Abstract
Hepatitis B virus (HBV) replication is inhibited in a noncytopathic manner by alpha/beta interferon (IFN-α/β) and IFN-γ. We demonstrate here that inhibitors of cellular proteasome activity can block this antiviral effect. These results suggest that a critical component of the IFN-induced antiviral response may be the proteasome-dependent degradation of viral or cellular proteins that are required for HBV replication.
Replication of hepatitis B virus (HBV) can be inhibited noncytopathically by various cytokines. The interferon-induced inhibition of HBV replication was first described in a 1.3-genome-length HBV transgenic mouse model (20). When injected with HBsAg-specific cytotoxic T lymphocytes, replication of HBV in the liver of the transgenic mice is strongly inhibited in a noncytopathic manner that is mediated primarily by gamma interferon (IFN-γ) and tumor necrosis factor alpha (19). The antiviral effect of the cytokines in this model occurs in two distinct phases. First, HBV DNA replicative intermediates disappear from the liver with no change in viral mRNA. Later, the level of HBV mRNA is downregulated by a posttranscriptional mechanism. Inhibition of HBV replication in the transgenic mice can also be induced by nonspecific stimuli that induce IFN-α/β [infection with lymphocytic choriomeningitis virus, murine cytomegalovirus, or adenovirus, injection with poly(I-C)] or IFN-γ (injection with interleukin 12 or α-galactosylceramide) (7, 8, 18, 27, 35). However, other than the observation that the antiviral effect of IFN-α/β is at the level of assembly or stability of pregenomic RNA containing HBV capsids (47), relatively little is known concerning the intracellular events that mediate this process.
The lack of understanding of the molecular mechanism of IFN-mediated inhibition of HBV replication is due in part to a lack of a suitable model system with which to study these events. To produce a model system in which the intracellular events that mediate the IFN-induced antiviral effect could be more easily studied, the 1.3-HBV transgenic mice were crossed with mice that express a constitutively active hepatocyte growth factor receptor (c-met) (1), and an immortalized hepatocyte cell line (HBV-Met) was established from the liver of a double-transgenic mouse (V. Pasquetto, S. F. Wieland, S. Uprichard, M. Tripodi and F. V. Chisari, unpublished data). These cells differentiate in culture when exposed to 2% dimethyl sulfoxide (DMSO), and upon differentiation express and replicate HBV from the integrated 1.3 transgene. Furthermore, HBV replication in the HBV-Met cell line is inhibited by either IFN-γ or IFN-β, similar to what is observed in vivo in the transgenic mice.
To better understand the cellular processes that mediate the antiviral effect, the changes in cellular gene expression that accompany the IFN-mediated inhibition of replication in the livers of the 1.3-HBV transgenic mice and in the HBV-Met hepatocyte cell line were examined by microarray analysis (S. F. Wieland et al., unpublished data). A number of genes were induced by both IFN-α/β and IFN-γ with kinetics that paralleled the antiviral effect. These included a variety of genes associated with the ubiquitin-proteasome pathway of protein degradation, such as the IFN-inducible proteasome catalytic subunits LMP2, LMP7 (5, 14, 28, 34, 40), and MECL-1 (17, 23, 38), the IFN-inducible proteasome activator PA28 (11, 33), and a ubiquitin-activating enzyme E1-like protein (UBE1L) (29, 30). The proteasome is a large multisubunit protease that degrades cytoplasmic and nuclear proteins that are targeted for degradation by polyubiquitination (46, 50). This process is important for the destruction of abnormal or damaged proteins, in the regulation of short-lived regulatory proteins, and in the generation of peptides for class I antigen presentation (10). Interestingly, the HBV X protein has been shown to bind to various proteasome subunits, and this interaction was demonstrated to be important for the transactivation activity of the X protein (2, 13, 24, 25, 45, 51).
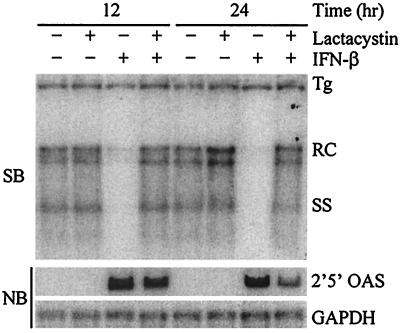
Because the expression of proteasome subunits was correlated with the IFN-induced inhibition of HBV replication, we hypothesized that the proteasome might play a role in mediating the antiviral effect. Therefore, we examined the requirement for proteasome activity in the IFN-induced antiviral effect in the HBV-Met cell line utilizing cell-permeable proteasome inhibitors (3, 31). Lactacystin is a Streptomyces metabolite that inhibits the proteasome irreversibly by covalent modification of the proteasome β-subunits (9, 12, 39). Lactacystin displays a high degree of specificity, since other than the proteasome, only cathepsin A activity is inhibited by lactacystin (41). HBV-Met cells (clone 1-1.4) were grown to confluence and cultured for 10 days in 2% DMSO to induce differentiation, HBV gene expression, and viral replication. The cells were then pretreated with 20 μM lactacystin (Calbiochem, San Diego, Calif.) for 1 h, after which 500 U of recombinant murine IFN-β/ml (22) (Provided by M. Moriyama, Toray Industries, Tokyo, Japan) was added to the cultures. The cells were then incubated for an additional 12 to 24 h, after which they were lysed with DNA lysis buffer (50 mM Tris-HCl [pH 8.0], 20 mM EDTA, 1% sodium dodecyl sulfate) or GTC solution (4.2 M guanidine isothiocyanate, 25 mM sodium citrate [pH 7.3], 0.5% Sarkosyl) for preparation of total DNA and RNA, respectively. Southern hybridization analysis was performed with a genome-length HBV probe on 20 μg of HindIII-digested DNA to determine the level of HBV DNA replicative intermediates. Northern blots were performed on 10 μg of total RNA for 2′5′-oligoadenylate synthetase (OAS) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts to serve as a control for IFN-mediated signaling and transcriptional induction. In the presence of lactacystin alone, viral replication was similar to that observed in control cultures, indicating that proteasome inhibition does not inhibit or enhance viral replication (Fig. 1). In the presence of IFN-β alone, viral replication was almost completely abolished by 12 h after addition of IFN-β (Fig. 1), even though expression of the HBV 3.5- and 2.1-kb mRNAs from the 1.3 transgene was unchanged compared to the control cultures (data not shown). However, if the cells were first treated with lactacystin, the antiviral effect of IFN-β was completely abolished. In both the presence and absence of lactacystin, expression of 2′5′-OAS was strongly induced by IFN-β. Thus, lactacystin blocked the IFN-β-induced antiviral effect even though IFN-β-mediated signaling and transcriptional induction remained functional.
FIG. 1.
Lactacystin blocks IFN-β-mediated inhibition of HBV replication. Differentiated HBV-Met (clone 1-1.4) cells were pretreated for 1 h with 20 μM lactacystin followed by addition of 500 U of IFN-β/ml. Cells were harvested at the indicated time points post-addition of IFN-β, and total DNA and RNA were prepared for Southern blot (SB) and Northern blot (NB) analysis of HBV DNA replicative intermediates and 2′5′-OAS/GAPDH expression. Tg, integrated transgene; RC and SS, relaxed-circle and single-stranded HBV DNA replicative forms.
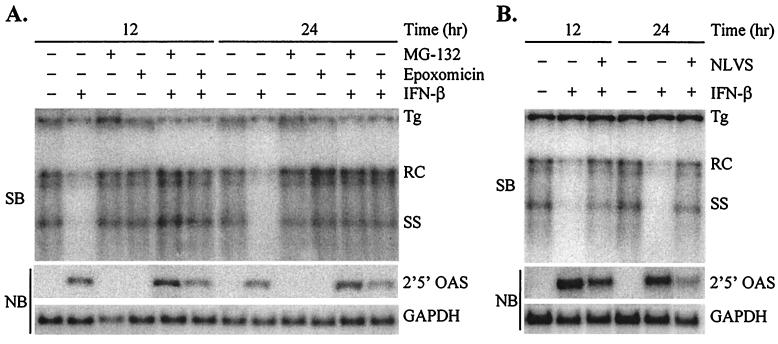
Although lactacystin is a well-characterized specific inhibitor of the proteasome, we wanted to test additional proteasome inhibitors to confirm that the effect on the IFN-β-induced antiviral effect was proteasome specific. MG-132 (carbobenzoxy-l-leucyl-l-leucyl-l-leucinal) is a tri-peptide aldehyde that functions as a reversible substrate-analog inhibitor of the proteasome (26, 32, 42, 43, 48, 49). MG-132 is less specific for the proteasome than lactacystin, as it can also inhibit a number of cellular cathepsins and calcium-dependent proteases. Epoxomicin is a natural product of Actinomycetes that, like lactacystin, inhibits the proteasome irreversibly by covalent modification (21, 36, 44). However, epoxomicin is more potent than lactacystin and has not been shown to inhibit any other cellular protease. Finally, NLVS (4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-Leu-vinylsulfone) is a derivative of a modified trileucine peptide aldehyde that inhibits the proteasome by irreversible covalent modification but may also inhibit cathepsin S activity (4). As was observed with lactacystin, MG-132 and epoxomicin did not affect HBV replication or IFN-β-mediated signal transduction but completely blocked the IFN-β-induced inhibition of HBV replication (Fig. 2A). Furthermore, NLVS also inhibited the IFN-β-mediated antiviral effect, similar to the other inhibitors, but with a moderate decrease in 2′5′-OAS induction (Fig. 2B). Therefore, four different proteasome inhibitors with various potencies, specificities, and biochemical mechanisms of inhibition all blocked the IFN-β-induced antiviral effect, suggesting that the effect of the inhibitors is indeed proteasome specific and that the antiviral effect of IFN-β is proteasome dependent.
FIG. 2.
Inhibition of antiviral effect by additional proteasome inhibitors. Differentiated HBV-Met cells were pretreated for 1 h with 50 μM MG-132 or 7.5 μM epoxomicin (A) or 50 μM NLVS (B) followed by addition of 500 U of IFN-β/ml. Cells were harvested at the indicated time points post-addition of IFN-β, and total DNA and RNA were prepared for Southern blot (SB) and Northern blot (NB) analysis of HBV DNA replicative intermediates and 2′5′-OAS/GAPDH expression. Tg, integrated transgene; RC and SS, relaxed-circle and single-stranded HBV DNA replicative forms.
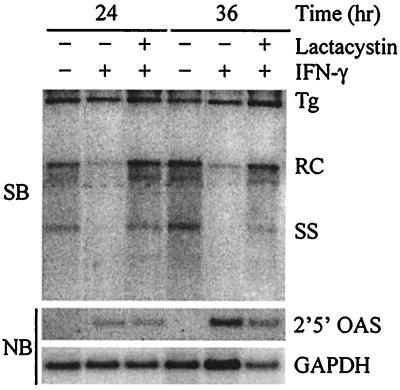
Since both IFN-α/β and IFN-γ inhibit HBV replication, we also wanted to determine if inhibiting the proteasome blocked the IFN-γ-mediated antiviral effect. As before, HBV-Met cells were incubated for 1 h with or without lactacystin, after which 1,000 U of recombinant murine IFN-γ (provided by S. Kramer, Genentech, South San Francisco, Calif.)/ml was added to the cells. In this experiment, cells were harvested at 24 and 36 h post-addition of IFN-γ since HBV is inhibited more slowly by IFN-γ in the HBV-Met cells than by IFN-β (data not shown). Like the IFN-β-mediated antiviral effect, the IFN-γ-induced inhibition of replication can also be blocked by inhibition of the proteasome (Fig. 3). Furthermore, 2′5′-OAS expression was also induced to similar levels by IFN-γ in the presence and absence of the inhibitor, indicating the IFN-γ-mediated signaling pathway remained functional. Thus, these results indicate that both the IFN-β- and IFN-γ-induced antiviral effects are proteasome dependent and may therefore occur through a common mechanism.
FIG. 3.
Lactacystin inhibits IFN-γ-induced antiviral effect. Differentiated HBV-Met cells were pretreated for 1 h with 20 μM lactacystin followed by addition of 1,000 U of IFN-γ/ml. Cells were harvested at the indicated time points post-addition of IFN-γ, and total DNA and RNA were prepared for Southern blot (SB) and Northern blot (NB) analysis of HBV DNA replicative intermediates and 2′5′-OAS/GAPDH expression. Tg, integrated transgene; RC and SS, relaxed-circle and single-stranded HBV DNA replicative forms.
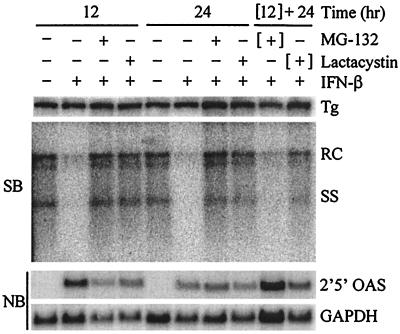
To provide further evidence that the inhibition of the antiviral effect is specific to an effect on the proteasome and not a general toxic effect due to the inhibitors, we determined whether the inhibition of the antiviral effect is reversible upon removal of MG-132 (a reversible inhibitor) and lactacystin (an irreversible inhibitor). Again, HBV-Met cells were pretreated with the inhibitors for 1 h followed by addition of 500 U of IFN-β/ml, and cells were harvested at 12 and 24 h for Southern blot analysis. In addition, two cultures were incubated for 12 h with MG-132 plus IFN-β or lactacystin plus IFN-β, after which the medium was removed and the cells were washed and further incubated in medium containing only 500 U of IFN-β/ml for an additional 24 h. As shown in Fig. 4, the inhibition of the antiviral effect by MG-132 is reversible upon removal of the inhibitor, while the lactacystin-mediated effect is not. Thus, the reversibility of the process is consistent with the known biochemical mechanisms of the two proteasome inhibitors, providing further evidence that their ability to block the antiviral effect of IFN-β is proteasome specific.
FIG. 4.
Reversible and irreversible inhibition of antiviral effect. Differentiated HBV-Met cells were pretreated for 1 h with 20 μM lactacystin or 50 μM MG-132 followed by addition of 500 U of IFN-β/ml. In the “[12] + 24”-h lanes, the cells were cultured with IFN-β and the proteasome inhibitor for 12 h, followed by a 24-h incubation in the presence of IFN-β alone. Cells were harvested at the indicated time points post-addition of IFN-β, and total DNA and RNA were prepared for Southern blot (SB) and Northern blot (NB) analysis of HBV DNA replicative intermediates and 2′5′-OAS/GAPDH expression. Tg, integrated transgene; RC and SS, relaxed-circle and single-stranded HBV DNA replicative forms.
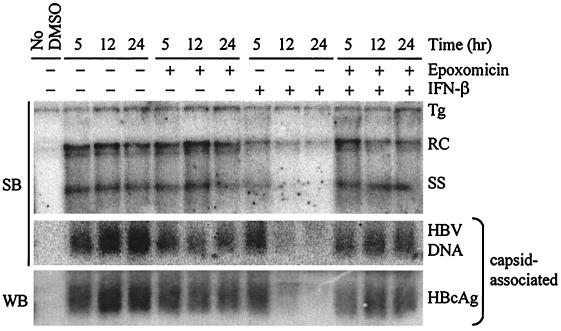
The inhibition of HBV replication by IFN-α/β is associated in vivo with a decline in cytoplasmic pregenomic RNA- and DNA-containing capsids (47). Therefore, we wanted to determine if proteasome inhibition would block the IFN-induced disappearance of capsid particles from the hepatocyte cytoplasm. Cytoplasmic protein extracts (cells lysed in 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1% NP-40) and total DNA were prepared from HBV-Met cells treated with epoxomicin (1 h of pretreatment) and/or IFN-β at 5, 12, and 24 h post-IFN-β addition. Southern blot analysis of the total DNA confirmed that IFN-β triggered the disappearance of DNA replicative intermediates and that this effect was blocked by epoxomicin (Fig. 5). The cytoplasmic protein extracts (100 μg) were electrophoresed at 50 V for 4 h on 0.6% agarose gels to resolve HBV capsid particles, which were transferred to a nitrocellulose membrane for immunoblot analysis with an anti-HBV core polyclonal antibody (Dako, Carpinteria, Calif.) or a nylon membrane for hybridization with an HBV probe to examine encapsidated HBV DNA. As shown in Fig. 5, IFN-β treatment results in a decrease in cytoplasmic HBV encapsidated DNA and capsid particle-associated core protein that temporally correlates with the decrease in DNA replicative intermediates. Furthermore, the disappearance of capsid particles that is induced by IFN-β is blocked by epoxomicin, consistent with what is observed with the replicative intermediates. This result is consistent with the interpretation that the effect of the proteasome inhibitors in blocking the antiviral effect is at the level of assembly or stability of cytoplasmic pregenomic RNA- or DNA-containing capsids, which is where the antiviral effect is known to occur in vivo.
FIG. 5.
Proteasome inhibition blocks IFN-β-induced disappearance of HBV capsids. Differentiated HBV-Met cells were pretreated for 1 h with 5 μM epoxomicin followed by addition of 500 U of IFN-β/ml and were harvested at the indicated time points post-addition of IFN-β for DNA and cytoplasmic protein preparations. The amount of DNA replicative intermediates was determined by Southern blot (SB) analysis, while the levels of viral capsids and encapsidated HBV DNA were assessed by agarose gel electrophoresis followed by Southern blot for HBV DNA or Western blot (WB) with an anti-HBV core polyclonal antibody. The “No DMSO” lane corresponds to HBV-Met cells that were cultured in the absence of 2% DMSO and therefore do not contain HBV DNA or HBcAg. Tg, integrated transgene; RC and SS, relaxed-circle and single-stranded HBV DNA replicative forms; HBcAg, HBV core antigen.
Relatively little is known about the cellular factors that are necessary for the IFN-induced inhibition of HBV replication. The results presented here indicate that proteasome activity is required for this antiviral effect. Furthermore, the fact that the IFN-β- and IFN-γ-mediated antiviral effects are both blocked by proteasome inhibition suggests that the inhibition of HBV replication induced by these two cytokines is mediated through similar if not identical mechanisms. By degrading viral proteins into peptides for class I antigen presentation, the proteasome is clearly an important, although indirect, component of the host immune response against most viruses. Our results indicate that the proteasome may also have a direct antiviral activity against HBV that is induced by both IFN-β and IFN-γ. This observation, therefore, may represent a novel mechanism by which IFN-induced proteins inhibit viral replication. Furthermore, the fact that the HBV X protein has been demonstrated to inhibit proteasome activity raises the possibility that one function of X may be to modulate this effect (24).
There are a number of possible events that could explain the proteasome dependence of the IFN-mediated inhibition of HBV replication. First, an IFN-induced increase in ubiquitination of proteins required for viral particle assembly or maturation could lead to their degradation, thus limiting viral replication. Second, it is possible that by stimulating the expression of the IFN-inducible proteasome activators and catalytic subunits, IFN enhances the rate of degradation of proteins necessary for replication. Third, there could be IFN-induced factors that target proteins required for replication to the proteasome for degradation in a ubiquitin-independent manner. In all of these cases, the critical proteins that are targeted for degradation by the proteasome could be either viral or cellular proteins. In addition, the effect may occur by inhibiting the assembly of nascent capsids or by the destabilization and degradation of capsids that have already formed.
It is also possible that one or more of the IFN-induced proteins with known antiviral activity (double-stranded-RNA-dependent protein kinase, RNase L, and Mx) (15, 16) mediates the inhibition of HBV replication in a proteasome-dependent manner. In addition, we cannot formally rule out the possibility that IFN induces a proteasome-sensitive, Jak/STAT-independent signal transduction cascade that mediates the antiviral effect. Finally, proteasome inhibition may have a number of other effects on the cell (such as depletion of free ubiquitin and the induction of a cellular stress response) that could possibly influence the IFN-induced antiviral effect (6, 37). Irrespective of which mechanism ultimately proves to be correct, determining the role of the proteasome in the IFN-mediated inhibition of HBV replication will lead to a better understanding of the factors that contribute to HBV persistence and pathogenesis.
Acknowledgments
This work was supported by grant CA40489 from the National Institutes of Health (F.V.C.). In addition, M.D.R. was supported by NIH fellowship AI49665 and by a fellowship from the Skaggs Institute.
We thank Toray Industries for providing the recombinant murine IFN-β and Genentech for providing the recombinant murine IFN-γ. We also thank Susan Uprichard and Amy Brideau-Andersen for helpful discussions and Bryan Boyd for technical assistance.
Footnotes
Manuscript no. 14416-MEM from the Scripps Research Institute.
REFERENCES
- 1.Amicone, L., F. M. Spagnoli, G. Spath, S. Giordano, C. Tommasini, S. Bernardini, V. De Luca, C. Della Rocca, M. C. Weiss, P. M. Comoglio, and M. Tripodi. 1997. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J. 16:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, O., A. Aronheim, and Y. Shaul. 2001. HBV X protein targets HIV Tat-binding protein 1. Virology 283:110-120. [DOI] [PubMed] [Google Scholar]
- 3.Bogyo, M., M. Gaczynska, and H. L. Ploegh. 1997. Proteasome inhibitors and antigen presentation. Biopolymers 43:269-280. [DOI] [PubMed] [Google Scholar]
- 4.Bogyo, M., J. S. McMaster, M. Gaczynska, D. Tortorella, A. L. Goldberg, and H. Ploegh. 1997. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA 94:6629-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, M. G., J. Driscoll, and J. J. Monaco. 1991. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature 353:355-357. [DOI] [PubMed] [Google Scholar]
- 6.Bush, K. T., A. L. Goldberg, and S. K. Nigam. 1997. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272:9086-9092. [DOI] [PubMed] [Google Scholar]
- 7.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1998. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J. Virol. 72:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J. Virology 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craiu, A., M. Gaczynska, T. Akopian, C. F. Gramm, G. Fenteany, A. L. Goldberg, and K. L. Rock. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 272:13437-13445. [DOI] [PubMed] [Google Scholar]
- 10.DeMartino, G. N., and C. A. Slaughter. 1999. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 274:22123-22126. [DOI] [PubMed] [Google Scholar]
- 11.Dubiel, W., G. Pratt, K. Ferrell, and M. Rechsteiner. 1992. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 267:22369-22377. [PubMed] [Google Scholar]
- 12.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, M., L. Runkel, and H. Schaller. 1995. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes 10:99-102. [DOI] [PubMed] [Google Scholar]
- 14.Glynne, R., S. H. Powis, S. Beck, A. Kelly, L. A. Kerr, and J. Trowsdale. 1991. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature 353:357-360. [DOI] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.Gordien, E., O. Rosmorduc, C. Peltekian, F. Garreau, C. Brechot, and D. Kremsdorf. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groettrup, M., R. Kraft, S. Kostka, S. Standera, R. Stohwasser, and P. M. Kloetzel. 1996. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur. J. Immunol. 26:863-869. [DOI] [PubMed] [Google Scholar]
- 18.Guidotti, L. G., P. Borrow, M. V. Hobbs, B. Matzke, I. Gresser, M. B. Oldstone, and F. V. Chisari. 1996. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc. Natl. Acad. Sci. USA 93:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virology 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada, M., K. Sugawara, K. Kaneta, S. Toda, Y. Nishiyama, K. Tomita, H. Yamamoto, M. Konishi, and T. Oki. 1992. Epoxomicin, a new antitumor agent of microbial origin. J. Antibiot. (Tokyo) 45:1746-1752. [DOI] [PubMed] [Google Scholar]
- 22.Higashi, Y., Y. Sokawa, Y. Watanabe, Y. Kawade, S. Ohno, C. Takaoka, and T. Taniguchi. 1983. Structure and expression of a cloned cDNA for mouse interferon-beta. J. Biol. Chem. 258:9522-9529. [PubMed] [Google Scholar]
- 23.Hisamatsu, H., N. Shimbara, Y. Saito, P. Kristensen, K. B. Hendil, T. Fujiwara, E. Takahashi, N. Tanahashi, T. Tamura, A. Ichihara, and K. Tanaka. 1996. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J. Exp. Med. 183:1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, J., J. Kwong, E. C. Sun, and T. J. Liang. 1996. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, T. J., M. A. Loo, S. Pind, D. B. Williams, A. L. Goldberg, and J. R. Riordan. 1995. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83:129-135. [DOI] [PubMed] [Google Scholar]
- 27.Kakimi, K., L. G. Guidotti, Y. Koezuka, and F. V. Chisari. 2000. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 192:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly, A., S. H. Powis, R. Glynne, E. Radley, S. Beck, and J. Trowsdale. 1991. Second proteasome-related gene in the human MHC class II region. Nature 353:667-668. [DOI] [PubMed] [Google Scholar]
- 29.Kok, K., R. Hofstra, A. Pilz, A. van den Berg, P. Terpstra, C. H. Buys, and B. Carritt. 1993. A gene in the chromosomal region 3p21 with greatly reduced expression in lung cancer is similar to the gene for ubiquitin-activating enzyme. Proc. Natl. Acad. Sci. USA 90:6071-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kok, K., A. Van den Berg, P. M. Veldhuis, M. Franke, P. Terpstra, and C. H. Buys. 1995. The genomic structure of the human UBE1L gene. Gene Expr. 4:163-175. [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 32.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280-27284. [DOI] [PubMed] [Google Scholar]
- 33.Ma, C. P., C. A. Slaughter, and G. N. DeMartino. 1992. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 267:10515-10523. [PubMed] [Google Scholar]
- 34.Martinez, C. K., and J. J. Monaco. 1991. Homology of proteasome subunits to a major histocompatibility complex-linked LMP gene. Nature 353:664-667. [DOI] [PubMed] [Google Scholar]
- 35.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 38.Nandi, D., H. Jiang, and J. J. Monaco. 1996. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J. Immunol. 156:2361-2364. [PubMed] [Google Scholar]
- 39.Omura, S., T. Fujimoto, K. Otoguro, K. Matsuzaki, R. Moriguchi, H. Tanaka, and Y. Sasaki. 1991. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J. Antibiot. (Tokyo) 44:113-116. [DOI] [PubMed] [Google Scholar]
- 40.Ortiz-Navarrete, V., A. Seelig, M. Gernold, S. Frentzel, P. M. Kloetzel, and G. J. Hammerling. 1991. Subunit of the ‘20S' proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature 353:662-664. [DOI] [PubMed] [Google Scholar]
- 41.Ostrowska, H., C. Wojcik, S. Omura, and K. Worowski. 1997. Lactacystin, a specific inhibitor of the proteasome, inhibits human platelet lysosomal cathepsin A-like enzyme. Biochem. Biophys. Res. Commun. 234:729-732. [DOI] [PubMed] [Google Scholar]
- 42.Read, M. A., A. S. Neish, F. W. Luscinskas, V. J. Palombella, T. Maniatis, and T. Collins. 1995. The proteasome pathway is required for cytokine-induced endothelial-leukocyte adhesion molecule expression. Immunity 2:493-506. [DOI] [PubMed] [Google Scholar]
- 43.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761-771. [DOI] [PubMed] [Google Scholar]
- 44.Sin, N., K. B. Kim, M. Elofsson, L. Meng, H. Auth, B. H. Kwok, and C. M. Crews. 1999. Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg. Med. Chem. Lett. 9:2283-2288. [DOI] [PubMed] [Google Scholar]
- 45.Sirma, H., R. Weil, O. Rosmorduc, S. Urban, A. Israel, D. Kremsdorf, and C. Brechot. 1998. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene 16:2051-2063. [DOI] [PubMed] [Google Scholar]
- 46.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 47.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 49.Wilk, S., and M. E. Figueiredo-Pereira. 1993. Synthetic inhibitors of the multicatalytic proteinase complex (proteasome). Enzyme Protein 47:306-313. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]