Abstract
Papillomaviruses normally replicate in stratified squamous epithelial tissues of their mammalian hosts, in which the viral genome is found as a nuclear plasmid. Two viral proteins, E1, a helicase, and E2, a transcriptional activator and plasmid maintenance factor, are known to contribute to the episomal replication of the viral genome. Recently, our laboratory discovered that papillomaviruses can also replicate in an E1-independent manner in mammalian cells (K. Kim and P. F. Lambert, Virology, in press; K. Kim and P. F. Lambert, submitted for publication). In this study, we describe experiments investigating the capacity of the human papillomavirus type 16 (HPV16) genome to replicate in yeast (Saccharomyces cerevisiae). The full-length HPV16 genome, when linked in cis to a selectable yeast marker gene, either TRP1 or URA3, could replicate stably as an episome in yeast. The replication of papillomavirus genomes in yeast is not limited to HPV16. Bovine papillomavirus type 1 and HPV6b, -11, -16, -18, and -31 were all capable of replicating in short-term assays over a period of 20 cell doublings. The long-term persistence of viral episomes did not require any one viral gene, as mutant genomes defective in single genes also replicated episomally. These results indicate that the viral episome can replicate in the absence of the E1 DNA helicase. Similarly, E2 was also not required for replication in yeast, and E2 mutant viral genomes were stably maintained in the absence of selection, indicating the existence of an E2-independent mechanism for plasmid maintenance. The episomal replication of papillomavirus genomes in yeast provides a genetically manipulatable system in which to investigate cellular factors required for episomal replication and may provide a novel means for generating infectious papillomavirus.
Human papillomaviruses (HPVs) infect and persist in differentiating epithelial cells present in the cutaneous, mucosal, and genital tissues. Papillomaviruses replicate as low-copy-number nuclear plasmids. The life cycles of these viruses are thought to occur in three distinct phases: establishment, in which there is an early amplificational event; maintenance, in which viral genomes are maintained extrachromosomally at a constant copy number; and an amplification stage, in which the viral copy number increases prior to encapsidation (17).
The replication of papillomaviruses involves two virally encoded proteins, E1, DNA helicase, and E2, a transcription and maintenance factor. It has generally been thought that replication of papillomaviruses is dependent upon the presence of E1 and E2 (11, 36, 46). However, our laboratory has recently observed that bovine papillomavirus type 1 (BPV-1) and HPV types 16, 18, and 31 (HPV16, -18, and -31) are capable of replicating extrachromosomally in human cells in the absence of functional E1 protein (21a; K. Kim and P. F. Lambert, submitted for publication). We have also found that in BPV-1, temperature-sensitive mutants of E1 are capable of stable replication at the nonpermissive temperature, further supporting the existence of an E1-independent mode of replication (21a). The contributions of E2 to stable replication of HPVs are not completely understood. Recent work has implicated E2 in the segregation of newly synthesized genomes, and this is thought to occur by E2 tethering viral genomes to host chromosomes during mitosis (2, 18, 26, 34, 42, 48).
Unlike more complex viruses, which carry their own polymerase genes, papillomaviruses rely largely on cellular replicative machinery, including polymerase α (Polα) (32). The E1 protein, a DNA helicase, binds to the p70 and p180 subunits of Polα (8). Thus, initiation at the E1-dependent origin in the long control region (LCR) is mediated by E1 recruitment of Polα (9, 32). This observation is supported by two-dimensional gel analysis (13). However, our recent studies have presented the possibility that an alternative mode of genome replication occurs which does not involve E1. This alternative mode is likely to require that cellular proteins substitute for E1 helicase activity. This led us to hypothesize that the cellular origin recognition complex (ORC) and replicative machinery mediate E1-independent replication, i.e., that papillomaviruses possess autonomously replicating sequence (ARS)-like elements.
Our prior investigation of the requirements for the viral trans-acting factor, E1, in stable replication led us to develop a novel replication assay system based on Saccharomyces cerevisiae. Our rationale for developing a yeast-based system was fourfold. (i) Cellular machinery likely to be required for papillomavirus replication in human cells is highly conserved in yeast. Although the specificities of human and yeast Polα enzymes differ (15), multiple components, such as proliferating cell nuclear antigen, minichromosome maintenance (MCM) proteins (24, 25), replication factor C (28), and replication protein A have functionally equivalent counterparts in yeast and mammalian cells (10, 21). (ii) ARSs in yeast share sequence features, such as A/T-rich clusters, with mammalian origins (7, 31). (iii) Despite greater complexity in mammalian cells, the ORC is functionally conserved from yeast to mammalian cells (7, 47). (iv) A yeast-based system would be relatively easy to manipulate while providing excellent genetic tools with which to analyze replication of the viral genome. Furthermore, another laboratory has successfully recreated complex replication programs of higher eukaryotic viruses, such as brome mosaic virus and Flockhouse virus, in S. cerivisiae (19, 35). Recent studies of EBNA1-dependent plasmid maintenance demonstrate that this activity can also be modeled in yeast (20). Yeast has also proven its utility in the study of papillomavirus E2-dependent transcriptional activation function (23, 30, 43). Thus, we tested whether HPV16 could replicate in yeast.
In this study, full-length HPV16 was linked in cis to selectable markers, and the recombinant plasmids were introduced into yeast. Based on these studies, we discovered that HPV16 contains intrinsic ARS- and centromere (CEN)-like activities. Furthermore, HPV16 was found to be capable of replicating in yeast in the absence of E1, suggesting that the mode of replication functional in yeast is analogous to the E1-independent mode of replication we recently discovered in mammalian cells (Kim and Lambert, submitted). Surprisingly, we also found that no individual gene, including the E2 gene, is required for stable replication in yeast, suggesting that a CEN-like element(s) in HPV16 DNA provides an E2-independent mechanism for the stable inheritance of episomes in daughter cells. Lastly, the capacity of papillomaviruses to replicate in yeast is a general phenomenon, as multiple papillomavirus genotypes, including BPV-1 and various anogenital papillomaviruses, were all capable of replicating in S. cerevisiae. Stable replication of HPVs in yeast provides obvious utility as a genetic tool in understanding the roles of cellular genes in these functions. In addition, the potential to model specific viral functions, such as genome amplification or encapsidation, in yeast makes this system potentially powerful.
MATERIALS AND METHODS
Yeast strains and transformation methods.
The yeast strain YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) used in the described experiments was a gift from Paul Ahlquist (University of Wisconsin--Madison [39]). In preliminary experiments, the HF7c yeast strain (MATaura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL-HIS3 URA3::[GAL4 17-mers]3 -CYC1-lacZ) was used (Clontech, Palo Alto, Calif.). Yeasts were grown on minimal media omitting tryptophan (Trp) or uracil (Ura), as needed, to select for plasmids. Plasmid DNAs were transformed into yeast using the standard lithium chloride method (38) or the EZ Yeast Transformation kit (Zymo Research, Orange, Calif.).
DNA constructions.
The pΔYac plasmid (23), which contains the yeast TRP1 gene, an ARS, and a CEN element, is a modification of pYac2 (6). pΔYac was modified to generate derivatives with either the ARS or the CEN element deleted. To create a CEN− derivative (pPA94), pΔYac was digested with SpeI and DraIII enzymes to remove a 978-bp fragment containing the CEN element. A linker (top, 5′-GTGCAGCTCGAGTCAA-3′; bottom, CTAGTTGACTCGAGCTGCACACA) with SpeI and DraIII ends was directly ligated into the gap. To create an ARS+ CEN− HPV16 construct, the BglII site adjacent to the ARS element of pPA94 was digested, and the entire HPV16 genome was excised from pEF399 with a BamHI digest. The BamHI-ended genome was ligated directly into the BglII site, resulting in an ARS+ CEN− vector containing HPV16, which was named pPA95. An ARS− CEN+ derivative of pΔYac (pKT273) was created by deleting 1,664 bp between the AatII and HpaI sites, which contains the ARS element. A BamHI fragment containing the entire HPV16 genome (E1TTL2 [Kim and Lambert, submitted]) was ligated into the BglII site of pKT273, resulting in the ARS− CEN+ HPV16 (E1TTL2) plasmid referred to as pKT274. Next, the TRP1 gene was excised from a cloning vector (pKT268) with SpeI and AatII digests and was cloned directly into the AatII and XbaI sites of pUC18. The resultant vector was ARS− CEN− and was named pKT270. By an identical strategy, the TRP1 gene was introduced into the pUC18 backbone of plasmids containing wild-type (wt) (pPA100) or mutant (E1TTL2 [pKT269], E2TTL [pPA101], E5fs [pKT304], E6TTL [pKT307], E7TTL [pKT303], L1TTL [pKT305], or L2TTL [pKT308]) HPV16 genome.
Two HPV16-containing constructs were created from which pUC18 could be deleted prior to the transformation of yeast. These vectors contain either TRP1 (pKT309) or URA3 (pPA103) biosynthetic markers. The pKT309 plasmid was created by digesting pEF399 with StuI and XbaI, which removed nucleotides 4496 to 6150 from the L2 and L1 open reading frames (ORFs). The 1.7-kb fragment was replaced with a StuI/SpeI fragment from pKT268 containing the TRP1 gene. To create pPA103, the pEF399 clone was first digested with PmlI, which cleaves just downstream of the L1 ORF (nucleotide 7266 of the genome). A blunt-ended linker containing an XhoI site (top, 5′-CGACTCGAGTGC-3′; bottom, 5′-GCACTCGAGTCG-3′) was ligated directly into the PmlI site, creating the construct referred to as pPA99. The URA3 gene was then amplified from pRS316 using 5′ (5′-GATCCTCGAGGCAGATTGTACTGAGAGTG-3′) and 3′ (5′-ACTGCTCGAGTAGTATACATGCATTTAC-3′) primers containing XhoI sites (underlined). The amplified product was digested with XhoI and ligated into the XhoI site of pPA99, creating pPA103. A BamHI digest of either pPA103 or pKT309 allows the 2.7-kb pUC18 backbone to be removed and the HPV16 DNA to be religated, still retaining the selectable marker. As a negative control for pPA103, the XhoI-digested PCR product containing the URA3 gene was cloned into the pUC18 SalI site, creating pPA104.
Colony formation assay.
Two-hundred nanograms of each of the described plasmids was transformed into YPH500 yeast. The transformants were plated on selective media and scored for the ability to form colonies after 3 days.
DNA isolation.
Yeast cells harboring HPV16 plasmids were grown in 25-ml cultures in selective media overnight to yield an optical density at 600 nm (OD600) over 1.0. Typically, 5 × 108 cells were harvested per 25-ml culture. The cells were pelleted and resuspended in 600 to 800 μl of lysis buffer (10 mM Tris [pH 8.0], 100 mM NaCl, 2% Triton X-100, and 1% sodium dodecyl sulfate). Acid-washed glass beads (0.5-mm diameter; Sigma, St. Louis, Mo.) were added to a volume of 300 μl. Six-hundred microliters of phenol-chloroform was added, and the mixture was vortexed for 1 to 2 min. The supernatants were recovered by centrifugation and transferred to a new tube. DNA was ethanol precipitated by the addition of 2.5 volumes of 100% ethanol. DNA samples were resuspended in double-distilled H2O at a concentration of 1 × 107 to 5 × 107 cell equivalents per μl.
DNA replication assays.
Plasmid-containing colonies were grown in continuous culture on solid media for 3 weeks prior to analysis. DNA was isolated from liquid cultures as described above. For the purpose of demonstrating that the plasmids had replicated in yeast, the DpnI resistance assay was performed. In this assay, bacterially methylated DNA is digested by DpnI and thus is distinguishable from DNA synthesized in eukaryotic cells, which is unmethylated at DpnI sites and therefore resistant to digestion (22). The DNA samples were digested with DpnI for 24 h. Approximately 108 cell equivalents of DNA was loaded on to a 1% agarose gel. The DNA was electrophoresed, transferred to nitrocellulose, and probed with radiolabeled HPV16 DNA (pEF399). Radiolabeling was achieved by use of the Rediprime kit (Amersham, Piscataway, N.J.) in accordance with the manufacturer's instructions. The replicated DNAs were visualized and quantified by use of a PhosphorImager (Molecular Dynamics, San Jose, Calif.).
Short-term DNA replication assays.
YPH500 yeast was transformed with 250 ng of plasmids containing the following full-length viral genomes: BPV-1 and HPV6b, -11, -16, -18, and -31. The transformants were pelleted, inoculated into 25 ml of yeast-peptone-dextrose medium, and cultured for 48 h (20 cell doublings). The approximate number of cells at the time of harvest was determined by spectrophotometer readings at OD600. Cells were collected by centrifugation, and the cell pellet was washed with phosphate-buffered saline. The cells were then lysed, and DNA was recovered as described previously. The samples were spiked with 5 ng of pUC18 DNA. The DNA was resuspended such that the final concentration was 1 × 107 to 5 × 107 cell equivalents per μl. Approximately 108 cell equivalents was digested with DpnI for 24 h. The samples were loaded onto a 1% agarose gel and electrophoresed. Southern blot analysis was performed using pUC18 as a probe so that all genomes would be equally detected.
Plasmid stability assay.
The plasmid stability experiments were performed as described by Kapoor et al. (20). Yeast colonies harboring episomal copies of pΔYac, pPA100 (wt HPV16), or pPA101 (E2TTL) were analyzed for plasmid stability in the absence of nutritional selection. Cultures were first grown in selective media to mid-log phase and diluted to an OD600 of 0.1 into new cultures containing nonselective media (with Trp). The cultures were grown for 0 to 17 cell generations. The cultures at either 0 or 17 generations were diluted to an OD600 of 0.1 and further diluted by 10-fold serial dilutions. Five-microliter aliquots of each dilution were spotted to selective (without Trp) and nonselective (with Trp) media. In order to determine the percentage of cells retaining plasmid, equal amounts of an appropriate dilution of the cultures were plated on selective and nonselective media and counted after 3 days of growth. The percentage of cells retaining plasmid was given by the ratio of the number of colonies present on selective medium versus the number on nonselective medium. The percent plasmid loss per cell generation was calculated by subtracting the percent plasmid retained after 17 generations from the percent plasmid retained at 0 generations and dividing by the total number of generations.
RESULTS
The HPV16 genome contains ARS- and CEN-like activities.
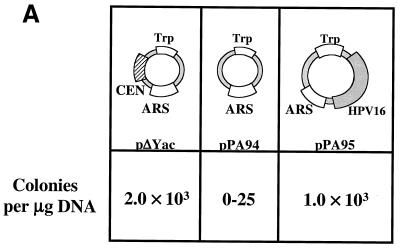
In order to analyze the cis-acting functions mediated by HPV16 genomes in yeast, a series of plasmids were generated which were either ARS+ CEN−, ARS− CEN+, or ARS− CEN− and which did or did not contain the full-length HPV16 genome. A Trp1 marker was present in the vector sequence of each plasmid in order to allow selection for yeast which could stably replicate a given plasmid. These plasmids were transformed into YPH500 yeast and allowed to grow for 3 days. Whereas a plasmid which was ARS+ CEN− (pPA94) formed colonies extremely poorly, an ARS+ CEN− plasmid containing the HPV16 genome (pPA95) or an ARS+ CEN+ control plasmid (pΔYac) efficiently formed colonies (Fig. 1A). These results support the concept that the HPV16 genome can restore maintenance function to an ARS+ CEN− plasmid.
FIG. 1.
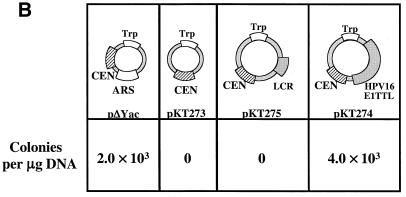
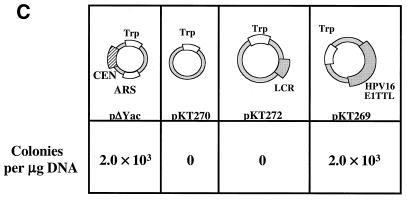
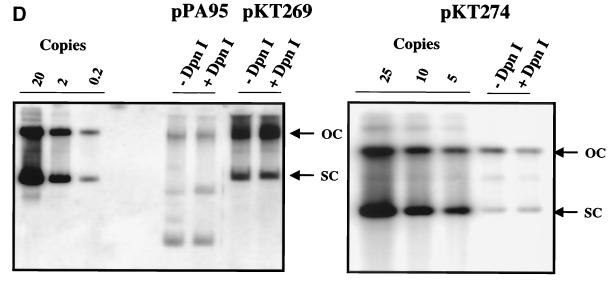
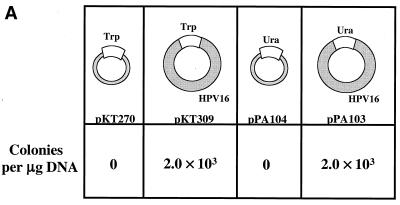
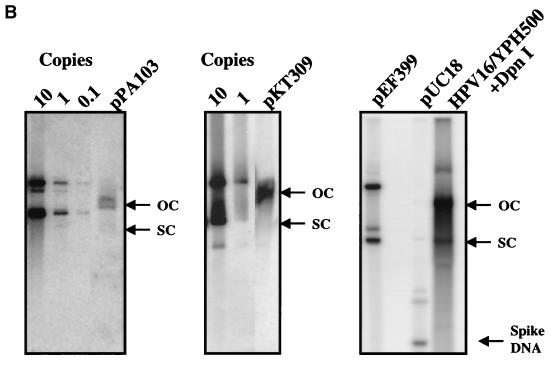
The HPV16 genome can substitute for both ARS and CEN activities of the pΔYac plasmid in yeast. (A) To test for maintenance function, plasmid derivatives of pΔYac were created which were ARS+ CEN− (pPA94) or ARS+ CEN− with the full HPV16 genome inserted (pPA95). (B) To test for ARS activity, plasmid derivatives of pΔYac were created which were ARS− CEN+ (pKT273) or were ARS− CEN+ but contained either the LCR (pKT275) or the full-length viral genome (pKT274 [E1TTL2]). (C) To test the ability of HPV16 sequences to provide both replication and maintenance functions, a puc18 plasmid was created containing only the Trp1 marker (pKT270). This vector was modified by the addition of either the LCR (pKT272) or the full-length genome (pKT269 [E1TTL2]). The plasmid constructs were transformed into YPH500 yeast and scored for the ability to form colonies on selective medium. The pΔYac plasmid was included as a positive control. The number of colonies obtained per microgram of plasmid DNA is indicated below each construct. (D) Episomal replication of HPV16-containing plasmids (pPA95, pKT269 [E1TTL2], and pKT274 [E1TTL2]). DNA isolated from yeast was electrophoresed on a 1% agarose gel and subjected to Southern blot analysis. The blot was probed with full-length HPV16. To the left of each blot are controls indicating the copy number per cell. Each DNA sample was subjected to DpnI digestion as indicated (+DpnI). The arrows to the right of each blot show the positions of open-circle (OC) and supercoiled (SC) plasmid forms.
Whereas an ARS+ CEN+ plasmid (pΔYac) efficiently formed colonies, an ARS− CEN+ plasmid (pKT273) was unable to form colonies (Fig. 1B). However, the addition of full-length HPV16 to an ARS− CEN+ plasmid (pKT274 [E1TTL2]) restored efficient colony formation. Interestingly, the LCR was not sufficient to rescue ARS function in the ARS− CEN+ plasmid (pKT275), indicating that the ARS-like function resides outside of the LCR and E1-dependent origin of replication in the HPV16 genome.
Not surprisingly, an ARS− CEN− plasmid (pKT270) lacked the ability to form colonies, in contrast to the control ARS+ CEN+ plasmid (pΔYac), while an ARS− CEN− plasmid containing the HPV16 genome (pKT269 [E1TTL2]) was capable of efficient colony formation (Fig. 1C). However, an ARS− CEN− plasmid containing the LCR (pKT272) was not sufficient to restore functions required for colony formation. These results suggest that both replication and maintenance functions can be provided by the HPV16 genome. Colonies derived from these experiments were further analyzed for evidence of episomal replication of the HPV16-containing plasmids.
Episomally replicating HPV16 plasmids were detected regardless of whether the backbone contained complementing yeast replication or maintenance elements (Fig. 1D). Therefore, HPV16 DNA is able to efficiently substitute for both ARS and CEN functions. The HPV16 plasmids were present in low copy numbers, approximately one to five copies per cell, depending on the construct. In these experiments, there was no evidence of integration of HPV16 DNA into the yeast genome, as indicated by the resolution of a single band upon digestion with NcoI (data not shown). Identical results were observed in an alternative yeast strain, HF7c (data not shown).
To confirm that replication and maintenance activities were dependent on HPV16 DNA and not on pUC18 or the marker DNA, HPV16 plasmids, containing either a TRP1 (pKT309) or a URA3 (pPA103) marker, were generated from which the pUC18 sequence could be excised prior to transfection into yeast. Both of these constructs displayed efficient colony formation (Fig. 2A), whereas negative control plasmids containing pUC18 and TRP1 (pKT270) or URA3 (pPA104) failed to form colonies. Both pPA103 and pKT309 replicated episomally in yeast, as detected by Southern blotting (Fig. 2B). Both of these plasmids have lower molecular weights than their parental plasmids (shown as copy controls), since the 2.7-kb pUC18 vector had been removed from each prior to the transformation of yeast. To assess further the role, if any, of nonviral sequences in supporting episomal replication, we transfected the full-length religated HPV16 genomes into yeast in short-term assays. Transformants were cultured for approximately 15 cell doublings and analyzed by Southern blotting. In these experiments, we discovered that HPV16, released from any vector or foreign sequence, replicated stably in yeast (Fig. 2B). In total, these results demonstrate that the HPV16 genome contains all the genetic elements necessary and sufficient for replication and maintenance in yeast.
FIG. 2.
HPV16 sequence is sufficient to allow stable episomal replication of plasmids in yeast. (A) Control plasmids with a pUC18 backbone were constructed with either a TRP1 (pKT270) or a URA3 (pPA104) marker. Corresponding HPV16-containing plasmids had either a TRP1 (pKT309) or a URA3 (pPA103) marker and were created such that the entire pUC18 sequence could be excised prior to transformation into yeast. Transformed yeasts were scored for their ability to form colonies on selective medium. The number of colonies obtained per microgram of plasmid DNA is indicated below each construct. (B) Episomal replication of pPA103 and pKT309 is indicated in the first two gels from the left. In each lane, 108 cell equivalents of DNA was loaded. The lanes on the left of each gel are controls for the copy number per cell. In the gel on the far right, YPH500 yeast was transfected with full-length HPV16 from which the pUC18 vector backbone had been excised. The yeast was grown for 36 h (approximately 15 cell doublings), and then cultures were harvested and DNA was isolated. As a control for the completion of DpnI digestion, 2 ng of pUC18 was added to each sample. DpnI digestion was carried out for 24 h, and samples were subjected to Southern blot analysis. The blot was probed with full-length pUC18-HPV16 (pEF399). The arrows to the right of each blot show the positions of open-circle (OC) and supercoiled (SC) HPV16 plasmids, respectively. In the rightmost blot, the position of the spiked pUC18 DNA (Spike DNA) is indicated by an arrow. Note that pPA103, pKT309, and HPV16 migrate as lower-molecular-weight species than their parental plasmids (shown as controls), since the 2.7-kb pUC18 sequences had been deleted prior to transformation of yeast.
No single ORF tested is required for replication of HPV16 in yeast.
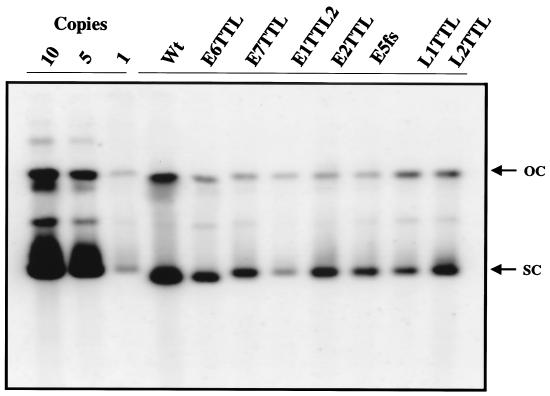
Several studies have indicated that virally encoded factors such as E1 and E2 are required for stable replication of papillomaviruses (11, 36). However, recent studies from our laboratory indicate that E1 is not strictly required for replication of BPV-1 and HPVs in mammalian cells (Kim and Lambert, submitted). Therefore, we wanted to formally test which ORFs, if any, were required for stable replication and maintenance of HPV16 in yeast. A series of plasmids were created which contained the TRP1 marker and full-length viral genomes bearing mutations--E1TTL, E2TTL, E5fs, E6TTL, E7TTL, L1TTL, and L2TTL--that lead to disruption of individual ORFs due to the introduction of stop codons (translation termination linker [TTL]) or frameshifts (fs) early within each gene. All of the mutant genomes, when transformed into yeast, formed colonies approximately as efficiently as the wt genome (data not shown). Southern blot analysis of yeasts containing these mutants revealed that all were capable of replication (Fig. 3).
FIG. 3.
No single ORF tested is required for episomal replication of HPV16 in yeast. Plasmid constructs which contain either the wt HPV16 genome (pPA100) or genomes with E1TTL2 (pKT269), E2TTL (pPA101), E5fs (pKT304), E6TTL (pKT307), E7TTL (pKT303), L1TTL (pKT305), or L2TTL (pKT308) mutations were transformed into YPH500 yeast. Five colonies were pooled and analyzed for stable episomal replication. DNA samples were DpnI digested prior to electrophoresis. The lanes on the left of the gel are controls for the copy number per cell. The blot was probed with full-length HPV16. The arrows to the right of the blot show the positions of open-circle (OC) and supercoiled (SC) plasmid forms. Note that the E1TTL genome replicated as efficiently as the wt in multiple experiments (Fig. 1D), and thus, the apparent reduction in replication efficiency of E1TTL observed in this experiment was not reproducible.
HPV16 genomes display mitotic stability similar to that of yeast CEN-containing plasmids.
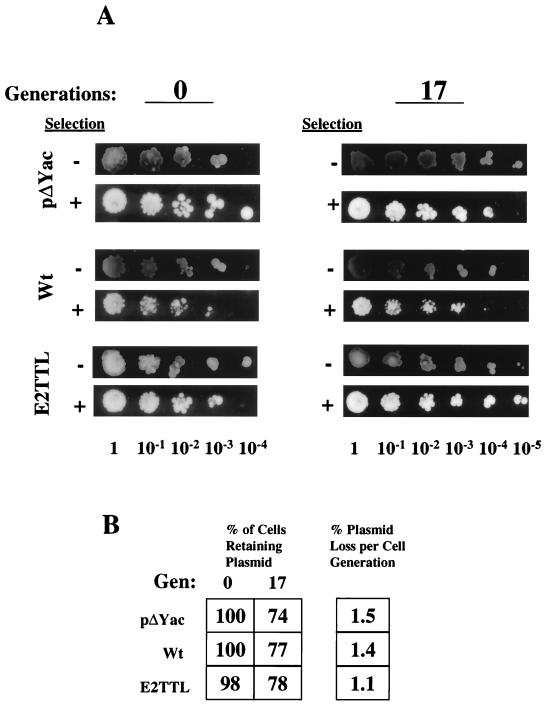
Newly synthesized papillomavirus genomes are efficiently partitioned to daughter cells during cytokinesis. Prior studies have suggested that the BPV-1 E2 protein contributes to maintenance, perhaps through its association with mitotic chromosomes (34, 42); however, it is not clear that this function of BPV-1 E2 is shared by HPV16 E2. Since HPV16 is capable of replication in yeast independent of any particular gene product, we questioned how stable these genomes were in the absence of E2. Therefore, we assessed the mitotic stabilities of wt HPV16, E2TTL, and pΔYac in the absence of selection (Fig. 4A). Yeasts harboring the plasmids were allowed to grow in the absence of selection for 0 to 17 cell generations. At each of the time points, serial dilutions of the cultures were plated on selective (without Trp) or nonselective (with Trp) medium. Clearly, at either 0 or 17 cell generations, the numbers of colonies formed on selective versus nonselective medium were very similar at each dilution tested. These results show that very little plasmid loss had occurred over 17 cell doublings. Each plasmid showed similar stability characteristics: for example, the percentage of plasmid retained after 17 cell generations in the absence of selection was 74% for pΔYac, 77% for wt HPV16, and 78% for E2TTL (Fig. 4B). Based on these results, the plasmid loss per cell generation was calculated to be 1.5% for pΔYac, 1.4% for wt HPV16, and 1.1% for E2TTL. Therefore, the HPV16 genome is as stable as a yeast CEN-containing plasmid, such as pΔYac. By quantitative Southern blot analysis of a plasmid loss assay, it was observed that episomal copies of HPV16 (pKT269) were retained after 10 cell doublings with a loss rate of 1% per cell generation (data not shown). These results support the concept that maintenance of HPV episomes occurs independently of E2 and thus is likely to be due to specific viral sequences.
FIG. 4.
HPV16 is as mitotically stable as an ARS+ CEN+ plasmid in yeast, and E2 appears not to be required for this function. Yeasts containing either pΔYac, wt HPV16 (pPA100), or HPV16 E2TTL (pPA101) were grown under nonselective conditions for 17 cell generations. (A) After the incubation period, the cell cultures were serially diluted from 1 to 10−5. Equal volumes of each of the dilutions were spotted onto selective (+) and nonselective (−) media. The result is a visual comparison which indicates the degree of plasmid stability. (B) To determine the percentage of cells retaining plasmids, cultures grown in the absence of selection were diluted to an appropriate extent and plated on selective and nonselective media. The percentage of cells retaining plasmids was determined by the ratio of the number of colonies present on the selective medium versus the number on the nonselective medium. The table indicates the percentage of cells retaining plasmids for each plasmid at 0 and 17 cell generations (Gen).
BPV-1 and multiple HPVs can replicate in yeast.
The results with HPV16 described above led us to question whether other papillomavirus genomes could replicate in yeast. In order to address this question, we transfected BPV-1 and HPV6b, -11, -16, -18, and -31 into YPH500 yeast and cultured the transformants for 20 cell doublings in rich medium (Fig. 5). Total DNA was then isolated, spiked with pUC18 DNA, DpnI digested, and analyzed by Southern blotting. Whereas bacterially synthesized pUC18 DNA was completely digested by DpnI, the papillomavirus DNAs recovered from yeast were DpnI resistant. These results suggest that features shared among these viral genomes allow the yeast replicative machinery to recognize origin sequences and successfully replicate these DNAs.
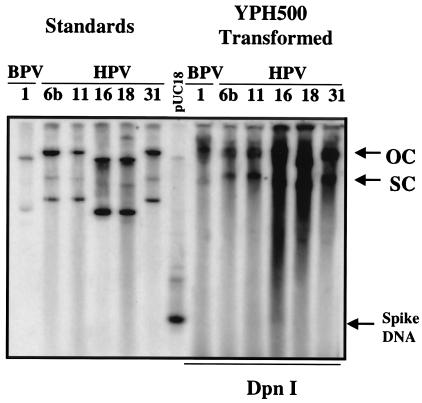
FIG. 5.
BPV-1 and Multiple HPVs are efficiently replicated in yeast. Yeast was transfected with pUC18-based plasmids containing BPV-1 and HPV6b, -11, -16, -18, and -31. The yeast was cultured in yeast-peptone-dextrose medium for 48 h (approximately 20 cell generations), and low-molecular-weight DNA was isolated. At the time of DNA isolation, each sample was spiked with 2 ng of pUC18 in order to monitor the completeness of DpnI digestion. DpnI digestion was carried out for 24 h, and DNA samples were subjected to Southern blot analysis using pUC18 as a probe. The lanes marked as standards were loaded to compare the molecular weights of the transfected plasmids. The arrows to the right of the blot show the positions of open-circle (OC) and supercoiled (SC) plasmids. The position of bacterially synthesized pUC18 DNA added to the samples (Spike DNA) is also shown by an arrow. Note that the mobility of the recovered plasmids in the lanes labeled “YPH500 Transformed” is retarded compared to the standards by the presence of yeast genomic DNA.
DISCUSSION
The results reported here establish the novel finding that papillomaviruses can replicate extrachromosomally in S. cerevisiae in the absence of complementing ARS or CEN elements (Fig. 1). Our experiments show that HPV16 plasmids replicate stably at a copy number between one and five, similar to a typical yeast ARS/CEN plasmid (5, 44).
Our experiments confirmed that replicative and maintenance functions reside in papillomavirus DNA. Use of either a TRP1 or a URA3 marker and removal of the pUC18 sequence had no effect on colony formation or on episomal replication of the constructs tested in yeast (Fig. 2). We found no evidence for integration or rearrangement of these constructs in yeast. Moreover, religated full-length HPV16, when introduced into yeast, was able to replicate in short-term assays over 15 cell doublings. In short, the behavior of HPV16 plasmids in yeast was as reliable as that of typical ARS/CEN plasmids.
Our experiments showing replication of HPV16 E1TTL (Fig. 1 and 3) support our previous findings that this trans-acting factor is not strictly required for replication of papillomavirus DNA in mammalian cells (21a; Kim and Lambert, submitted). The major roles of E1 helicase in papillomavirus replication are in the recruitment of Polα and in unwinding of the origin (14, 32). We hypothesize that in the absence of E1, these functions must be provided by cellular helicases. E1 hexamers form a clamp around DNA reminiscent of several cellular helicases, for example, members of the RecQ family, Werner's and Bloom's syndrome gene products (WRN and BLM, respectively), and the MCM family of proteins (12, 27, 29, 33). A detailed mapping analysis of the E1-independent origin sites in HPV16 utilized in yeast and in human cells will lead to a better understanding of the functional significance of this mode of replication.
The HPV16 mutant genomes in all of the ORFs were capable of episomal replication in yeast (Fig. 3). Of particular interest is the fact that E2 was not required for the replication of HPV16 in yeast, given the potential importance of E2 in the replication and maintenance of viral genomes (18, 26, 36, 45). Interestingly, we have found that exogenous E2 expression in yeast enhances the copy number of HPV16 episomes 5- to 10-fold (data not shown). This result suggests that E2 might function in yeast to support stable replication of papillomavirus genomes; however, we do not know whether this effect of E2 is due to its transcription, replication, or maintenance properties or a combination thereof.
Analysis of the mitotic stability of HPV16 in yeast revealed that viral episomes are as stable as a typical ARS/CEN plasmid (pΔYac). The mitotic stability of wt HPV16 and E2TTL plasmids was assessed using an assay described by Kapoor et al. (20). The rate of plasmid loss for an ARS/CEN plasmid is approximately 1 to 2% per cell generation (16). Very similar to this expectation, we measured a loss rate of 1.5% for pΔYac. Interestingly, both wt HPV16 and E2TTL plasmids were very stable, having loss rates of 1.4 and 1.1% per cell generation, respectively. These data indicate that viral episome maintenance in yeast occurs independently of E2. The majority of findings for mammalian cells indicate that E2 has an important role in the partitioning of plasmids (2, 34, 42, 45). It is possible that maintenance of HPV16 genomes in yeast is mediated by a fortuitous interaction with a cellular factor. Alternatively, there may exist cis-acting CEN-like elements in the HPV16 genome that heretofore have not been detected in mammalian cells. This possibility is supported by evidence that a fragment of the L2 ORF of BPV-1 can functionally substitute for a CEN element in a yeast vector (4).
When linked to a selectable marker, HPV16 plasmids essentially function as minichromosomes in yeast. Multiple studies have taken advantage of episomal replication properties of plasmids in yeast and in mammalian cells. The development of the yeast artificial chromosome vectors represented a great advancement in manipulating large DNA fragments in yeast (6). Stable plasmids, such as those based on Epstein-Barr virus OriP (49), have been employed to isolate human DNA fragments which contain ARS-like activity (22). Similarly, a vector described by Simpson et al. (40, 41) is a hybrid between an ARS/CEN yeast artificial chromosome plasmid and an Epstein-Barr virus OriP plasmid. This vector is capable of stable episomal replication in both yeast and human cells while harboring more than 600 kb of foreign DNA. In future studies, we will analyze the utility of our HPV plasmids as yeast-human cell shuttle vectors and define the minimal sequences required for episomal replication in yeast and human cells.
Our analyses also revealed that multiple HPVs and BPV-1 were capable of replication in yeast. In these experiments, DpnI-resistant DNA was recovered from yeast after 20 cell doublings (Fig. 5). During the course of our study, we learned that another group had independently discovered that BPV-1 can replicate in S. cerevisiae (50). Our data support the idea that sequence elements required for origin recognition in yeast are conserved among multiple viruses. This raises the obvious question of whether papillomavirus “origins” active in yeast can also function in human cells? Though our data do not address this question completely, it is interesting that, as we found in yeast (Fig. 1), the E1-independent replication element(s) lies outside of the LCR in mammalian cells (Kim and Lambert, submitted). There is some evidence that ARS sequences have conserved determinants from yeast to humans (1). Despite sequence differences, a comparison of replication initiation sites of yeast ARS1 with the simian virus 40 origin finds that leading- and lagging-strand initiation sites flanking the origin map in similar patterns (3), suggesting conserved recognition machinery. Furthermore, the constituents of the ORC in mammalian cells and budding yeast have significant homology and are thought to retain mechanistic similarity (47). Recognition of A/T-rich clusters in the origin by ORC proteins which contain A/T hook domains is likely to play a key role in site recognition in yeast and mammalian cells, though the precise specificities may differ slightly (7). Thus, it is plausible that yeast replicative machinery recognizes sequences shared among papillomaviruses and that the same sites are potentially utilized in both yeast and human cells.
The system we have described represents a significant advance which will aid in our understanding of papillomaviruses. This system is ideal for analyzing the requirements for episomal replication, in particular, the contributions of E1 and E2. Regulated expression of these genes may allow the development of a model for genome amplification. Furthermore, yeasts provide superb genetic tools with which to dissect the roles of cellular and viral trans-acting factors involved in extrachromosomal replication. Recent work has demonstrated that yeasts expressing HPV16 L1 and L2 allow the formation of virus-like particles (37). Using our system, we foresee the possibility of packaging full-length HPV16 into virions.
Acknowledgments
Peter C. Angeletti and Kitai Kim contributed equally to this work.
We thank Paul Ahlquist for important discussions of this work. We thank Ian Frazier for discussing his laboratories' studies prior to publication.
This work was supported by grants from NCI (22443 and 07175). Peter C. Angeletti was supported by NIH postdoctoral training grant T32 CA09075. Kitai Kim is a Cremer Scholar.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 3.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 4.Blangy, A., G. F. Carle, V. Pierrefite, M. Rassoulzadegan, and F. Cuzin. 1992. Mammalian and viral DNA sequences which interfere with the maintenance of a centromeric vector in yeast. Biochem. Biophys. Res. Commun. 187:737-743. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D. T., G. F. Carle, and M. V. Olson. 1987. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236:806-812. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, R. Y., and T. J. Kelly. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA 96:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conger, K. L., J. S. Liu, S. R. Kuo, L. T. Chow, and T. S. Wang. 1999. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 9.Del Vecchio, A. M., H. Romanczuk, P. M. Howley, and C. C. Baker. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePamphilis, M. L. 1998. Initiation of DNA replication in eukaryotic chromosomes. J. Cell. Biochem. Suppl. 31:8-17. [PubMed] [Google Scholar]
- 11.DiMaio, D., and J. Settleman. 1988. Bovine papillomavirus mutant temperature sensitive for transformation, replication and transactivation. EMBO J. 7:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 13.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 15.Francesconi, S., W. C. Copeland, and T. S. Wang. 1993. In vivo species specificity of DNA polymerase alpha. Mol. Gen. Genet. 241:457-466. [DOI] [PubMed] [Google Scholar]
- 16.Hieter, P., C. Mann, M. Snyder, and R. W. Davis. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40:381-392. [DOI] [PubMed] [Google Scholar]
- 17.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, P. M. Howley, et al., (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 18.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 20.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 21a.Kim, K., and P. F. Lambert. Requirement for E1 of BPV-1 in the establishment and maintenance phase of the viral life cycle. Virology, in press.
- 22.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert, P. F., N. Dostatni, A. A. McBride, M. Yaniv, P. M. Howley, and B. Arcangioli. 1989. Functional analysis of the papilloma virus E2 trans-activator in Saccharomyces cerevisiae. Genes Dev. 3:38-48. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. K., and J. Hurwitz. 2000. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 275:18871-18878. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. K., and J. Hurwitz. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei, M., Y. Kawasaki, M. R. Young, M. Kihara, A. Sugino, and B. K. Tye. 1997. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 11:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X., and P. M. Burgers. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae RFC3 gene, an essential component of replication factor C. Proc. Natl. Acad. Sci. USA 91:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohaghegh, P., J. K. Karow, R. M. Brosh, Jr., V. A. Bohr, and I. D. Hickson. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrissey, L. C., J. Barsoum, and E. J. Androphy. 1989. Trans activation by the bovine papillomavirus E2 protein in Saccharomyces cerevisiae. J. Virol. 63:4422-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newlon, C. S., and J. F. Theis. 1993. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3:752-758. [DOI] [PubMed] [Google Scholar]
- 32.Park, P., W. Copeland, L. Yang, T. Wang, M. R. Botchan, and I. J. Mohr. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. USA 91:8700-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 34.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 35.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabson, M. S., C. Yee, Y. C. Yang, and P. M. Howley. 1986. Bovine papillomavirus type 1 3′ early region transformation and plasmid maintenance functions. J. Virol. 60:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi, J. L., L. Gissmann, K. Jansen, and M. Muller. 2000. Assembly of human papillomavirus type 16 pseudovirions in Saccharomyces cerevisiae. Hum. Gene Ther. 11:1165-1176. [DOI] [PubMed] [Google Scholar]
- 38.Schiestl, R. H., M. Dominska, and T. D. Petes. 1993. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol. Cell. Biol. 13:2697-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson, K., and C. Huxley. 1996. A shuttle system for transfer of YACs between yeast and mammalian cells. Nucleic Acids Res. 24:4693-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, K., A. McGuigan, and C. Huxley. 1996. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol. 16:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanway, C. A., M. P. Sowden, L. E. Wilson, A. J. Kingsman, and S. M. Kingsman. 1989. Efficient activation of transcription in yeast by the BPV1 E2 protein. Nucleic Acids Res. 17:2187-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stinchcomb, D. T., K. Struhl, and R. W. Davis. 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282:39-43. [DOI] [PubMed] [Google Scholar]
- 45.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vashee, S., P. Simancek, M. D. Challberg, and T. J. Kelly. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276:26666-26673. [DOI] [PubMed] [Google Scholar]
- 48.Yang, L., and M. Botchan. 1990. Replication of bovine papillomavirus type 1 DNA initiates within an E2-responsive enhancer element. J. Virol. 64:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, K.-N., and I. H. Frazer. 2002. Replication of bovine papillomavirus type 1 (BPV1) DNA in Saccharomyces cerevisiae following infection with BPV1 virions. J. Virol. 76:3359-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]