Abstract
Retroviral virions each contain two identical genomic RNA strands that are stably but noncovalently joined in parallel near their 5′ ends. For certain viruses, this dimerization has been shown to depend on a unique RNA stem-loop locus, called the dimer initiation site (DIS), that efficiently homodimerizes through a palindromic base sequence in its loop. Previous studies with Moloney murine leukemia virus (Mo-MuLV) identified two alternative DIS loci that can each independently support RNA dimerization in vitro but whose relative contributions are unknown. We now report that both of these loci contribute to the assembly of the Mo-MuLV dimer. Using targeted deletions, point mutagenesis, and antisense oligonucleotides, we found that each of the two stem-loops forms as predicted and contributes independently to dimerization in vitro through a mechanism involving autocomplementary interactions of its loop. Disruption of either DIS locus individually reduced both the yield and the thermal stability of the in vitro dimers, whereas disruption of both eliminated dimerization altogether. Similarly, the thermal stability of virion-derived dimers was impaired by deletion of both DIS elements, and point mutations in either element produced defects in viral replication that correlated with their effects on in vitro RNA dimerization. These findings support the view that in some retroviruses, dimer initiation and stability involve two or more closely linked DIS loci which together align the nascent dimer strands in parallel and in register.
Retroviral virions each contain two copies of the single-stranded viral genomic RNA that are stably but noncovalently linked to form a homodimer (17, 42). The dimeric genome is believed to promote retroviral replication and recombination by allowing reverse transcriptase to switch from one viral RNA strand to the other whenever it encounters a break in its template. Indirect evidence suggests that the dimerized chromosomes are bound together by direct RNA-RNA linkages along their entire lengths; this scenario could explain, for example, how such dimers remain intact even when both RNA strands are extensively nicked and all virion proteins have been removed (39). However, the nature and locations of the imputed dimer contacts are almost entirely unknown. Indeed, the only site of contact that has been characterized in detail is a short region, known as the dimer linkage site (DLS), that occurs near the 5′ ends of many or all retroviral genomes (17, 42). The DLS was first defined by electron microscopic examination of dimers extracted from virions, and its appearance and location are well conserved among diverse retroviruses (2, 22, 28, 37, 47). It is typically the most stable site of linkage between strands (often the only such site that can survive denaturation in 75% formamide) and hence may be the first to form during dimer assembly in vivo. Viruses carrying mutations in the DLS have been found to contain anomalously dimerized or monomeric RNA (as judged by nondenaturing gel electrophoresis) and also to have impaired replication (3, 12, 24, 30, 31, 35, 40, 41, 43), suggesting that accurate DLS-mediated dimerization is critical for the retroviral life cycle.
Synthetic RNAs that contain retroviral 5′ sequences can dimerize efficiently in vitro, and this fact has provided a model for dissecting the molecular basis of DLS assembly. Such studies have revealed that in several retroviruses, DLS dimerization depends upon a unique RNA stem-loop, called the dimer initiation site (DIS), that includes a palindromic (i.e., self-complementary) base sequence in its loop. DIS-mediated dimerization begins when the DIS loops of two strands anneal to form a metastable “kissing-loop” complex; in a subsequent isomerization step, the DIS stems in this complex melt and then reanneal to form a single, linear double helix that stably links the two strands. In protein-free artificial systems, isomerization does not occur unless the kissing-loop complex is heated briefly to 50 or 60°C; in the virus, however, it is catalyzed efficiently at room temperature by the viral nucleocapsid protein (6, 16, 44). Although their sequences differ widely, analogous DIS stem-loops have been identified in a growing number of retroviruses, suggesting that they may be a universal feature of these viruses.
Efforts to map the DIS locus of one prototypical retrovirus, Moloney murine leukemia virus (Mo-MuLV), have yielded divergent results (15, 38, 44). Early attention centered on a potential stem-loop at residues 283 to 298 that has a palindromic sequence and was shown to be sufficient to direct RNA dimerization in vitro. This candidate DIS, together with two additional stem-loops immediately downstream, also form part of the viral packaging signal (12, 34, 35). Recently, however, other authors identified a second candidate DIS stem-loop a short distance upstream (at residues 204 to 228) that reportedly can mediate efficient dimerization independently of the known downstream elements (38). While each of the two proposed DIS elements has the features necessary, in theory, to support kissing-loop interactions, the actual folded conformations of these elements and the pathways by which they may dimerize have not been directly explored. Additionally, as neither locus has been tested for dimerization activity in vivo, their biological relevance is as yet unproven, and it is not yet clear to what extent Mo-MuLV RNA dimerization depends on one, the other, or both.
To address these questions, we have carried out studies by using Mo-MuLV RNA sequences that encompass both of the candidate DIS loci. Through systematic mutagenesis and antisense interference, we have probed the functional organization of these loci as well as the contributions that each makes to RNA dimerization in vitro. In addition, we have also compared the effects of mutating these elements individually in virions. Our findings reveal a novel bipartite organization for the signals that initiate genomic dimer assembly in this virus.
MATERIALS AND METHODS
RNA synthesis.
DNA templates for in vitro transcription were synthesized by PCR from the vector pINA10 (a kind gift from E. Barklis), which comprises a G418 resistance gene flanked by 5′ and 3′ sequences from Mo-MuLV (35). Each of the forward primers (5′-GAAGATCTTCTAATACGACTCACTATAGGTGGGGGCTCGTCCGGGATC-3′ and 5′-GAAGATCTTCTAATACGACTCACTATAGGCCTGCGTCGGTACAGTTAGC-3′) incorporated a T7 promoter sequence and a BlgII restriction site and, when paired with the reverse primer (5′-CCCAAGCTTGGGCTAATTCTCAGACAAATACAGAAACACAGTCAGAC-3′), amplified residues 146 to 621 and residues 270 to 621, respectively, of the Mo-MuLV genome, where residue 1 is the first transcribed base. PCR products were purified by using a QIAquick kit (Qiagen) and redissolved in RNase-free water. Transcription reaction mixtures (20 μl) contained 1 μg of DNA template; 0.5 mM each unlabeled ATP, GTP, and CTP; 50 μM unlabeled UTP; and 2.5 μM α-32P-labeled UTP. Reactions were performed by using a T7 MAXIscript kit (Ambion). Transcripts were purified by electrophoresis on 5% polyacrylamide-8 M urea gels in TBE buffer (90 mM Tris-borate, 2 mM EDTA), ethanol precipitated, and resuspended in RNase-free water.
RNA dimerization assay.
For the standard dimerization assay, 8-μl samples containing one or more labeled RNAs (each at approximately 1.5 nM in water) were heat denatured at 95°C for 3 min, immediately snap-cooled on ice, and combined with 2 μl of cold D5X buffer (500 mM NaCl, 250 mM Tris [pH 7]). The samples were incubated at various temperatures (0 to 70°C) for 1 h, chilled, and analyzed by electrophoresis on nondenaturing 5% polyacrylamide gels in TBE buffer. After being dried, gels were analyzed by using a PhosphorImager (Molecular Dynamics). Antisense inhibition studies were performed in the same way, except that antisense DNA oligonucleotides were added (at a final concentration of 60 μM) before heat denaturation. For each sample, the fraction of RNA in dimeric form was determined from phosphorimager signal intensities as the ratio of dimeric RNA to the sum of both the monomeric and the dimeric species.
Cell cultures, transfections, and infections.
GP+E86 murine ecotropic packaging cells (ATCC CRL 9642) and NIH 3T3 murine fibroblasts (ATCC CRL 1658) were maintained in Dulbecco's modified Eagle's medium with a high glucose concentration (4.5 g/liter) and 10% calf serum (standard medium). Wild-type or mutant pINA10 DNAs (20 μg) were transfected into 5.0 × 105 GP+E86 cells in each 60-mm polystyrene dish by calcium phosphate coprecipitation along with 0.25 μg of the luciferase-encoding vector pGL3 (Promega) as an internal control. Viral titers were estimated by luciferase assays of the transfected cells as described previously (26, 27), and aliquots of culture supernatants were used to infect 2.5 × 105 NIH 3T3 cells in the presence of 8 μg of Polybrene/ml. Infectious titers were determined with these cells by G418-resistant colony formation assays as described previously (26, 27). Producer cell lines were made by stably transfecting GP+E86 cells with pINA10 vectors containing either wild-type or mutated DIS sequences and then selecting neomycin-resistant colonies by growth in 500 μg of Geneticin (Gibco-BRL)/ml; several hundred colonies were pooled and used to produce virus for subsequent experimentation.
Nondenaturing gel electrophoresis.
Northern blot analysis of virion RNA on native agarose gels was performed as described by Fu and colleagues (13, 14). Briefly, 5 × 106 cells stably transfected with a neomycin resistance cassette (neor) in 40 ml of standard medium containing 500 μg of Geneticin/ml were seeded into T-175 tissue culture flasks (Falcon). Virus supernatants were collected 2 days later and passed through 0.45-μm-pore-size filters. Virus was pelleted from 70 ml of total culture volume for each of the viral constructs by ultracentrifugation in an SW28 rotor (Beckman) at 25,000 rpm for 30 min at 4°C. Virion RNAs were then extracted with virion lysis buffer (0.05 M Tris, 0.01 M EDTA, 0.1 M NaCl, 1% sodium dodecyl sulfate, 0.05 mg of yeast RNA/ml, 0.1 mg of proteinase K/ml) and resuspended in R buffer (0.01 M Tris, 0.001 M EDTA, 1% sodium dodecyl sulfate, 0.05 M NaCl) as described previously (13). A 0.1 volume of resuspended virion RNAs was incubated at various temperatures for 10 min and placed on ice before being loaded onto 1% agarose gels. RNAs were separated in 1× Tris-acetate-EDTA buffer in a cold room (4°C) at 450 V, stained with ethidium bromide (1 μg/ml), and visualized. Nondenaturing Northern blotting was performed essentially as previously described (13). A TurboBlotter system was used to transfer RNAs onto Nytran supercharged nylon membranes as described by the manufacturer (Schleicher & Schuell). RNAs were UV cross-linked by using a Stratalinker (Stratagene) and hybridized with 106 cpm of riboprobe containing an antisense sequence of RNA-4 (see Fig. 2A), which is transcribed from the SP6 promoter of the pSP72-MuLVψ vector (unpublished data). Monomeric and dimeric RNAs were quantified by phosphorimaging.
FIG. 2.
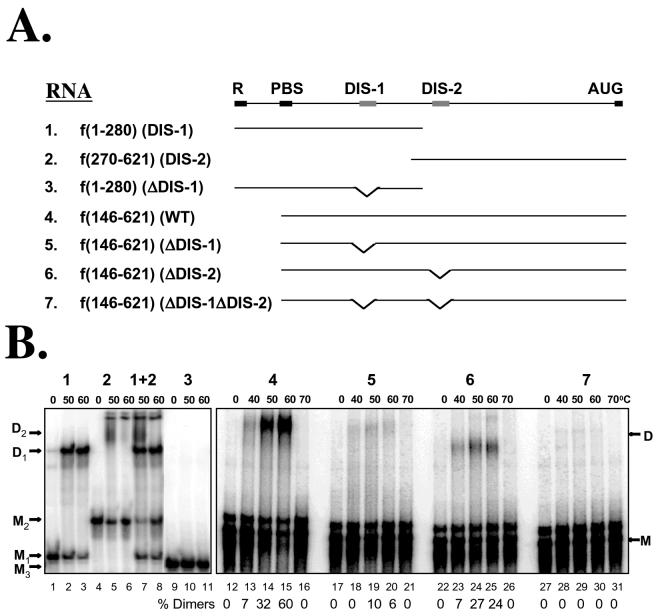
DIS-1 and DIS-2 each contribute independently to Mo-MuLV RNA dimerization in vitro. (A) Various radiolabeled synthetic RNA fragments containing DIS-1, DIS-2, or both (RNA-1 through RNA-7) were synthesized and tested in vitro. Terminal residues and major features of each are listed. PBS, primer binding site; SD, splice donor. (B) Temperature dependence of in vitro dimer formation. Each RNA was analyzed electrophoretically after preincubation at the temperatures listed above the lanes; RNA-1 and RNA-2 were also tested in combination (lanes 7 and 8). Positions of presumed monomers (M) and dimers (D) are indicated. For lanes 12 to 31, the percentage of dimers in each lane (quantified by phosphorimaging) is shown below the lanes.
RESULTS
Formation and stability of Mo-MuLV 5′ RNA dimers in vitro.
DIS elements are defined experimentally by their ability to promote efficient dimerization of a retroviral RNA in its native, highly folded conformation. For in vitro DIS assays, synthetic RNA is first heat denatured and then snap-cooled on ice in order to simulate native monomeric folding. Studies of human immunodeficiency virus type 1 RNA indicate that, if maintained at 40°C or below, these monomers form kissing-loop dimers but that such dimers are too labile to survive electrophoresis on TBE gels. Transient heating to 50 or 60°C, however, provides the energy necessary for isomerization into linear form (see above), yielding dimers that are stable on TBE gels (4, 10, 23, 36, 46, 48). We applied this same protocol in the current study, synthesizing and refolding portions of the Mo-MuLV 5′ sequence, briefly incubating these RNAs at various temperatures, and then examining the resultant complexes by TBE electrophoresis. As detailed below, our findings verified the existence of two closely linked DIS elements, one at residues 204 to 228 and the other at residues 283 to 298, denoted here as DIS-1 and DIS-2, respectively (Fig. 1). The dimer-sized RNA complexes that we detected electrophoretically were proven to be dimers in the course of our work (see Fig. 5); for brevity, we refer to these as dimers throughout.
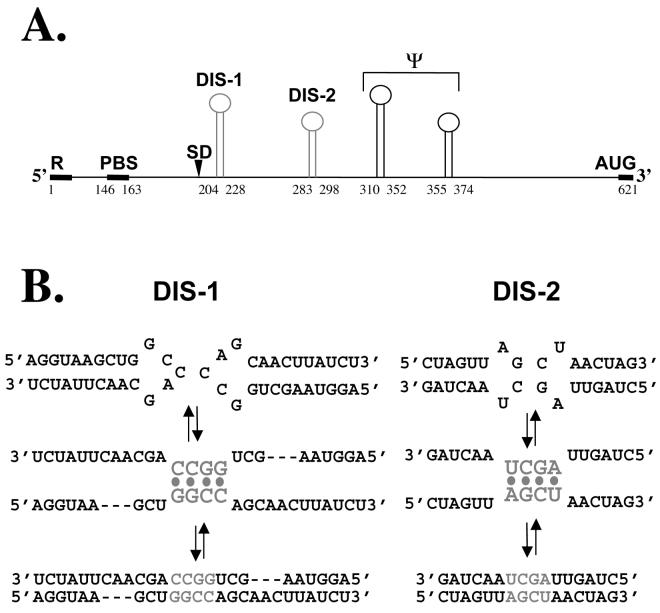
FIG. 1.
Candidate DIS loci in the Mo-MuLV genome. (A) Schematic depiction of the Mo-MuLV 5′ leader region, extending from the first transcribed base (residue 1) in the R region through the gag start codon (to residue 621). Locations of the primer binding site (PBS), major 5′ splice donor (SD), and palindromic portions of four putative stem-loop structures are shown. Of the latter, DIS-1 and DIS-2 are considered in this study; the two downstream stem-loops appear to serve as the core RNA packaging signal (ψ) (see reference 35). (B) Hypothetical pathways of kissing-loop dimerization for DIS-1 and DIS-2. Each initially homodimerizes through four central palindromic bases in and around its loop, forming a transient kissing-loop complex which then can isomerize into linear form.
FIG. 5.
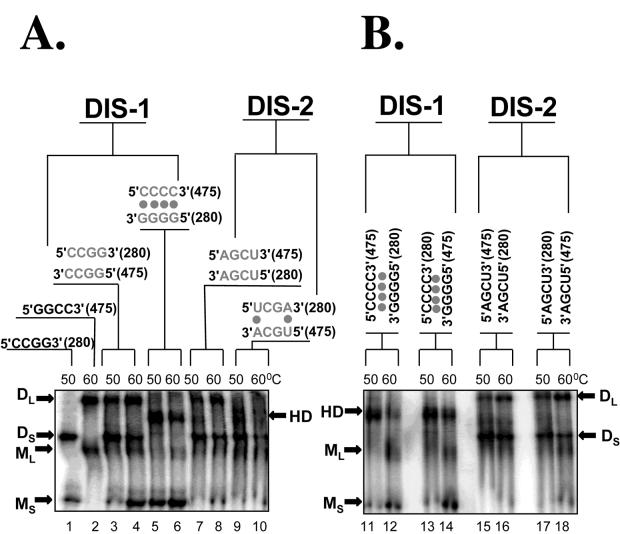
Heterodimer formation provides direct evidence for a complementary loop-loop interaction at each DIS locus. Mutant RNAs were synthesized in which the four central palindromic nucleotides of DIS-1 or DIS-2 (Fig. 1B) were replaced by the indicated sequences in the context of either RNA-1 (280 bases) or RNA-4 (475 bases). These were then assayed for dimerization alone or in combination (at 1.5 nM each) after preincubation at the temperatures indicated. Above lanes in which two different RNAs were combined, the sequences of their central bases are shown in antiparallel alignment, with dots indicating any expected base pairing. M and D, monomers and homodimers, respectively, of the long (L) and short (S) RNAs; HD, heterodimers. Panels A and B were derived from two separate representative experiments done with different combinations of mutants.
We began by confirming that RNAs containing either DIS-1 or DIS-2 alone could dimerize spontaneously in vitro. We transcribed two nonoverlapping portions of the Mo-MuLV genome, which are schematized in Fig. 2A: the first (RNA-1) extended from the 5′ R region through DIS-1 (i.e., residues 1 to 280), and the second (RNA-2) extended from immediately upstream of DIS-2 through the start codon of gag (residues 270 to 621). As shown in Fig. 2B, each exhibited temperature-dependent homodimer formation. When melted, snap-cooled, and maintained on ice, RNA-1, containing DIS-1, migrated exclusively as a monomer (M1); however, when heated to 50 or 60°C, it formed a discrete, dimer-sized complex (D1) that accounted for more than half of the input RNA (Fig. 2B, lanes 1 to 3). Precise deletion of the DIS-1 sequence from this RNA abolished all apparent dimerization (Fig. 2B, lanes 9 to 11). The somewhat longer RNA-2, which contained DIS-2, also was monomeric at a low temperature (M2) but formed a dimer (D2) when heated (Fig. 2B, lanes 4 to 6). When the two RNAs were mixed, snap-cooled, and incubated together, they each efficiently homodimerized but failed to produce an intermediate-sized complex that would suggest heterodimer formation (Fig. 2B, lanes 7 and 8). These data were in accord with earlier reports ascribing DIS activity to each of these regions in Mo-MuLV and further showed that the two regions do not heterodimerize appreciably.
We next evaluated a longer Mo-MuLV transcript (RNA-4) that extended from the primer binding site through the start codon of gag (i.e., residues 146 to 621) and so encompassed both DIS-1 and DIS-2. We also created three variants of this sequence (RNA-5, RNA-6, and RNA-7) from which DIS-1, DIS-2, or both had been precisely deleted (Fig. 2A). As shown in Fig. 2B, the wild-type RNA-4 remained exclusively monomeric (M) when kept on ice (lane 12) but formed a dimer (D) when incubated at 50 to 60°C (lanes 13 to 15). This dimer was most abundant in samples prepared at 60°C, where it accounted for more than half of the total RNA, but it dissociated back to monomers when heated to 70°C (Fig. 2B, lane 16). The mutants lacking either DIS-1 or DIS-2 alone, by comparison, yielded reduced amounts of dimer-sized complexes (accounting for no more than 10 to 30% of the input RNA) that were approximately equally abundant after preheating to either 50 or 60°C (Fig. 2B, lanes 17 to 26). When both DIS-1 and DIS-2 were deleted, no evidence of dimerization was detected over the range of temperatures that we tested (Fig. 2B, lanes 27 to 31). Taken together, these results indicate that DIS-1 and DIS-2 each can support dimerization independently but that dimers form most efficiently and stably when both loci are present. Deleting either locus alone significantly reduces the formation and/or thermal stability of the resulting dimers, and deleting both eliminates in vitro dimerization altogether.
Functional mapping of DIS loci by antisense inhibition.
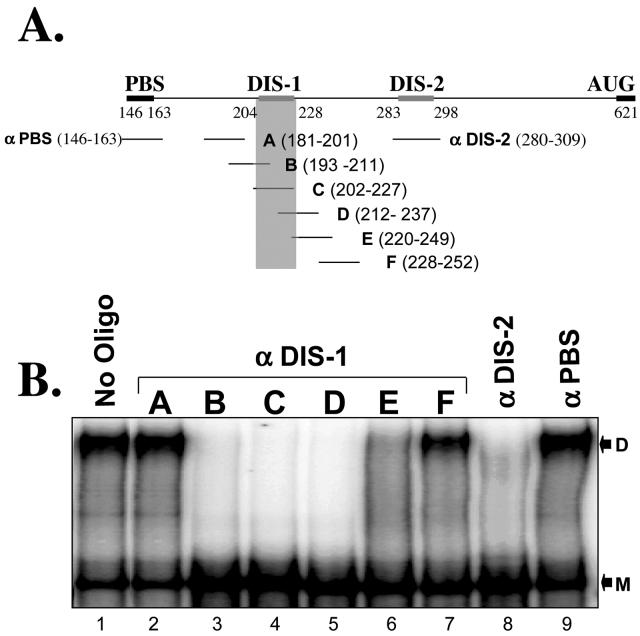
As an alternative means to delineate sequences required for Mo-MuLV dimer formation, we tested whether antisense DNA oligomers that targeted various regions of the Mo-MuLV leader sequence could serve as inhibitors of dimer formation (Fig. 3A). Our efforts focused mainly on DIS-1, which has been less extensively characterized than DIS-2. We synthesized a series of DNA oligonucleotides, each 18 to 29 bases long (Table 1), which were complementary to regions of Mo-MuLV RNA in and around the DIS loci (Fig. 3A). Each was then combined, in approximately a 1,000-fold molar excess, with the dimerization-competent RNA-4 (residues 146 to 621). The mixture was heat denatured, snap-cooled, warmed to 60°C, and tested for dimerization. As shown in Fig. 3B, RNA-4 dimerized efficiently in the absence of oligonucleotides (lane 1) and in the presence of DNAs complementary to sequences immediately upstream or downstream of DIS-1 (lanes 2 and 7) or to the primer binding site (lane 9). In contrast, oligonucleotides complementary to part (Fig. 3B, lanes 3, 5, and 6) or all (lane 4) of DIS-1 completely blocked dimer formation, as did the single oligonucleotide that targeted DIS-2 (lane 8). These results functionally verify the extent and location of DIS-1 and confirm that both DIS-1 and DIS-2 are specifically required for the formation and/or stability of Mo-MuLV dimers in vitro.
FIG. 3.
In vitro dimerization of full-length RNA (RNA-4) is inhibited by antisense DNA oligonucleotides complementary to the DIS sequences. (A) Antisense DNA oligomers complementary to various regions of the Mo-MuLV 5′ UTR were designed; their properties are detailed in Table 1. PBS, primer binding site. (B) α-32P-labeled RNA-4 (1.5 nM) was incubated with 60 μM concentrations of various unlabeled antisense oligomers at 60°C for 1 h and then analyzed on a 5% nondenaturing polyacrylamide gel. Dimerization of RNA-4 was inhibited only by oligomers that overlapped portions of the predicted palindromes in DIS-1 or DIS-2. D, dimeric RNA; M, monomeric RNA. αDIS-1, αDIS-2, and αPBS, antisense oligonucleotides complementary to DIS-1, DIS-2, and PBS sequences, respectively.
TABLE 1.
Antisense oligonucleotides
| Oligonucleotide (bases)a | Sequence (5′-3′)b | Melting temp (°C)c |
|---|---|---|
| αPBS (146-163) | ATCCCGGACGAGCCCCC | 74 |
| αDIS-2 (280-309) | CAGATACAGAGCTAGTTAGCTAACTAGTAC | 75 |
| A (181-201) | CGGTGGTGGGTCGGTGGTCCCTGGG | 86 |
| B (193-211) | GCTTACCTCCCGGTGGTGG | 74 |
| C (202-227) | GATAAGTTGCTGGCCAGCTTACCTC | 76 |
| D (212-237) | GACAGACACAGATAAGTTGCTGGCCA | 76 |
| E (220-249) | ACTAGACAATCGGACAGACACAGATAAGTT | 75 |
| F (228-252) | GACACTAGACAATCGGACAGACACA | 74 |
Numbering is with respect to the Mo-MuLV viral mRNA (GenBank accession no. JO2255).
Sequences are antisense in polarity with respect to the viral mRNA.
As estimated by the percent GC method.
Mutational evidence of base pairing in the DIS-1 and DIS-2 stems.
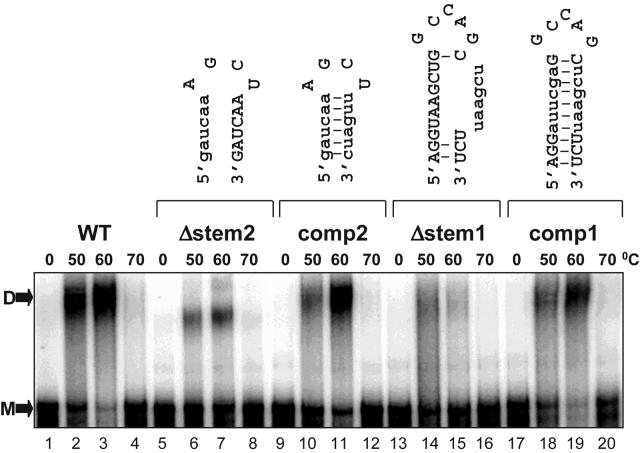
We used a mutational approach to assess whether the putative stem regions of DIS-1 and DIS-2 fold as predicted. Into the dimerization-competent RNA-4 sequence (residues 146 to 621), we first introduced point mutations designed to disrupt all or most predicted base pairing of the DIS-1 and DIS-2 stems (mutants Δstem1 and Δstem2, respectively) and then created compensatory mutations that could restore base pairing (mutants comp1 and comp2). As shown in Fig. 4, we found that disrupting either stem alone substantially inhibited dimer yield and stability but that compensatory mutations restored dimerization to nearly wild-type levels. These results suggest that the stems of DIS-1 and DIS-2 each form and contribute to dimer formation in vitro.
FIG. 4.
The predicted stems of DIS-1 and DIS-2 form and are essential for in vitro dimerization. Wild-type RNA-4 (WT) and four mutant forms of this RNA were tested for dimerization after preincubation at the temperatures indicated. The mutations were designed to either disrupt (Δstem1 and Δstem2) or maintain (comp1 and comp2) base pairing in the predicted stems of DIS-1 and DIS-2, producing the hypothetical secondary structures shown at the top. D, dimer; M, monomer.
Evidence for homologous loop-loop interactions.
To determine conclusively whether the slower-migrating species seen on electrophoresis resulted from loop-mediated DIS dimerization, we used a heterodimerization assay described previously (4, 27). In this assay, two RNAs of different lengths and each containing a candidate DIS locus are combined and tested for dimerization; the detection of a novel intermediate-sized complex distinct from either homodimer implies that the two RNAs have heterodimerized. For the present study, we prepared synthetic Mo-MuLV transcripts that were 280 or 475 bases long and that each spanned both DIS-1 and DIS-2; we also prepared variants of these transcripts that carried specific point mutations in the four central bases of each DIS loop palindrome to test their effects on dimer formation. As shown in Fig. 5A, we found that a short (280-base) RNA containing the wild-type DIS-1 loop palindrome (CCGG) homodimerized efficiently (lane 1), as did a long (475-base) RNA containing the related palindrome GGCC (lane 2). When these two were combined, each homodimerized independently, but no heterodimers were observed at either 50 or 60°C (Fig. 5A, lanes 3 and 4). In contrast, when we combined two other RNAs carrying the nonpalindromic DIS-1 loop sequences CCCC and GGGG, each failed to homodimerize, but abundant heterodimers (HD) were observed (Fig. 5A, lanes 5 and 6). Similarly, RNAs bearing either the wild-type sequence AGCU or the variant palindrome UCGA in the DIS-2 loop formed homodimers but not heterodimers (Fig. 5A, lanes 7 and 8), whereas the two partially complementary DIS-2 variants UCGA and UGCA were able to support heterodimer formation (lanes 9 and 10). Identical results were obtained when the loop sequences in the long and short RNAs were reversed (Fig. 5B). Together, these results suggest that dimerization normally depends on homologous base pairing at the DIS-1 and DIS-2 loops, as would be expected if each of these loci homodimerized through loop-loop interactions.
Effects of DIS mutations in Mo-MuLV virions.
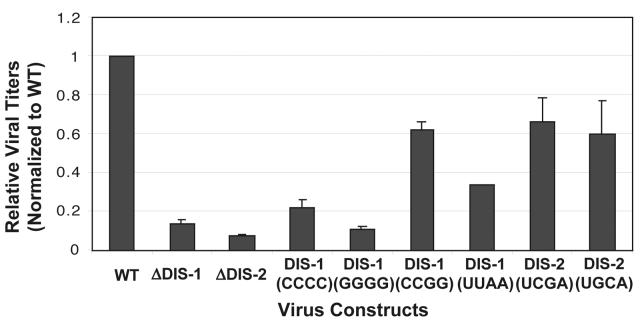
We next explored the effects of mutating the DIS loci on the properties of Mo-MuLV virions. For this purpose, mutations were introduced into the candidate DIS sequences in pINA10, an Mo-MuLV-derived retroviral vector carrying a neor gene. The resulting mutant vectors were transfected into the packaging cell line GP+E86, which is capable of incorporating wild-type pINA10 transcripts into Mo-MuLV virion-like particles that are then capable of transducing the selectable neor marker into susceptible target cells. Enumeration of the resulting neomycin-resistant colonies provides a measure of the transduction efficiency of the virus-like particles. As summarized in Fig. 6, we found that precisely deleting either DIS-1 or DIS-2 alone reduced transduction by approximately 90% compared to the results obtained with wild-type pINA10. Indeed, simply altering two of the four central bases of the DIS-1 loop palindrome to create the nonpalindromic sequence CCCC or GGGG inhibited transduction to nearly the same degree as deleting DIS-1 entirely. In contrast, four other mutations that preserved the palindromic natures of the DIS-1 or DIS-2 loops caused somewhat lesser deficits, in most cases inhibiting transduction by only 30 to 40%. Thus, in general, the relative impact of each mutation on transduction efficiency in vivo correlated with its effect on RNA dimerization in vitro.
FIG. 6.
Relative titers of Mo-MuLV virus-like particles carrying DIS mutations. Particles were harvested from the supernatants of cells transfected with derivatives of the vector pINA10 that contained either wild-type (WT) or mutant forms of the Mo-MuLV leader region. The samples were assayed for transducing titers by using a colony-forming assay (see Materials and Methods). The mutations tested included precise deletions of DIS-1 or DIS-2 (ΔDIS-1 or ΔDIS-2, respectively) or substitutions of the indicated four-base sequences for the four central bases of the palindrome in DIS-1 or DIS-2. Titers were normalized to that of the WT vector, which averaged 5.7 × 105 CFU/ml in the experiment shown here. Data were from three independent assays. Error bars show standard deviations.
To determine whether deleting the DIS loci affects RNA dimerization in these virion-like particles, we extracted RNA from particles carrying both DIS-1 and DIS-2 mutations, incubated it at various temperatures, and then analyzed it by nondenaturing gel electrophoresis and Northern blotting. As shown in Fig. 7, wild-type RNA containing both DIS loci migrated primarily as dimer-sized complexes (D) that dissociated into apparent monomers (M) at a melting temperature of 60°C (lanes 1 to 3). The RNA lacking both DIS loci, in contrast, yielded dimers with a melting temperature of less than 55°C. These results closely parallel those that we obtained for the in vitro dimerization of the same mutant constructs (Fig. 2) and confirm the contributions of these DIS loci to Mo-MuLV RNA dimerization in vivo.
FIG. 7.
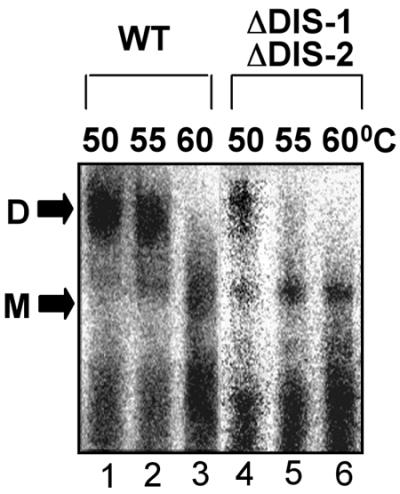
Nondenaturing Northern blots of virion-derived RNAs. Deproteinized RNAs extracted from Mo-MuLV virion-like particles containing either wild-type (WT) pINA10 or a mutant that lacked both DIS-1 and DIS-2 (ΔDIS-1 ΔDIS-2) were preheated to the indicated temperatures for 10 min, electrophoresed on a 1% agarose gel at 4°C, blotted onto a nylon membrane, and probed for pINA10 sequences. All lanes shown are from a single blot, but exposure times for the two constructs varied to compensate for differences in image intensity; these differences may reflect disparities in RNA packaging, virion release, or other, undetermined factors. Results shown are typical of three independent experiments. D, dimer; M, monomer.
DISCUSSION
The importance of Mo-MuLV as both a model retrovirus and a versatile gene transfer vector has prompted numerous studies of the cis-acting dimerization signals in its genome. Early electron microscopic images of dimers from Mo-MuLV virions identified the DLS as the most stable point of contact between RNA strands, suggested that it linked the strands in parallel (i.e., 5′ to 5′), and indicated its location as approximately 400 bases from the 5′ end (2, 22, 28, 37, 47). Later, artificial DLS dimers formed from synthetic Mo-MuLV sequences showed a similar morphology, a finding that helped confirm the validity of the in vitro dimerization assay (44). The in vitro DLS could also be mapped more precisely to residues 215 to 420, a region previously shown to encompass critical packaging signals (29, 45), and this finding raised the possibility that RNA dimerization and packaging are mechanistically linked (44). Subsequent research on both processes has tended to focus primarily on this region of the genome.
Chemical probing, computerized structure prediction, and mutational analyses all indicate that Mo-MuLV residues 215 to 420 can fold into at least three separate stem-loops (1, 7, 15, 51). The most 3′ of these, which we term DIS-2 (other authors refer to it as stem-loop B or H1), is centered around residues 283 to 298 and has been shown to be essential for dimerization in vitro (7, 15, 16) as well as to enhance the efficiency of genomic packaging into Mo-MuLV virions (12, 34, 35). The palindromic sequence of its loop, together with thermodynamic, kinetic, and chemical evidence that it undergoes a conformational change during dimerization (51), suggests that DIS-2 dimerizes through a kissing-loop pathway similar to that described for other retroviruses (4, 8, 9, 11, 19, 20, 25, 26, 33, 39). Two other presumed stem-loops downstream of DIS-2 (at residues 310 to 352 and residues 355 to 374) appear to function primarily in packaging (12, 34, 35) but also reportedly enhance the kinetics of DIS-2-dependent dimer formation (7), perhaps by homodimerizing weakly through their loops (21). Similarly, the inclusion of additional sequences farther downstream (residues 364 to 565) was also found to stabilize DIS-2-dependent dimers (16), though the exact sequences involved have not been further characterized.
Nearly all of the foregoing results were derived from studies of RNAs that lacked most sequences upstream of DIS-2. Oroudjev and colleagues (38), in contrast, recently reported that those upstream sequences alone could dimerize efficiently in vitro and that this scenario depended on a hypothetical stem-loop at positions 204 to 228. This upstream element, which we term DIS-1, is well conserved among murine C-type retroviruses, including those that lack any apparent ortholog of DIS-2. Data from Mikkelsen and coworkers (32) further suggested that DIS-1 might serve as a hot spot for template switching by reverse transcriptase, an activity similar to that reported for DIS-2 of Mo-MuLV and for DIS elements in certain other retroviruses. In light of those findings, it became unclear whether DIS-1 or DIS-2 was the primary initiator of Mo-MuLV dimerization.
Our study reconciles and extends the earlier reports by showing that both proposed DIS loci contribute to dimer formation. By analyzing longer RNAs that span the entire 5′ untranslated region and so encompass both DIS-1 and DIS-2, we found that precisely deleting either locus alone partially inhibits in vitro dimer formation but that deleting both eliminates dimerization altogether (Fig. 2B). We observed that each locus contributes independently not only to the yield but also to the thermal stability of in vitro dimers: whereas the wild-type dimer was stable at 60°C, dimers lacking either DIS-1 or DIS-2 or the loop sequences of these two structures were generally more abundant at 50°C than at 60°C (Fig. 5 and data not shown). These findings are in accord with published melting data for shorter RNAs that contained one or the other DIS (38, 44). Our data can also account for the earlier observation that deleting DIS-2 along with two downstream stem-loops reduced in vitro dimer yield by only about 40%, provided that upstream sequences remained intact (7).
Although the detailed structures of DIS-1 and DIS-2 remain unknown, the results of our site-directed mutagenesis studies help confirm that each folds as predicted and that their folded structures are important for efficient dimer formation. Mutations that disrupt the proposed stem structures markedly reduced dimerization in our in vitro assay, whereas compensatory mutations designed to restore those stems yielded nearly wild-type levels of dimerization (Fig. 4). Similarly, a mutational analysis of the predicted loop regions indicated that the in vitro DIS activity of either locus depends upon Watson-Crick complementarity between the loops (Fig. 5 and data not shown). To our knowledge, these are the first reported results of point mutagenesis for either locus, and they strongly support the assumption that each dimerizes independently through a kissing-loop pathway.
We also tested the effects of DIS mutations on RNA dimerization within Mo-MuLV virion-like particles in vivo by using a truncated recombinant Mo-MuLV genome in a previously validated cell culture model (33). As shown in Fig. 7, we found that deproteinated RNA extracted from these particles was present primarily in the form of a dimer that melted at 55 to 60°C; these temperatures are comparable to the melting temperatures previously reported for genomic dimers extracted from authentic Mo-MuLV (13, 28). In agreement with our in vitro data, we found that deleting both DIS-1 and DIS-2 reduced dimer stability, so that melting was essentially complete at 55°C. It is not surprising that this double deletion did not abolish in vivo dimer formation completely, since dimer contacts are believed to occur along the entire length of a genomic strand once it has been encapsidated (39). Similar instability has been reported for some DIS mutants of human immunodeficiency virus type 1 (4, 5, 18, 24). In our series, mutations that inhibited dimer formation in vitro also tended to reduce viral titer in vivo (Fig. 6), but is not clear to what extent these infectivity defects can be attributed to aberrant dimerization, since the mutated regions also influence packaging, reverse transcription, and possibly other aspects of Mo-MuLV replication (12, 34, 35).
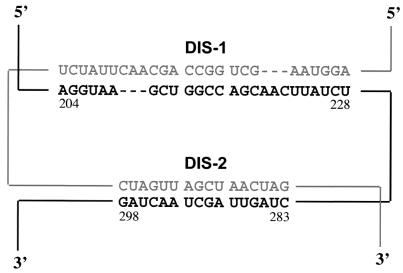
Our findings indicate that two separate stem-loop elements, situated 55 bases apart in the Mo-MuLV leader region, each contribute to genomic dimer initiation through independent, homologous kissing-loop interactions. Although this may be the first documented example of dimer initiation through dual kissing loops, the notion that two or more adjacent loci can act in concert to achieve dimer linkage is not unprecedented and may prove to be a common theme among retroviruses (11, 26, 49, 50). In murine sarcoma virus, for example, the genomic strands are thought to associate through specific G-rich elements, which then facilitate dimerization through an adjacent kissing loop (25, 26). Even in Mo-MuLV itself, additional elements downstream of the bipartite DIS can enhance the kinetics and stability of dimer formation (7, 16). Although not yet proven, it seems likely that the two consecutive stretches of interstrand base pairing created by the bipartite DLS of Mo-MuLV (Fig. 8) may appear as a single DLS structure when viewed at the resolution of the electron microscope. This topology illustrates how two or more locally antiparallel linkages can give rise to a globally parallel genomic dimer.
FIG. 8.
Proposed model of the globally parallel genomic dimer formed by two locally antiparallel DIS loci in Mo-MuLV.
Acknowledgments
We thank E. Barklis for the generous gift of pINA10 vectors, J. L. Clever for technical advice and many stimulating discussions, and D. Miranda and Y. Liang for help with artwork.
This research was supported by NIH grants AI37036 and AI40317 to T.G.P. H.L. was supported in part by UCSF AIDS postdoctoral training grant NIH AI07395.
REFERENCES
- 1.Alford, R. L., S. Honda, C. B. Lawrence, and J. W. Belmont. 1991. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology 183:611-619. [DOI] [PubMed] [Google Scholar]
- 2.Bender, W., Y.-H. Chien, S. Chattopadhyay, P. K. Vogt, M. B. Gardner, and N. Davidson. 1978. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 25:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout, B., and J. L. van Wamel. 1996. Role of the dimer initiation site hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darlix, J.-L., C. Gabus, M.-T. Nugeyre, F. Clavel, and F. Barre-Sinoussi. 1990. cis-elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 216:689-699. [DOI] [PubMed] [Google Scholar]
- 7.deTapia, M., V. Metzler, M. Mougel, B. Ehresmann, and C. Ehresmann. 1998. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374). Biochemistry 37:6077-6085. [DOI] [PubMed] [Google Scholar]
- 8.Dirac, A. M., H. Huthoff, J. Kjems, and B. Berkhout. 2001. The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem. 276:32345-32352. [DOI] [PubMed] [Google Scholar]
- 9.Doria-Rose, N., and V. M. Vogt. 1998. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 72:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, Y. X., W. Fu, A. J. Winter, J. G. Levin, and A. Rein. 1995. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J. Virol. 69:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, J., and S. P. Goff. 1998. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology 244:133-145. [DOI] [PubMed] [Google Scholar]
- 13.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard, P. M., B. Bonnet-Mathoniere, D. Muriaux, and J. Paoletti. 1995. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry 34:9785-9794. [DOI] [PubMed] [Google Scholar]
- 16.Girard, P. M., H. de Rocquigny, B. P. Roques, and J. Paoletti. 1996. A model of psi dimerization: destabilization of the C278-G303 stem-loop by the nucleocapsid protein (NCp10) of MoMuLV. Biochemistry 35:8705-8714. [DOI] [PubMed] [Google Scholar]
- 17.Greatorex, J., and A. Lever. 1998. Retroviral RNA dimer linkage. J. Gen. Virol. 9:2877-2882. [DOI] [PubMed] [Google Scholar]
- 18.Haddrick, M., A. L. Lear, A. J. Cann, and S. Heaphy. 1996. Evidence that a kissing loop structure facilitates genomic RNA dimerization in HIV-1. J. Mol. Biol. 259:58-68. [DOI] [PubMed] [Google Scholar]
- 19.Jossinet, F., J. S. Lodmell, C. Ehresmann, B. Ehresmann, and R. Marquet. 2001. Identification of the in vitro HIV-2/SIV RNA dimerization site reveals striking differences with HIV-1. J. Biol. Chem. 276:5598-5604. [DOI] [PubMed] [Google Scholar]
- 20.Katoh, I., H. Kyushiki, Y. Sakamoto, Y. Ikawa, and Y. Yoshinaka. 1991. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J. Virol. 65:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, C. H., and I. Tinoco, Jr. 2000. A retroviral RNA kissing complex containing only two G·C base pairs. Proc. Natl. Acad. Sci. USA 97:9396-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kung, H. J., S. Hu, W. Bender, J. M. Bailey, N. Davidson, M. O. Nicolson, and R. M. McAllister. 1976. RD-114, baboon, and woolly monkey viral RNAs compared in size and structure. Cell 7:609-620. [DOI] [PubMed] [Google Scholar]
- 23.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 24.Laughrea, M., L. Jette, J. Mak, L. Kleiman, C. Liang, and M. A. Wainberg. 1997. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 71:3397-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lear, A. L., M. Haddrick, and S. Heaphy. 1995. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology 212:47-57. [DOI] [PubMed] [Google Scholar]
- 26.Ly, H., D. P. Nierlich, J. C. Olsen, and A. H. Kaplan. 1999. Moloney murine sarcoma virus genomic RNAs dimerize via a two-step process: a concentration-dependent kissing-loop interaction is driven by initial contact between consecutive guanines. J. Virol. 73:7255-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ly, H., D. P. Nierlich, J. C. Olsen, and A. H. Kaplan. 2000. Functional characterization of the dimer linkage structure RNA of Moloney murine sarcoma virus. J. Virol. 74:9937-9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maisel, J., W. Bender, S. Hu, P. H. Duesberg, and N. Davidson. 1978. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J. Virol. 25:384-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann, R., and D. Baltimore. 1985. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J. Virol. 54:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen, J. G., A. H. Lund, M. Duch, and F. S. Pedersen. 1998. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J. Virol. 72:6967-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monie, T., J. Greatorex, and A. M. Lever. 2001. Oligonucleotide mapping of the core genomic RNA dimer linkage in human T-cell leukaemia virus type-1. Virus Res. 78:45-56. [DOI] [PubMed] [Google Scholar]
- 34.Mougel, M., and E. Barklis. 1997. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 71:8061-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougel, M., Y. Zhang, and E. Barklis. 1996. cis-Active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 70:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 37.Murti, K. G., M. Bondurant, and A. Tereba. 1981. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 37:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oroudjev, E. M., P. C. Kang, and L. A. Kohlstaedt. 1999. An additional dimer linkage structure in Moloney murine leukemia virus RNA. J. Mol. Biol. 291:603-613. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz-Conde, B. A., and S. H. Hughes. 1999. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J. Virol. 73:7165-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486-27493. [PubMed] [Google Scholar]
- 41.Paillart, J. C., L. Berthoux, M. Ottmann, J. L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paillart, J. C., R. Marquet, E. Skripkin, C. Ehresmann, and B. Ehresmann. 1996. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie 78:639-653. [DOI] [PubMed] [Google Scholar]
- 43.Paillart, J. C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93:5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prats, A.-C., C. Roy, P. A. Wang, M. Erard, V. Housset, C. Gabus, C. Paoletti, and J. L. Darlix. 1990. cis Elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 64:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartzberg, P., J. Colicelli, and S. P. Goff. 1983. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J. Virol. 46:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi, K. I., S. Baba, P. Chattopadhyay, Y. Koyanagi, N. Yamamoto, H. Takaku, and G. Kawai. 2000. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA 6:96-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrent, C., T. Bordet, and J. L. Darlix. 1994. Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus. J. Mol. Biol. 240:434-444. [DOI] [PubMed] [Google Scholar]
- 50.Torrent, C., C. Gabus, and J. L. Darlix. 1994. A small and efficient dimerization/packaging signal of rat VL30 RNA and its use in murine leukemia virus-VL30-derived vectors for gene transfer. J. Virol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tounekti, N., M. Mougel, C. Roy, R. Marquet, J.-L. Darlix, J. Paoletti, B. Ehresmann, and C. Ehresmann. 1992. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 223:205-220. [DOI] [PubMed] [Google Scholar]