviruses is shown in gray type, and the SG start site is indicated. A series of deletions were made from the 5′ end of SGP-2; arrows designating each deletion point from the first nucleotide that was retained. Transcripts from constructs with deletions in bold type (B438, B439, B472, and B473) had a wt phenotype (produced CPE and numerous plaques following transfection), those in a gray box (B440 and B441) had an attenuated phenotype (delayed CPE and few plaques), and that in italic type (B442) was lethal. The deletion in a hatched box (B474) had a phenotype intermediate between wt and attenuated.
TABLE 2.
Phenotypes associated with SGP-2 mutations in dsRobo/GFP
| Mutation CPE at daya: | Results for:
|
||||
|---|---|---|---|---|---|
| dsRobo402/GFP
|
No. of NRobo402 plaquesc | ||||
| GFP expressiona | CPE at daya: | No. of plaquesa | RNAsb | ||
| SGP-2 | Widespread | 5 | TNTC | P0, 2 SG RNAs | TNTC |
| B438 | Widespread | 5 | TNTC | P0, 2 SG RNAs | TNTC |
| B439 | Widespread | 5 | TNTC | P0, 2 SG RNAs | TNTC |
| B472 | Widespread | 5 | TNTC | P0, 2 SG RNAs | TNTC |
| B473 | Widespread | 5 | TNTC | P0, 2 SG RNAs | TNTC |
| B474 | Single cells | 6.5 | 12 | P1, 1 SG RNA | ∼50 |
| B440 | None | 8.5 | 5 | P2, 1 SG RNA | 0 |
| B441 | None | 8.5 | 3 | P3, 1 SG RNA | 0 |
| B442 | None | None | 0 | None | 0 |
Duplicate plates of cells were transfected with transcripts from dsRobo402/GFP constructs with the indicated SGP-2 deletion mutations. Following transfection, one plate was maintained with medium, while the second was overlaid with plaque assay agar. Plates were examined daily for GFP expression and the appearance of CPE (the earliest day posttransfection on which CPE were detected is indicated). The plates overlaid with plaque assay agar were stained on day 7 posttransfection, and plaques were counted (TNTC, too numerous to count).
In an independent experiment, at the time of appearance of CPE, transfection plates were radiolabeled with 32PO4, and virus-specific RNAs were analyzed. The culture fluid was passaged to fresh cells up to three times, and RNA analysis was done similarly at each passage. The passage (P) number at which virus-specific RNAs were first detected (P0, transfection) and the number of SG RNAs observed are indicated.
Phenotype of the SGP-2 mutation when transferred to NRobo402.
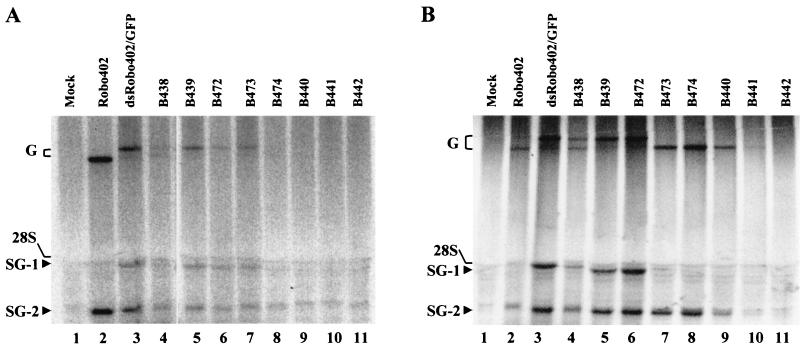
In explanation of these results, we reasoned that transcripts from deletion constructs B438, B439, B472, and B473 gave rise to viable virus while transcripts from deletion constructs B474, B440, B441, and B442 had severely impaired viability or were nonviable. Some of these deletion constructs might have been capable of limited RNA replication, and occasional recombination between SGP-1 and SGP-2 might have occurred, resulting in deletion of the GFP gene and restoration of a virus with a single, functional SGP that could direct the expression of the SP-ORF (explaining the small number of plaques formed). Such recombination between the SGPs resulting in deletion of the GFP gene was documented with dsRobo402/GFP (15). To test this hypothesis, viral RNAs produced in transfected cells were metabolically labeled and analyzed by agarose gel electrophoresis. As shown in Fig. 3A, as expected, dsRobo402/GFP as well as mutants containing deletions B438, B439, B472, and B473 synthesized two SG mRNAs. (As can be seen in both panels, background bands exist in the vicinity of both SG RNAs. In the case of SG-1, there were three distinct background bands; the upper one was due to the 28S RNA. SG-2 migrated slightly more rapidly than the single distinct background band.) However, a double genomic band was present. The more intense band was larger than the native RUB genomic RNA, as expected for a RUB genome with a GFP insert; however, a band similar in size to the native genomic RNA was also detectable. This finding indicates the occurrence of deletions through recombination restoring the native genomic RNA. No viral RNAs were detected in cells infected with mutants containing deletions B474, B440, B441, and B442, presumably because the titers of virus in transfection culture fluids were too low for detectable viral RNA synthesis within the experimental time frame. After a single passage, a single SG RNA was detected in cells infected with B473, B474, and B440; these viruses also synthesized only a standard-sized genomic RNA (Fig. 3B). In addition, after one passage, dsRobo402/GFP, B438, B439, and B472 maintained the synthesis of two SG RNAs; however, the doublet nature of the genomic RNA was more pronounced.
FIG. 3.
Virus-specific RNAs produced by dsRobo402/GFP constructs with SGP-2 mutations. (A) Passage 0. Vero cells were mock transfected (Mock; lane 1) or transfected with transcripts from Robo402 (lane 2), dsRobo402/GFP (lane 3), B438 (lane 4), B439 (lane 5), B472 (lane 6), B473 (lane 7), B474 (lane 8), B440 (lane 9), B441 (lane 10), or B442 (lane 11). Five days posttransfection, cells were metabolically labeled with 32PO4 in the presence of actinomycin D. Following 8 h of incubation, total cell RNA was extracted and subjected to agarose gel electrophoresis. (B) Passage 1. Vero cells were mock infected (Mock; lane 1) or infected at a multiplicity of infection of ∼1 PFU/cell with Robo402 virus (lane 2) or stocks of dsRobo402/GFP (lane 3), B438 (lane 4), B439 (lane 5), B472 (lane 6), B473 (lane 7), B474 (lane 8), B440 (lane 9), B441 (lane 10), or B442 (lane 11) viruses harvested from transfection plates. Two days postinfection, cells were metabolically labeled with 32PO4 for 8 h; this step was followed by total cell RNA isolation and agarose gel electrophoresis. G, genomic RNAs (the wt genome and the larger genome of dsRobo402/GFP virus with a GFP insert); 28S, 28S cell rRNA, which causes a background blob; SG-1 and SG-2, SG RNAs synthesized from SGP-1 and SGP-2, respectively (in Robo402 virus, the single SG RNA is the same size as SG-2). A background band that migrated slightly more slowly than SG-2 can be seen.
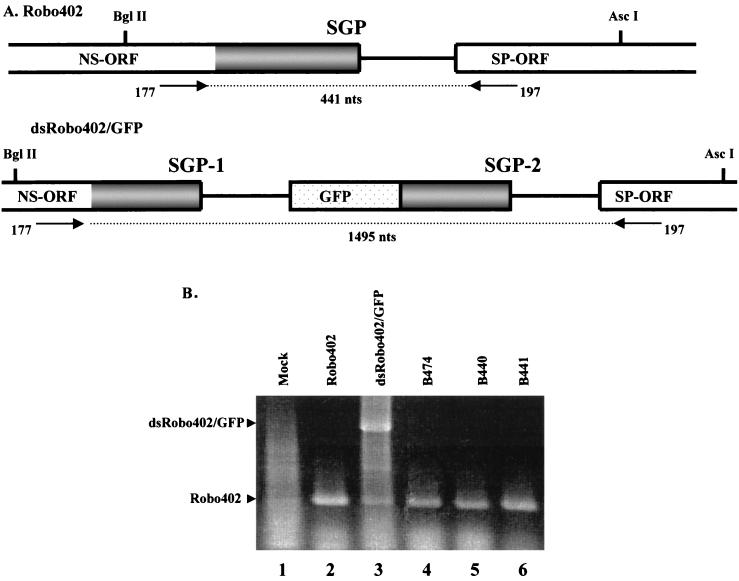
As an additional approach to detecting recombination between the SGPs of dsRobo402/GFP and its derivatives, virion RNA was isolated from the medium of plates transfected with either Robo402, dsRobo402/GFP, or the three constructs that appeared to be viable only through recombination, B474, B440, and B441. This RNA was used as a template for RT-PCR with primers specific for the SGP region. As shown in Fig. 4A, the product expected in this assay for a single SGP was 441 nt, and that expected for a double SGP was 1,495 nt. The major band produced from B474, B440, and B441 was similar in size to the band produced from Robo402, indicating homologous recombination between the two SGPs in these constructs (Fig. 4B). Interestingly, while dsRobo402/GFP yielded a larger RT-PCR band, as expected, a band of a size similar to the Robo402 band was also present, indicating the occurrence of homologous recombination with this construct as well.
FIG. 4.
Study of homologous recombination between the SGPs in dsRobo402/GFP SGP-2 mutants. (A) Schematic diagram of regions of the genomes of Robo402 (wt) and dsRobo402/GFP containing the SGPs, the oligonucleotide primers (177 and 197) used for RT-PCR, and the expected sizes (nucleotides) of the amplification products from each template. (B) Cells were mock transfected (Mock; lane 1) or transfected with Robo402 (lane 2), dsRobo402/GFP (lane 3), B474 (lane 4), B440 (lane 5), or B441 (lane 6) in vitro transcripts. Virion RNAs were extracted from the medium and used as templates for RT-PCR with primers 177 and 197. RT-PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The bands expected for amplification from the dsRobo402/GFP and Robo402 templates are labeled.
Mapping the SGP in RUBrep/GFP and Robo402.
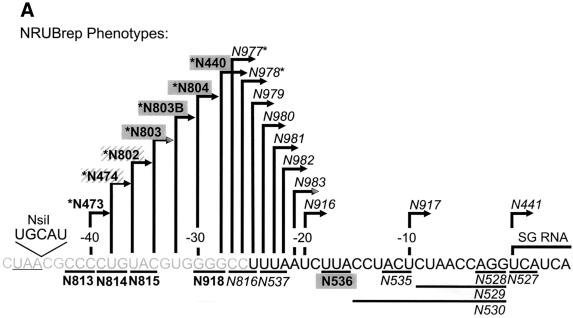
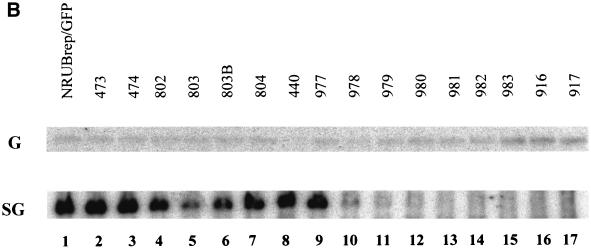
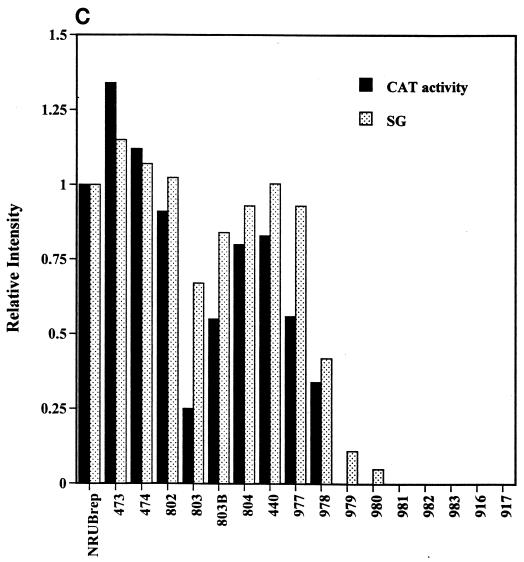
The results obtained with dsRobo402/GFP indicated that SGP-2 could have a deletion as large as 133 nt (B473; 4 nt downstream from the end of the NS-ORF, nt −40 with respect to the SG start site) without impairment of the phenotype (viability and synthesis of SG RNA-2). Since sequences at the 3′ end of the NS-ORF are thus not involved in SGP function, mapping was extended by using the replicon vectors RUBrep/GFP and RUBrep/CAT. To facilitate the introduction of mutations into the SGP of these vectors, a unique NsiI site was created immediately downstream from the NS-ORF termination codon, producing NRUBrep/GFP and NRUBrep/CAT (Fig. 1). A nested series of deletions were produced in these vectors that extended from the NsiI site through nt −40 to nt −20 with respect to the SG start site; the deletions across nt −40 to −20 differed by 1 or 2 nt (Fig. 5A). Transcripts from this series of constructs in RUBrep/GFP were used to transfect Vero cells, and SG RNA synthesis was determined by Northern analysis. As shown in Fig. 5B, constructs with deletions through nt −28 (construct N977, which maintains the junction UTR beginning at nt −27) produced a pronounced SG RNA band. Following N977, construct N978 (nt −26) synthesized a markedly reduced amount of SG RNA, constructs N979 (nt −25) and N980 (nt −24) synthesized little, if any, SG RNA, and constructs N981 (nt −23) through N917 (nt −10) synthesized no detectable SG RNA. Of note is that the constructs that synthesized no SG RNA still produced a genomic RNA and thus were capable of replication. When CAT activity was measured in cells transfected with transcripts from the RUBrep/CAT equivalent of these constructs (Table 3), constructs through N978 (nt −26) expressed detectable CAT activity; in NRUBrep/GFP, this was the largest deletion that produced a definitive SG RNA band (Fig. 5B). When SG RNA synthesis by the NRUBrep/GFP constructs was quantitated by densitometry and compared to CAT expression by the corresponding NRUBrep/CAT constructs, there was a good correlation (Fig. 5C). Interestingly, the N803 deletion (deletion to nt −34) exhibited lower levels of SG RNA synthesis and CAT expression than did the subsequent four larger deletions; the levels of SG RNA synthesis and CAT expression were also lower with the N803B deletion (nt −32) than with the longer deletions, but not to the same degree as with the N803 deletion.
FIG. 5.
SGP mapping in NRUBrep and NRobo402. (A) The sequence from the NS-ORF termination codon through the SG start site is shown; the NsiI site is underlined, and the region that shares homology with the SGP of alphaviruses is shown in gray type. A series of 5′ deletions were made in NRUBrep/CAT, NRUBrep/GFP, and NRobo402 extending from the NsiI site; arrows designating each deletion point from the first nucleotide that was retained. Deletions that maintained the synthesis of an SG RNA in both NRUBrep/GFP (assayed by Northern blotting) and NRUBrep/CAT (assayed by CAT activity) are marked by asterisks. Additionally, a series of short deletions (underlined) were created in NRobo402. Transcripts from NRobo402 constructs with deletions shown in bold type had a wt phenotype (produced CPE within 3 days posttransfection), those in a hatched box had an intermediate phenotype (produced CPE within 4 days posttransfection), those in a gray box had an attenuated phenotype (produced CPE within 5.5 days posttransfection), and those in italic type were lethal. (B) Vero cells were transfected with transcripts from NRUBrep/GFP or NRUBrep/GFP constructs containing the SGP deletions shown in panel A. Four days posttransfection, total RNA was extracted and examined by Northern hybridization by using a 32P-labeled GFP-specific probe. The regions of the Northern blot containing the replicon genomic (G) RNAs and SG RNAs are shown. (C) Comparative synthesis of SG RNA and CAT expression by 5′ SGP deletions mutants. SG RNA synthesis by NRUBrep/GFP constructs containing 5′ SGP deletions (panel B), as quantitated by densitometry, and CAT expression by NRUBrep/CAT constructs with the same series of mutations (Table 3) were normalized to SG RNA synthesis and CAT expression by NRUBrep/GFP and NRUBrep/CAT, respectively, and are displayed as a bar graph. The mutations are listed in order of increasing deletion size.
TABLE 3.
Phenotypes associated with SGP deletions in NRUBrep and NRobo402 CPE at dayc:
| Deletion | Results for:
|
|||
|---|---|---|---|---|
| NRUBrep
|
NRobo402
|
|||
| SG RNAa | CAT activityb | CPE at dayc: | Titerc | |
| None (wild type) | + | 8,883 | 3 | 2.2 × 105 |
| N473 | + | 11,890 | 3 | 2.4 × 105 |
| N474 | + | 9,915 | 4 | 5.2 × 104 |
| N802 | + | 8,032 | 4 | 1.8 × 105 |
| N803 | + | 2,223 | 5.5 | 1.7 × 104 |
| N803B | + | 4,904 | 5.5 | 1.1 × 105 |
| N804 | + | 7,071 | 5.5 | 1.3 × 105 |
| N440 | + | 7,332 | 5.5 | 1.4 × 105 |
| N977 | + | 5,002 | None | 0 |
| N978 | ± | 2,984 | None | 0 |
| N979 | ? | 0 | None | 0 |
| N980 | ? | 0 | None | 0 |
| N981 | − | 0 | None | 0 |
| N982 | − | 0 | None | 0 |
| N983 | − | 0 | None | 0 |
| N916 | − | 0 | None | 0 |
| N917 | − | 0 | None | 0 |
| N441 | − | 0 | None | 0 |
| N442 | − | 0 | None | 0 |
Vero cells were transfected with transcripts from NRUBrep/GFP containing the indicated deletion mutations. At 4 days posttransfection, total intracellular RNA was extracted and subjected to Northern analysis using a GFP-specific probe (Fig. 5B). The synthesis of SG RNA by each mutant is indicated (+, present; ±, present but faint; ?, not clear [very faint, not clear if present]; −, absent).
Vero cells were transfected with transcripts from NRUBrep/CAT containing the indicated deletion mutations. At 4 days posttransfection, cells were lysed and assayed for CAT activity. CAT activity is the amount (in cpm) of [3H]acetyl-CoA partitioning into the aqueous phase following reaction in the presence of chloramphenicol (15). The activity given is the average from two independent experiments.
Vero cells were transfected with transcripts from Robo402 containing the indicated deletion mutations. Following transfection, cells were examined daily for the appearance of CPE. The initial day posttransfection on which CPE were detected is indicated; at that time, the culture fluid was harvested, and the virus titer was determined by plaque assay.
To correlate SGP function with virus replication, the series of SGP deletion mutations were introduced into NRobo402, a Robo402 equivalent of NRUBrep containing the unique NsiI site at the 5′ end of the junction UTR. As shown in Fig. 5A and as tabulated in Table 3, deletions through N440 (nt −28) gave rise to viable virus which produced plaques, although the phenotype associated with deletions past nt −40 (N473) was attenuated in that the appearance of CPE following transfection was delayed. Thus, the phenotype associated with deletions in NRobo402 correlated with the phenotype associated with deletions in dsRobo402/GFP, since B473 and N473 exhibited a wild-type (wt) phenotype, B474 and N474 exhibited an intermediate phenotype, N440 exhibited an attenuated phenotype, N441 and N442 failed to yield viable virus, and B440 and B441 appeared to yield virus only through homologous recombination. The results of introduction of all of the initial deletion mutations in dsRobo402/GFP into NRobo402 are shown in Table 2. The titers to which viruses recovered from the deletion series in NRobo402 replicated were also determined. As shown in Table 3, most of these viruses replicated to approximately 105 PFU/ml, with the exception of N803 (∼104 PFU/ml), which produced a reduced amount of SG RNA. It should be noted that in this experiment, infected cultures were incubated until CPE appeared and so the harvest time was extended for some of the mutants, indicating a slower replication rate.
A series of 3-nt internal deletions were also made across the SGP in NRobo402 from nt −42 through nt +3 with respect to the SG start site (Fig. 5A). With one exception, constructs with deletions upstream from nt −28 yielded viable virus with a wt phenotype in terms of CPE induction and plaque formation, while constructs with deletions downstream from nt −28 (including deletion of one of the UCA repeat triplets at the 5′ end of the SG RNA) did not yield viable virus. The one exception was a construct with a deletion of nt −16 to −18, which yielded a virus with an attenuated phenotype (delayed production of CPE and no plaque formation following transfection). Thus, with this one exception, the effect of these internal deletion mutations correlated with that of the 5′-terminal deletion mutations.
Sequencing of SGPs produced by recombination.
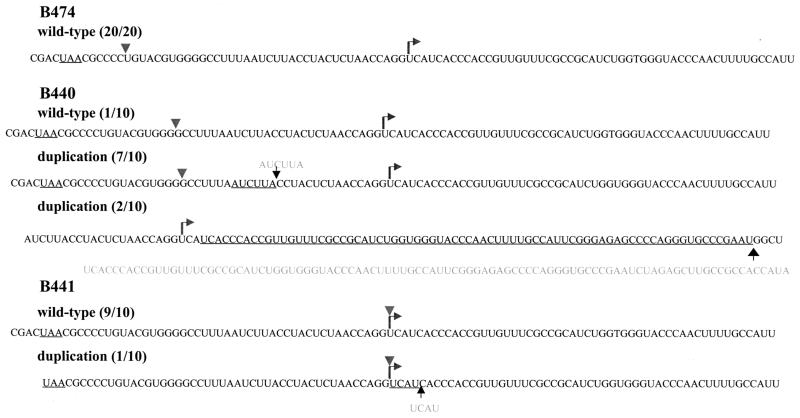
Since we found that homologous recombination between the SGPs in some of the dsRobo402/GFP SGP-2 deletion constructs generated viruses with a single functional SGP, we decided to use RT-PCR to amplify such recombined SGPs followed by sequencing to analyze whether novel functional SGP sequences were generated in the process. To this end, plaques were picked from plates transfected with B474, B440, and B441 transcripts and subsequently overlaid with agar. After one round of amplification of each plaque, RT-PCR was done by using the SGP-specific primers described previously (Fig. 4A). With all of the amplified plaques, an amplification product of the size expected for native virus with a single SGP (∼450 nt) was produced, consistent with the occurrence of homologous recombination. The sequences of the amplification products of these plaques are summarized in Fig. 6. With both B474 and B441, the preponderance of amplification products had the wt sequence, except for one plaque from B441 which had a UCAU insertion at the SG start site. However, of the 10 plaques amplified from B440 transfection plates, 1 had the wt sequence, 2 had a large duplication downstream from the SG start site followed by an insertion of a nonviral sequence, and 7 had a duplication of the AUCUUA sequence that resides between nt −16 and −21 from the SG start site in the wt SGP.
FIG. 6.
Sequencing of SGPs resulting from homologous recombination in dsRobo402/GFP viruses. Following transfection of Vero cells with B474, B440, and B441 transcripts, individual plaques were isolated and amplified by one passage in Vero cells. Total intracellular RNAs isolated from the amplification plates were used in a PCR with oligonucleotide primers 177 and 197, designed to amplify the SGP region (Fig. 4). Amplification products were sequenced directly without subcloning. For each construct, the number of plaques recovered with each sequence is indicated. For each sequence, the deletion site in the original construct is marked with an arrowhead, the SG start site is denoted with an arrow, duplicated sequences are underlined, and the duplication is shown in gray type. The long duplication in 2 out of 10 B440 clones sequenced was followed by nonviral sequences.
DISCUSSION
The goal of this study was to begin mapping the RUB SGP. To this end, we initiated this study by using a double-SG RUB vector, dsRobo402/GFP, which allowed us to leave the native SGP intact while manipulating the duplicated SGP, SGP-2. Since SGP-2 directed the expression of the SP-ORF, while the native SGP-1 directed the expression of the GFP gene (the reporter gene used), mutated SGP-2 constructs were screened for viability. SGP-2 contained 127 nt at the 3′ end of the NS-ORF in addition to the entire junction region (including the 47 nt of the junction region upstream from the SG start site). When deletions from the 5′ end of SGP-2 deleted this region of the NS-ORF, the resulting constructs were phenotypically similar to parental dsRobo402/GFP in both replication and RNA synthesis, indicating that this region is not necessary for SGP function. However, 5′ deletions of more than 9 nt into the junction region resulted in substantially reduced viability (delayed appearance of CPE following transfection and few plaques formed). That such constructs exhibited any viability at all was due to homologous recombination between SGP-1 and the deletion-containing SGP-2, resulting in the removal of the GFP gene and the restoration of a functional SGP directing the expression of the SP-ORF. This notion was evidenced by the following findings obtained from plates transfected with transcripts from these constructs. (i) Virion RNA from the culture fluid yielded RT-PCR products of the sizes expected for the wt junction region. (ii) The virus that eventually grew had a wt-sized genome and synthesized a single SG RNA species. (iii) Virus amplified from purified plaques generally had a junction region with the wt sequence (but could also have rearrangements of the wt sequence). Additionally, constructs with wt viability exhibited two genomic RNA bands, one of wt size and one larger than wt size, again consistent with deletion of the GFP gene through homologous recombination between the SGPs restoring the wt genome.
The propensity of the dsRobo402/GFP vector to undergo such recombination compromised its utility in mapping the SGP. Interestingly, when a similar vector was used for comparing relative strengths of SIN SGP mutations (16), homologous recombination, as evidenced by double genomic RNA bands, was not observed in passage 1 virus. In contrast, we observed double genomic bands with the RUB vector during passage 0. In the SIN study, the efficiency of reference and mutagenized SGPs included in the same vector could be directly compared by the intensities of the SG RNA bands synthesized from each. This approach would not be possible with dsRobo402/GFP, since the amount of homologous recombination evident at passage 1 would be enough to skew the production of the SG RNA species. Since we found that the 3′ NS-ORF region included in SGP-2 in dsRobo402 was not necessary for the function of this SGP, it could be deleted from the vector, reducing the region of homology over which homologous recombination might occur and possibly increasing the stability of expression of foreign genes by the vector.
Since the 3′-terminal 127 nt of the NS-ORF included in SGP-2 were not necessary for its function, RUBrep replicon and Robo402 infectious clone derivatives (NRUBrep and NRobo402) with a unique NsiI site at the exact 3′ end of the NS-ORF (5′ end of the junction region) were constructed and used for SGP mapping without the possibility of homologous recombination occurring to confuse the results. With both NRUBrep/GFP and NRUBrep/CAT, 5′-terminal deletions through nt −26 from the SG start site resulted in constructs that both synthesized SG RNA and expressed CAT activity, while deletions that extended closer to the SG start site did not. Thus, the minimal SGP can be defined as extending from nt −26 through at least the SG start site. Interestingly, while the level of SG RNA synthesis generally decreased with increasing size of these deletions, deletions to nt −35 to −33 resulted in a reduced level of SG RNA synthesis that did not follow this correlation. This result indicates either that these nucleotides exert a negative effect on SG RNA synthesis that is overcome by their deletion or the presence of upstream nucleotides or that without the upstream nucleotides, a sequence context occurs or a secondary structure is formed that is inhibitory to SG RNA synthesis. With NRobo402, viable virus was recovered from constructs with deletions extending through nt −28 from the SG start site, although only virus from constructs with deletions extending through nt −40 had a wt phenotype. Consistent with these findings, short internal deletions between nt −42 and −28 and the SG start site yielded viable virus with a wt phenotype, while internal deletions between nt −28 and the SG start site were lethal, with the exception of one that yielded virus with an attenuated phenotype.
The 3′ boundary of the RUB SGP was not mapped but minimally appears to include the UCA doublet at the beginning of the SG RNA (or to nt +6 with respect to the SG start site), since deletion of one of these two UCAs was lethal in NRobo402, although SG RNA synthesis was not assayed. Thus, the minimal SGP can be defined at this stage as extending from nt −26 through the SG start site and possibly extending to nt +6. Since the sequence following SGP-1 in dsRobo402/GFP ends with the junction region (nt +76 with respect to the SG start site) at the beginning of the MCS and this SGP is fully functional (15), it appears that the SGP does not extend beyond the 3′ end of the junction region. The minimal SGP in alphaviruses has been mapped to nt −19 through +5. This minimal alphavirus SGP was defined in terms of promoter activity. The optimal SGP necessary for wt levels of SG RNA synthesis and virus replication is larger (nt −98 to +14) (10, 16); this is also the case for brome mosaic virus (3). Recently, such optimizing sequences were mapped to nt −40 to −20 and nt +6 to +14 with respect to the SG start site in SIN (25). With respect to the upstream sequences, this is also the case for RUB, since NRobo402 constructs with the minimal 5′ SGP were not viable and inclusion of 12 nt upstream from the minimal sequence was necessary for the production of a virus with a wt phenotype and maximal promoter activity.
The positioning of the SGP within the sequences immediately upstream from the SG start site is therefore similar in RUB and the alphaviruses; however, the minimal RUB SGP (nt −26 to +6) does not contain the region of nucleotide sequence homology with the alphavirus SGP (present just upstream from the minimal SGP between nt −48 and −23). While the reason for the conservation of this region of nucleotide sequence homology in the RUB genome is not known, it is possible that this sequence did function as the SGP in an RUB ancestor closely related to the alphaviruses and that its function in this capacity has been superseded by the current SGP sequence. Such an event would not be unlikely, since SGPs exhibit a large degree of diversity among positive-polarity RNA viruses in the alpha-like virus superfamily and completely different SGPs can be present in the same virus genome (7, 12). As discussed above, part of this sequence is the region upstream from the minimal SGP that serves to optimize SGP function, a function which it may subsequently have acquired. Although this sequence shares ∼60% homology with the consensus alphavirus SGP sequence, it has diverged significantly, since it functions only minimally as an alphavirus SGP when introduced into an alphavirus vector (<5% of the activity of native alphavirus SGP) (6).
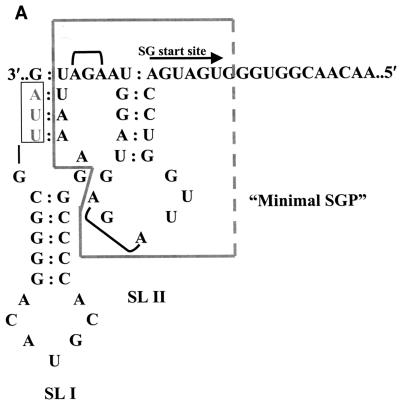
The current model of SGP function is that the negative-strand complement of the SGP is recognized by the viral RDRP or some viral component or host factor that interacts with the viral RDRP, which then initiates SG RNA synthesis in a primer-independent fashion at the SG start site (12). RDRPs of plant virus members of the alpha-like virus superfamily accurately initiate SG RNA synthesis in vitro on a single-stranded RNA (or ssDNA) template consisting of the negative-strand SGP sequences and the SG start site (5, 20). Conflicting results have been obtained as to whether SGP function is based on primary sequence alone or whether secondary structures are critical. The predicted secondary structure of the RUB junction region in the negative polarity contains two stem-loop structures located 3′ terminal to the SG start site (Fig. 7A); of these, only the second (SLII) is within the minimal SGP and therefore may function in SGP activity. A third stem-loop structure that is much larger than either of the first two is located 5′ terminal to the SG start site in the negative sense and contains a complement of the SP-ORF initiation codon (data not shown); whether it functions in SGP activity was not assessed by the mutagenesis experiments done in this study.
FIG. 7.
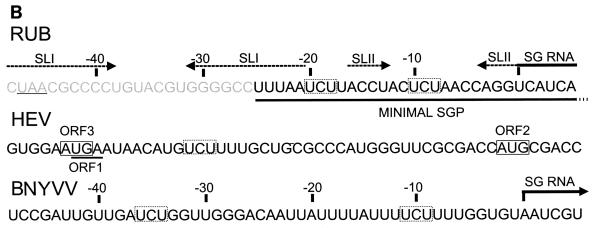
RUB SGP sequence analysis. (A) Secondary structure of the negative-polarity complement of the RUB junction region. The sequence shown extends from nt 6381 through 6453. Included in this region are the NS-ORF termination codon (small box) and the SG start site (arrow). Two thermodynamically stable stem-loops (SLs), denoted SLI and SLII, were predicted. Two AGA triplets (UCU in the positive sense) conserved in the SGPs of alphavirus family members are indicated. The minimal SGP defined in this study is outlined by a large box with a broken line on one side. (B) Alignment of the junction UTRs of RUB, HEV, and RNA3 of BNYVV. In the RUB sequence, the SG start site, the minimal SGP defined in this study, the UCU triplets conserved in SGPs of alphavirus superfamily members, and the sequences forming the stems of SLI and SLII are indicated; the sequence sharing homology with alphavirus SGPs is shown in gray type. In the HEV sequence, the UGA terminating ORF1 is underlined, the AUGs initiating ORF2 and ORF3 are boxed, and the conserved UCU triplet is indicated by a box with broken lines. In the BNYVV sequence, the SG start site and the conserved UCU triplets are highlighted.
From comparisons of SGP sequences among members of the alpha-like viruses and in vitro studies, a consensus superfamily SGP primary sequence has been recognized. In negative polarity, this sequence is 3′-AGANNCNG(N)5A(N)3-5XA-5′, where X is the SG start site and is usually a U or a C (20). Other than the presence of two AGA triplets (Fig. 7), the RUB SGP does not fit this consensus, including the X nucleotide, which is an A in RUB (a G also appears to suffice; W.-P. Tzeng, unpublished observations). Interestingly, the majority of the recombinant viruses recovered from one of the dsRobo402/GFP 5′ deletion mutants with minimal viability, the mutant with deletion B441, contain a duplicated AUCUUA sequence that contributes an additional AGA triplet in the negative polarity (this duplication is immediately upstream from SLII and thus does not affect this secondary structure).
Among alphavirus superfamily members, RUB is most closely related to beet necrotic yellow vein virus (BNYVV) and hepatitis E virus (HEV) on the basis of amino acid homology in the helicase and replicase regions of the nonstructural proteins (8, 24). The SGP of BNYVV RNA3 has been mapped to between nt −16 and +100 to +208 with respect to the SG start site (1), and there is no large-scale homology between the SGPs of RUB and BNYVV (or with those of alphaviruses). Interestingly, however, the BNYVV SGP has two AGA triplets (in the negative polarity) within 40 nt of the SG start site, one of which is at nt −9 to −11 of the SG start site, very close to the position of one of the AGA triplets in the RUB SGP (nt −8 to −10) (Fig. 7B). While the HEV SGP has not been mapped, there is one AGA triplet in the junction region between ORF1 (the NS-ORF) and ORF2 (the SP-ORF) (Fig. 7B). This conservation suggests that these AGA triplets may be important in SGP function in these three viruses and leads to the prediction that the SG start site of HEV is at nt 18 or 19 upstream from the start of ORF2 (there may be a second SG RNA synthesized by HEV from which ORF3, which overlaps both ORF1 and ORF2 and encodes a protein of unknown function, is translated).
Acknowledgments
This research was supported by grant AI-21389 from NIAID, NIH. W.-P.T. received Research Program Enhancement support from the GSU Office of Research and Sponsored Programs.
REFERENCES
- 1.Balmori, E., D. Gilmer, K. Richards, H. Guilley, and G. Jonard. 1993. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie 75:517-521. [DOI] [PubMed] [Google Scholar]
- 2.Chen, M.-H., and T. K. Frey. 1999. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73:3386-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vitro synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey, T. K. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haasnoot, P. C. J., F. T. Brederode, R. C. L. Olsthoorn, and J. F. Bol. 2000. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertz, J. M., and H. V. Huang. 1992. Utilization of heterologous alphavirus junction sequences as promoters by Sindbis virus. J. Virol. 66:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koev, G., and W. A. Miller. 2000. A positive-strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 74:5988-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 9.Lai, M. M. 1990. Coronavirus: organization, replication and expression of genome. Annu. Rev. Microbiol. 44:303-333. [DOI] [PubMed] [Google Scholar]
- 10.Levis, R., S. Schlesinger, and H. V. Huang. 1990. The promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol. 64:1726-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Pugachev, K. V., E. S. Abernathy, and T. K. Frey. 1997. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J. Virol. 71:562-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugachev, K. V., E. S. Abernathy, and T. K. Frey. 1997. Genomic sequence of the RA27/3 vaccine strain of rubella virus. Arch. Virol. 142:1165-1180. [DOI] [PubMed] [Google Scholar]
- 15.Pugachev, K. V., W.-P. Tzeng, and T. K. Frey. 2000. Development of a rubella virus vaccine expression vector: use of a picornavirus internal ribosome entry site increases stability of expression. J. Virol. 74:10811-10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raju, R., and H. V. Huang. 1991. Analysis of Sindbis virus promoter recognition in vivo using novel vectors with two subgenomic mRNA promoters. J. Virol. 65:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Sawicki, S. G., and D. L. Sawicki. 1990. Cornavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 20.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng, W.-P., M.-H. Chen., C. A. Derdeyn, and T. K. Frey. 2001. Rubella virus DI RNAs and replicons: requirement for nonstructural proteins acting in cis for amplification by helper virus. Virology 289:63-73. [DOI] [PubMed] [Google Scholar]
- 23.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Artervirus discontinuous mRNA transcription is guided by base paring between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver, S. C., A. Hagenbaugh, L. A. Bellew, S. V. Netesov, V. E. Volchkov, G.-J. Chang, D. K. Clarke, L. Gousset, T. W. Scott, D. T. Trent, and J. J. Holland. 1993. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197:375-390. [DOI] [PubMed] [Google Scholar]
- 25.Wielgosz, M. M., R. Raju, and H. V. Huang. 2001. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J. Virol. 75:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]