Abstract
The major immediate-early (MIE) gene of human cytomegalovirus (HCMV) expresses IE86, IE72, IE55, and IE18 mRNA by differential splicing. Reverse transcription-PCR with IE72-specific primers generated an 0.65-kb cDNA from HCMV-infected fibroblast RNA, which does not correspond to any known MIE cDNA. Nucleotide sequencing revealed that the 0.65-kb cDNA is from exons 1, 2, and 3 and part of exon 4, indicating that it is derived from a novel alternatively spliced mRNA of the MIE gene. The cDNA encodes a 172-amino-acid polypeptide, termed IE19, which corresponds to an IE72 variant with an internal deletion from Val86 to Pro404 and appears as a band at 38 kDa on a sodium dodecyl sulfate-polyacrylamide gel. IE19 mRNA was expressed at a low level in the immediate-early, early, and late period of viral infection. IE19 was localized in nuclei, and a transient-expression assay revealed that IE19 enhances IE72-dependent activation of the HsOrc1 promoter, which is identified here as an IE72 target promoter. Another MIE protein, IE86, activated the same promoter but only weakly compared to IE72, and coexpression of IE19 did not alter the IE86-mediated transcriptional activation. In addition, IE19 did not enhance the IE72-dependent activation of the HCMV UL54 promoter. These results suggest that IE19 is a transcriptional coactivator that works with IE72.
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that causes pneumonitis, encephalitis, retinitis, hepatitis, and gastroenteritis in immunosuppressed or immunocompromised individuals. Fetal infection causes hearing loss and mental retardation (2, 21). However, HCMV rarely causes diseases upon primary infection in healthy people. After HCMV infection, a sequential cascade of the viral gene expression is initiated in cells to facilitate efficient synthesis of viral proteins and replication of the DNA genome (42). The expression of viral genes is classified into three periods, immediate-early, early, and late, based upon this cascade (13, 41, 58, 66, 67).
The major immediate-early (MIE) gene is furthest upstream in the regulatory cascade of virus genes and does not require de novo protein synthesis for its transcription (24, 42, 43, 47). The MIE gene has a single promoter region and five exons, with two poly(A) additional sequences in exon 4 and exon 5 (55, 56). Alternative splicing of the MIE transcript produces at least four different mRNAs, which encode IE72/IE1 (72 kDa), IE86/IE2 (86 kDa), IE55/IE2 (55 kDa), and IE18/IE2 (18 kDa) (3, 20, 29, 55-58). IE72 and IE86 are major products of the MIE gene in the immediate-early period of HCMV infection and are potent transcriptional activators for several viral and cellular genes. These MIE-responsive genes include the genes for HCMV UL4, UL54, UL83, and UL112-113 (10, 14, 28, 39, 59) and cellular genes of DNA polymerase α, dihydrofolate reductase, NF-κB, transforming growth factor β1, and interleukin 1β (18, 40, 64, 65, 71, 72). IE55 and IE18 are minor products of the MIE gene, and their functions in HCMV infection are so far unknown (29). It should be noted that IE55 expression is restricted in the late period of infection and that IE18 expression in HCMV-infected human fibroblasts is detected only under cycloheximide blockage (29, 60). However, IE18 is relatively abundant in normally infected macrophages.
Although previous reports suggest that IE72 and IE86 bind DNA directly, IE72 and IE86 activate genes by interacting with cellular transcription factors. It has been reported elsewhere that IE72 associates with E2F (38, 40) and that IE86 interacts with Sp1, Spi-1/PU.1, AP-1, and Egr-1 (9, 32, 38, 50, 52, 53, 65, 70, 71). In addition, both of them associate with basic transcription factors such as TATA box-binding protein (TBP)-associated factors (TAFIIs) including TAFII110, TAFII130, and TAFII40 (37). IE86 also binds directly to TBP (7, 15, 17, 25, 26, 36, 54). This would suggest that IE72 and IE86 are links between various sequence-specific DNA-binding transcriptional regulators and the basic transcriptional machinery.
IE72 and IE86 activate genes with promoters containing E2F sites (40, 64). E2F regulates many genes required for DNA replication including those of the replication enzymes and the replication origin-binding complex proteins (prereplicative complex) (12, 19, 23, 33). E2F sites are often found around transcription initiation sites, and the E2F/Rb complex bound to these sites represses nearby transcription (23, 44). In the G1 and S phases of the cell cycle, phosphorylation of Rb results in dissociation of the E2F/Rb complex and loss of the transcriptional suppression. It is reported elsewhere that IE72 phosphorylates E2F and releases the promoter from the E2F/Rb-imposed suppression (46). In addition, the transcription of these replication-related genes, which is regulated by E2F, is often driven by TATA-less promoters that contain several Sp1 sites. Because IE72 and IE86 activate genes through the Sp1 site, IE72 and IE86 could regulate these replication-related genes through these sites. The dhfr gene is the first gene identified as being activated by IE72 through the E2F site, but little is known about the role of IE72 and IE86 in the expression of other E2F-regulated replication-related genes. HsOrc1 is the human homolog of the Saccharomyces cerevisiae replication origin recognition complex protein, Orc1p, which is essential for the initiation of cellular DNA replication and cell growth (4-6, 16). Like the dhfr gene, the HsOrc1 promoter is also TATA-less and regulated by E2F (45). Thus, the HsOrc1 promoter might be regulated by IE72 upon HCMV infection.
Earlier studies suggested expression of unidentified splicing variants of MIE proteins in HCMV-infected cells. Stenberg et al. (58) examined MIE proteins expressed in HCMV (Towne)-infected human foreskin fibroblast cells by using antibodies to viral peptides and found that a minor 38-kDa protein reacted with antibodies to an IE72-specific peptide from amino acids (aa) 383 to 420. Kerry et al. (29) examined an MIE cDNA library to isolate low-abundance cDNAs and showed that a subpool of MIE cDNAs contained an 0.65-kb cDNA hybridizing to an exon 1-specific probe. However, these MIE-related proteins and cDNAs were not characterized any further. On preparing IE72 cDNA by reverse transcription-PCR (RT-PCR) from HCMV-infected human embryonic lung fibroblast (HEL) cells, we obtained a cDNA shorter than that expected for IE72 cDNA. In this study, we report that this short cDNA encodes IE19, a novel splicing variant of MIE proteins, and that IE19 is a transcriptional coactivator that works with IE72 in the activation of the HsOrc1 promoter, a novel target of IE72.
MATERIALS AND METHODS
Cell culture and viral infection.
HEL cells and the human glioblastoma cell line U373MG were cultured in Dulbecco's modified Eagle's essential medium containing 10% fetal calf serum (FCS) and 1 mM sodium pyruvate. HeLa cells were cultured in Eagle essential medium containing 10% FCS. For infection of HCMV (Towne), HEL cells were incubated with the virus in a culture medium containing 2% FCS for 2 h. These cells were then washed and cultured in fresh medium containing 2% FCS.
RNA.
HEL cells were infected with HCMV (Towne) at a multiplicity of infection (MOI) of 1 or 5 and harvested at given time points. Total RNA of HCMV-infected cells was prepared using ISOGEN (Nippon Gene). Poly(A)+ RNA was prepared with an mRNA purification kit (Pharmacia).
RT-PCR.
RT-PCR was performed using the Superscript One-Step RT-PCR system (Lifetech). Total RNA (1 μg) or poly(A)+ RNA (0.2 μg) was used for a single reaction. Nucleotide sequences of the oligonucleotide primers used for RT-PCR are shown in Table 1. The reverse transcriptase reaction was performed at 55°C for 30 min. To amplify the IE72, IE19, and IE86 cDNAs, each sample was denatured at 94°C for 1 min, annealed at 55°C for 1 min, and extended at 72°C for 6 min, and this cycle was repeated 40 times. For RT-PCR analysis of mRNA expression, each sample was denatured at 94°C for 30 s, annealed at 50°C for 30 s, and extended at 72°C for 2 min, and this cycle was repeated 20, 30, or 40 times. RT-PCR products were subjected to agarose gel electrophoresis and transferred to a Hybond N+ membrane (Amersham) for Southern hybridization analysis. The 32P-labeled IE72-specific probe was the PstI-EcoRI fragment of exon 4. Hybridization was performed at 65°C overnight, and a final wash was performed in a solution containing 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate (SDS) at 55°C for 1 h. Hybridized signal was quantified with a BAS2000 Image analyzer (Fuji).
TABLE 1.
Primers and probes
| Procedure (figure) and oligonucleotide | Type | Sequence | Nucleotide coordinates in HCMV AD169 |
|---|---|---|---|
| RT-PCR (Fig. 1) | |||
| E2 (IE72/IE86) | Primer (sense) | 5′-GACACGATGGAGTCCTCTGC-3′ | 172771-172752 |
| E4R (IE72) | Primer (antisense) | 5′-AATAGTTACTGGTACTGGTCAGCCT-3′ | 171000-171019 |
| E5R (IE86) | Primer (antisense) | 5′-TTTCACTTACTGAGACTTGT-3′ | 169358-169377 |
| E1(+1) | Primer (sense) | 5′-TCAGTCGCCTGGAGACGCCAT-3′ | 173730-173709 |
| E1(−60) | Primer (sense) | 5′-AATGGGCGGTAGGCGTGTAC-3′ | 173790-173771 |
| E1(−130) | Primer (sense) | 5′-GGAGTTTGTTTTGGCACCAA-3′ | 173860-173841 |
| RT-PCR (Fig. 5) | |||
| IE72/IE9 | Primer (sense) | 5′-TGGTCAGCCTTGCTTCTAGT-3′ | 171010-171029 |
| Primer (antisense) | 5′-TAACAGTCAGCTGAGTCTGG-3′ | 172529-172501 | |
| Probe | 5′-TCTCAGACACTGGCTCAGAC-3′ | 171112-171132 | |
| UL112-113 | Primer (sense) | 5′-AACGTCACGAAGAACGGCGA-3′ | 161151-161170 |
| Primer (antisense) | 5′-TTGAGAAAGGCCACCGCTTC-3′ | 161680-161661 | |
| Probe | 5′-TGCTCCGTCTCGTTCTTGCC-3′ | 161590-161571 | |
| β-Actin | Primer (sense) | 5′-CAAGAGATGGCCACGGCTGCT-3′ | |
| Primer (antisense) | 5′-TCCTTCTGCATCCTGTCGGCA-3′ | ||
| RT-PCR (Fig. 6) | |||
| HsOrc1 | Primer (sense) | 5′-CCTTCTCGGAGATCACCTCA-3′ | |
| Primer (antisense) | 5′-TCTTCGTCACTGCTAGAGTC-3′ |
Northern blot hybridization.
Poly(A)+ RNA (18 μg) was subjected to agarose electrophoresis in a 1% formaldehyde gel with 1× morpholinepropanesulfonic acid (MOPS) buffer. RNA was then transferred to the Hybond N+ membrane and hybridized with the 32P-labeled HsOrc1 cDNA in the 50% formamide hybridization buffer at 42°C overnight. A final washing was performed in a solution containing 0.2× SSC and 1% SDS at 50°C for 1 h. After removal of the hybridized probe, the same membrane was analyzed with the 32P-labeled chicken β-actin cDNA.
Cloning of IE cDNAs.
The 0.65-kb IE19 cDNA amplified with E1(+1) and E4R primers and the 0.5-kb IE19 cDNA amplified with E2 and E4R primers were cloned into pGEM-T Easy vector (Promega). Nucleotide sequences of these cDNAs were determined by the dye-terminator PCR method (Perkin-Elmer). The IE72 cDNA amplified with E1(+1) and E4R primers and the IE86 cDNA amplified with E1(+1) and E5R primers were cloned similarly.
Plasmids.
Mammalian expression plasmids for IE19 (pME-IE19), IE72 (pME-IE72), and IE86 (pME-IE86) were constructed by inserting each IE cDNA into the expression vector pME18S. These MIE cDNAs were prepared by RT-PCR as described above. To construct the plasmid pME-IE19FLAG, which expresses IE19 tagged with FLAG at its carboxyl terminus, the stop codon of IE19 was deleted and a BamHI site was inserted by a PCR-assisted method. The modified cDNA was cloned into pME18FLAG, a derivative of pME18S. The luciferase reporter plasmid (pHsOrc1Luc) containing the promoter and upstream regulatory region from nucleotide (nt) −1053 to +182 of the HsOrc1 gene and the luciferase reporter plasmid (pPolLuc) containing the UL54 promoter were described previously (34, 45). The pHsOrc1Luc with a mutation of the E2F site, pHsOrc1LucΔE2F, was originally designated pHsOrc1Luc(−E2F) and has been described previously (45). To introduce a mutation at the Sp1P site of the HsOrc1 promoter by PCR, the entire pHsOrc1Luc plasmid was amplified with two oligonucleotide primers with the mutated sequences 5′-CAGGCCACGCCGATTGGCGCG-3′ and 5′-CAGCCCAGACCGTCCCTTCGT-3′. These primers were designed to anneal the different strands at the E2F site in a tail-to-tail manner. A mutation of Sp1D was introduced similarly with another set of primers, 5′-CAGGCGAGCTAGTTGGTGTCG-3′ and 5′-CAGCACTCCGATCACCTAAGA-3′. Each of the amplified linear plasmids was self ligated. The nucleotide sequence of the promoter region was confirmed by sequencing.
Western blot analysis.
Cells were collected and solubilized with the sample buffer for SDS-polyacrylamide gel electrophoresis (PAGE). The solubilized proteins were separated by SDS-PAGE with a 10% polyacrylamide gel and transferred to an Immobilon-P membrane (Millipore) with a semidry transfer apparatus with transfer buffer containing 0.1 M Tris, 0.192 M glycine, 0.1% SDS, and 20% methanol. Before the transfer, the membrane was first soaked with methanol for 1 min and with the transfer buffer at room temperature for 2 h. Electrophoretic transfer was performed at a constant current of 100 mA (5 V) for 1 h. The blotted membrane was soaked with 5% skim milk in phosphate-buffered saline containing 0.1% Tween 20 (PBST). To detect MIE proteins, the membrane was reacted with monoclonal antibody (MAb) 810 diluted 1:200 (Chemicon) in PBST containing 3% bovine serum albumin and subsequently with the horseradish peroxidase-conjugated anti-mouse immunoglobulin antibody diluted 1:1,000 (Amersham) in PBST containing 2% skim milk. Enhanced chemiluminescence substrate was used to detect the second antibody. To detect IE19FLAG fusion protein, the anti-FLAG antibody M2 (Sigma) was used.
Indirect immunofluorescence method.
The expression plasmids pME-IE19, pME-IE72, and pME-IE86 were transfected into HLF cells with Lipofectamine reagent (Lifetech). One day after transfection, cells were fixed with 4% paraformaldehyde in PBS and reacted with MAb 810 followed by fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibody (Cappel). Stained cells were examined with a fluorescence microscope equipped with a charge-coupled device camera (Zeiss).
Reporter assay.
The MIE expression plasmid(s) (pME-IE19, pME-IE72, and/or pME-IE86), the luciferase reporter plasmid, and pRSVLacZ were transfected into U373MG cells in a 60-mm-diameter dish by the calcium phosphate DNA precipitation method. The cells were harvested 2 days after transfection, and cell extracts were prepared by freeze-thawing them three times in 100 μl of 0.1 M potassium phosphate (pH 7.8). Luciferase activity in the extracts (20 μl) was then analyzed as described previously (48), and the β-galactosidase activity was determined using o-nitrophenyl-β-d-galactopyranoside as a substrate. Luciferase activity was normalized to β-galactosidase activity.
Nucleotide sequence accession number.
The accession no. of the nucleotide sequence of IE19 cDNA is AB057730.
RESULTS
Identification of a novel cDNA amplified with IE72-specific primers.
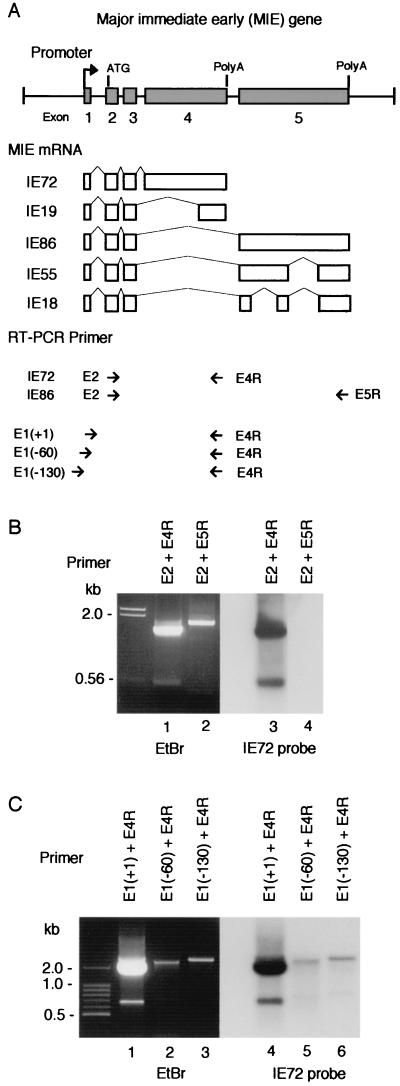
To amplify the complete coding regions of IE72 and IE86 mRNAs by RT-PCR, we used three oligonucleotide primers covering the initiation codon in exon 2 (E2) and the stop codons of exon 4 (E4R) and exon 5 (E5R) (Fig. 1A). RNA was prepared from HCMV (Towne)-infected HEL cells at 72 h postinfection (hpi). When a pair of IE72-specific primers (E2 and E4R) were used for RT-PCR, the IE72 cDNA (1.7 kb) was amplified (Fig. 1B, lane 1). Sequence analysis confirmed that the cDNA contained the complete coding sequence of IE72. In addition, the same RT-PCR produced 0.5- and 1.8-kb cDNAs. The 1.8-kb cDNA could be derived from unspliced precursor mRNA of the MIE gene. However, the length of the shorter cDNA does not fit that of any known MIE mRNAs. Because Southern hybridization analysis indicated that the cDNA contains at least part of MIE exon 4 (Fig. 1B, lane 3), we determined the nucleotide sequence and found that the cDNA is from exon 2 and exon 3 and the 3′ part of exon 4 (Fig. 2A). This suggests that the 0.5-kb cDNA is a product of novel splicing variants of MIE mRNA, which encodes IE19 (see below). To identify the transcription initiation site of the IE19 mRNA, we performed another RT-PCR analysis with three primers, E1(+1), E1(−60), and E1(−130), which annealed to the common transcription initiation site of IE72 and IE86 mRNAs (+1) or the upstream region of IE19 mRNA around nt −60 or nt −130 (Fig. 1A) (55, 62). In addition to the 1.85-kb IE72 cDNA, an 0.65-kb IE19 cDNA was amplified with E1(+1) and E4R primers (Fig. 1C, lane 1). Sequence analysis showed that the 0.65-kb cDNA generated with E1(+1)/E4R RT-PCR contains the 0.5-kb IE19 cDNA and a short 5′ region identical to the 5′ untranslated region of IE72 mRNA (Fig. 2A). This indicates that the first intron of the MIE gene was spliced out for IE19 mRNA. When E4R and E1(−60) or E1(−130) were used for RT-PCR, no cDNA derived from IE19 mRNA was detected (lanes 2 and 3). A small amount of cDNA longer than 2 kb was generated with each primer combination (lanes 2 and 3). These cDNAs could be derived from the read-through transcripts of the upstream genes. The results suggest that the transcription initiation site of the novel mRNA is very similar or identical to the initiation site of IE72 mRNA. We also amplified IE86 cDNA by RT-PCR with E2 and E5R primers but did not detect any splicing variant of IE86 in this RNA sample (Fig. 1B, lane 2).
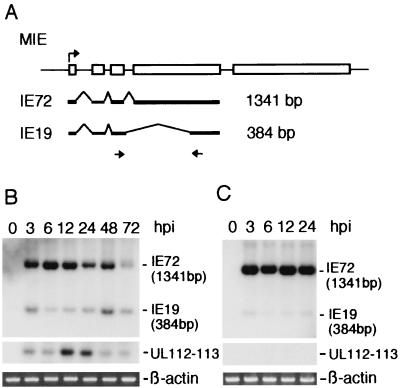
FIG. 1.
RT-PCR analysis of MIE mRNA. (A) MIE gene and the primers used for RT-PCR analysis. (Top) Major exon-intron structure of the MIE gene. Numbered solid boxes indicate each exon. The initiation codon (ATG) is located in exon 2. Both exon 4 and exon 5 have a stop codon and a poly(A) additional signal. (Middle) Alternative splicing to produce MIE mRNAs. (Bottom) Short arrows show position and direction of RT-PCR primers. The name of each primer is indicated. E1(+1) primer covers the transcription initiation site, and E1(−60) and E1(−130) primers cover to nt −60 and to nt −130 in the 5′-flanking region of the initiation site, respectively. (B and C) Results of RT-PCR analysis. RT-PCR was performed with total RNA (1 μg) of HCMV-infected HEL cells prepared at 48 hpi and with the primers indicated in each lane. RT-PCR products were examined by agarose gel electrophoresis (ethidium bromide [EtBr]) and Southern blot hybridization with an IE72-specific probe (IE72 probe). The positions of nucleotide markers are indicated. The nucleotide sequences of these RT-PCR primers and probes are shown in Table 1.
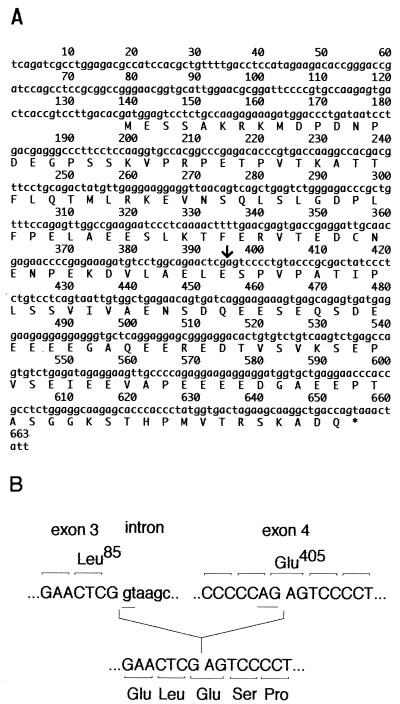
FIG. 2.
IE19 cDNA. (A) Nucleotide sequence of the 0.65-kb IE19 cDNA. The cDNA encodes a polypeptide of 172 aa. An arrow indicates the joining site of exon 3 and exon 4 of IE19 mRNA. (B) Nucleotide sequence of the MIE gene at the splicing donor site of exon 3 and the acceptor sites inside exon 4. After splicing, the GAG codon of Glu405 at the splicing acceptor site within exon 4 was regenerated in the IE19 mRNA.
The 0.65-kb cDNA encodes a 19-kDa nuclear protein, IE19.
The nucleotide sequence of the 0.65-kb IE19 cDNA is shown in Fig. 2A. The 0.5-kb IE19 cDNA amplified by E2 and E4R primers stretches from nt 133 to 663. Sequence analysis revealed that MIE exons 1, 2, and 3 were spliced in IE19 mRNA as in other MIE mRNAs, and the sequence from nt 395 to 663 corresponded to the 3′ region of exon 4. However, the rest of exon 4 was missing in the IE19 mRNA (Fig. 1A and 2A). This indicates that IE19 mRNA is derived from a novel splicing variant of MIE mRNA. The GT-AG rule was conserved at a presumed acceptor site of IE19 mRNA in exon 4, and the reading frame is the same as that of IE72 mRNA (Fig. 2B). We also performed RT-PCR with a pair of primers outside this open reading frame because the E4R primer has a stop codon in it and confirmed that IE19 and IE72 mRNAs share the stop codon (data not shown). Therefore, the IE19 mRNA encodes a polypeptide of 172 aa that corresponds to an IE72 derivative with an internal deletion from aa 88 to aa 404. The calculated molecular mass of the polypeptide is 19 kDa, and therefore, this protein was designated IE19.
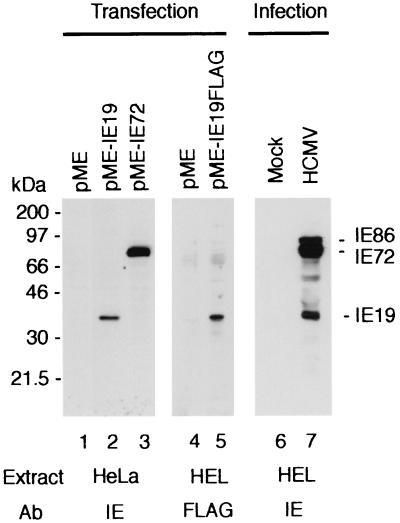
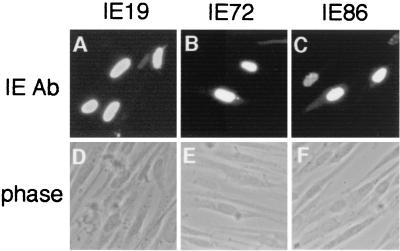
The expression plasmid carrying the 0.65-kb IE19 cDNA, pME-IE19, was transfected into HeLa cells, and proteins were examined by Western blot analysis with the anti-IE protein MAb 810, which recognizes the amino-terminal domain of MIE proteins. A 38-kDa protein appeared in cells transfected with pME-IE19 (Fig. 3, lane 2). The 38-kDa protein was not detected in cells transfected with control vector or the IE72 expression plasmid pME-IE72 (lanes 1 and 3). We also transfected an expression plasmid for the FLAG-tagged IE19 protein into HEL cells and confirmed the expression of a 38-kDa protein with anti-FLAG antibody (lane 5). These results suggest that the IE19 cDNA encodes the 38-kDa IE72-related protein. Western analysis using MAb 810 confirmed that IE19 is expressed in the HCMV-infected HEL cells (lane 7). Indirect immunofluorescence analysis of the pME-IE19-transfected HEL cells showed that IE19 localized predominantly in nuclei (Fig. 4). There was no apparent difference in the subcellular localizations of IE19, IE72, and IE86 in the analysis.
FIG. 3.
Expression of IE19 protein. The expression vector for IE19 (pME-IE19) or IE72 (pME-IE72) or the control vector (pME) was transfected into HeLa cells, and the expression of each IE protein was examined by Western blotting using anti-IE protein MAb 810 (lanes 1 to 3). The plasmid vector expressing IE19-FLAG fusion protein (pME-IE19FLAG) or the control plasmid was transfected into HEL cells. Expression of FLAG protein was examined by Western blotting with anti-FLAG antibody (lanes 4 and 5). HEL cells infected with HCMV (Towne) at an MOI of 5 were harvested at 24 hpi, and the expression of IE proteins was examined with MAb 810 (lanes 6 and 7). The positions of IE86, IE72, and IE19 proteins are indicated.
FIG. 4.
Subcellular localization of IE19 protein. The expression vector pME-IE19 (A and B), pME-IE72 (C and D), or pME-IE86 (E and F) was transfected into HEL cells, and the localization of IE proteins was examined by indirect fluorescence with the anti-IE protein MAb 810.
Expression of IE19 mRNA during HCMV infection.
We compared the expression levels of IE19 and IE72 mRNAs after HCMV infection by RT-PCR. IE19 mRNA was detected as early as 3 hpi and remained at a low level from the immediate-early (3 and 6 hpi) to the late (48 and 72 hpi) period of infection, while IE72 mRNA reached its highest level in the immediate-early period and decreased significantly in the late period (Fig. 5B). We also confirmed the expression of an early gene, UL112-113, at 12 and 24 hpi and synthesis of the virus DNA at 48 and 72 hpi (Fig. 5B and data not shown). When the infected cells were pretreated with cycloheximide, the expression of IE19 was reduced but not eliminated whereas UL112-113 transcription was totally inhibited (Fig. 5C). This suggests that de novo protein synthesis was not essential for efficient transcription of IE19 mRNA in the immediate-early period.
FIG. 5.
Expression of IE19 mRNA. (A) The MIE gene and its transcripts. RT-PCR primers and the predicted sizes of amplified cDNAs are also shown. (B) Expression of IE19, IE72, and UL112-113 mRNAs was examined by RT-PCR. HEL cells were infected with HCMV (Towne) at an MOI of 5 and harvested at 0, 3, 6, 12, 24, 48, and 72 hpi for RNA preparation. Total RNA (1 μg) was used for each RT-PCR. The PCR cycle was repeated 30 times. The products were examined by Southern blot hybridization with an oligonucleotide probe specific for IE72 and UL112-113 mRNAs. As an internal control, expression of β-actin mRNA was also examined by RT-PCR. An ethidium bromide staining image of the agarose gel is shown for β-actin. (C) HEL cells were cultured with cycloheximide (200 μg/ml) for 2 h before infection and maintained in the cycloheximide-containing medium after infection. Viral infection and analysis of total RNA were performed as described for panel A. The nucleotide sequences of these RT-PCR primers and probes are shown in Table 1.
IE19 cooperates with IE72 in the activation of the HsOrc1 promoter.
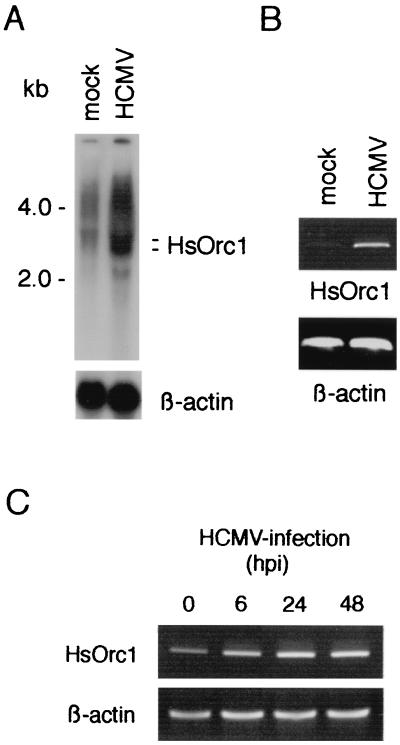
Because IE19 was related to IE72 and was localized in nuclei, we examined the effects of IE19 expression on activation of the E2F-regulated gene. For this analysis, we used the promoter of the HsOrc1 gene because it was a typical E2F-regulating promoter (45). The HsOrc1 gene encodes a key subunit of the origin recognition complex formed at the replication initiation sites in G1 phase (4, 35). Expression of the HsOrc1 gene is suppressed in serum-starved G0 cells and activated in the G1 and S phase in growing cells (45). The HsOrc1 gene was mostly suppressed in the mock-infected cells because these cells were cultured in the low-serum medium for viral infection (Fig. 6A). However, HsOrc1 mRNA greatly accumulated in HCMV-infected cells under the same culture conditions at 72 hpi, suggesting that viral protein(s) enhances the HsOrc1 expression. We confirmed that the level of β-actin mRNA was not affected by HCMV infection. The accumulation of HsOrc1 mRNA in HCMV-infected cells was also confirmed by RT-PCR (Fig. 6B). We next examined the time course of the HsOrc1 gene activation after HCMV infection and found that HsOrc1 mRNA had increased at 6 hpi and accumulated further by 48 hpi (Fig. 6C). Thus, HCMV infection activated the HsOrc1 gene during the immediate-early, early, and late period of infection.
FIG. 6.
Activation of the HsOrc1 gene by HCMV infection. (A) Expression of HsOrc1 mRNA in HCMV-infected cells was examined by Northern blot hybridization. Poly(A)+ mRNA (18 μg) of HCMV-infected or mock-infected HEL cells (48 hpi) was applied to each lane. An HsOrc1 cDNA probe was used for the hybridization. (B) Expression of HsOrc1 mRNA was examined by RT-PCR. The same preparation of poly(A)+ mRNA (0.2 μg) used in Northern analysis (A) was examined by RT-PCR with HsOrc1-specific primers. As a control, expression of β-actin mRNA was also examined. The PCR cycle was repeated 20 times. (C) Kinetics of the HsOrc1 gene activation was examined by RT-PCR. Total RNA of HCMV-infected HEL cells (0.2 μg) was used for RT-PCR. The PCR cycle was repeated 40 times. The RNA samples are described in the legend to Fig. 5B. The nucleotide sequences of these RT-PCR primers and probes are shown in Table 1.
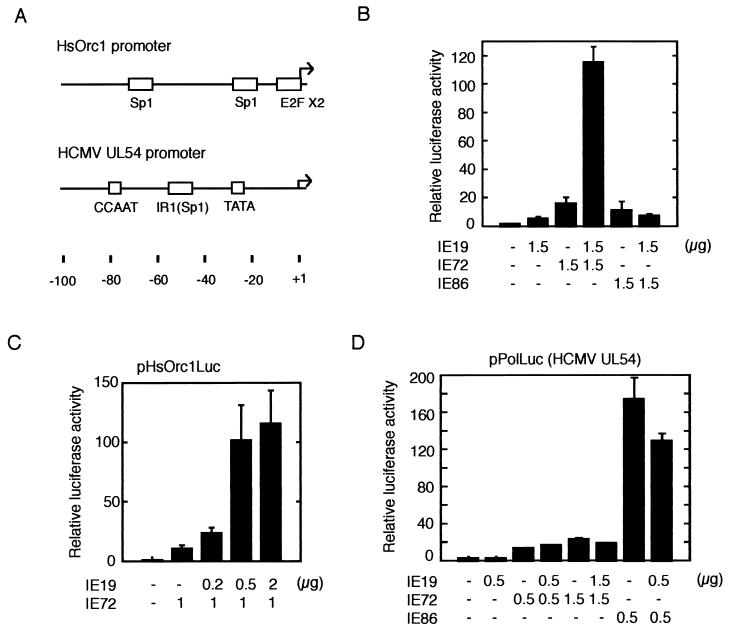
Given the finding that HCMV infection activated the HsOrc1 gene, we examined the effect of IE19 or IE72 expression on the HsOrc1 promoter by a luciferase reporter gene assay (45). The reporter gene (pHsOrc1Luc), which was driven by the HsOrc1 promoter and its upstream region, was transfected with the IE72 and/or IE19 expression plasmid into the glioblastoma cell line U373MG along with the internal control plasmid pRSVLacZ. At 2 days after transfection, the luciferase activity expressed in transfected cells was measured and normalized to the β-galactosidase activity in the same cells. The HsOrc1 promoter showed very low activity in growing U373MG cells, and expression of IE72 activated the promoter by 16-fold (Fig. 7B). IE86 activated the HsOrc1 promoter weakly (10-fold), and IE19 showed only a 5-fold activation. However, when IE19 was expressed together with IE72, the HsOrc1 promoter was activated 115-fold over the basal activity, indicating that IE19 works cooperatively with IE72. This enhancing effect of IE19 on the IE72-mediated promoter activation showed clear dose dependency for IE19 input (Fig. 7C). When 1 μg of IE72 expression plasmid was transfected, cotransfection of 0.2 μg of the IE19 plasmid resulted in a significant enhancement of the HsOrc1 promoter (2-fold), and 0.5 μg of the IE19 plasmid resulted in an almost maximum effect (10-fold). The effect of cotransfection of IE19 and IE86 was also examined, but IE19 did not show any enhancement (Fig. 7B).
FIG. 7.
Activation of HsOrc1 promoter by IE19 and IE72. (A) Promoter regions of the HsOrc1 and UL54 genes are shown. Arrows indicate the transcription initiation sites. For the HsOrc1 gene, only a major initiation site is shown here. Sp1, E2F, CCAAT, and TATA elements are indicated by open boxes. Nucleotide numbers relative to the initiation site are indicated. (B) Activation of the HsOrc1 promoter by IE19, IE72, and/or IE86 was examined using the HsOrc1 luciferase reporter plasmid (pHsOrc1Luc), which contains the HsOrc1 promoter region from −1053 to +182. The expression plasmid for IE72, IE86, and/or IE19 (1.5 μg each) was transfected into the human glioblastoma cell line U373MG with pHsOrc1Luc (1 μg) and an internal control plasmid, pRSVLacZ (1 μg). The total amount of transfected plasmids was adjusted with an additional control plasmid. Cells were harvested 2 days after transfection, and the luciferase and β-galactosidase activities were measured. The luciferase activity was normalized to the β-galactosidase activity in the same extract and shown as activity relative to that for the vector plasmid-transfected sample. Three experiments were performed to obtain the average activity with standard error. (C) Activation of the HsOrc1 promoter was dose dependent on the IE19 expression plasmid. Various amounts of pME-IE19 (0, 0.2, 0.5, and 2 μg) were cotransfected with a fixed amount of pME-IE72 (1 μg) in an HsOrc1 reporter assay. Results are shown as for panel B. (D) Activation of the UL54 promoter was examined using the UL54 luciferase reporter plasmid. Expression plasmid(s) (pME-IE72, pME-IE86, and/or pME-IE19) was transfected with pPolLuc (1 μg) in a similar reporter assay. Results are shown as for panel B.
To elucidate whether IE19 enhances IE72-dependent activation of another promoter, we chose the HCMV UL54 promoter. In contrast to the HsOrc1 promoter, which is TATA-less and regulated by E2F, the UL54 promoter is a typical promoter, containing TATA and CCAAT elements (Fig. 7A). It was previously shown that IE72 and IE86 activate the HCMV UL54 promoter through the upstream Sp1 site (30, 34). We examined the activity of the UL54 promoter by using the reporter plasmid pPolLuc (34). Transfection of IE72 expression plasmid (0.5 μg) activated the UL54 promoter about 12-fold; however, cotransfection of IE19 plasmid (0.5 μg) did not enhance the IE72-dependent promoter activation (Fig. 7D). The lack of response to IE19 was not due to saturated activation by IE72, because transfection of 1.5 μg of IE72 expression plasmid enhanced the promoter further (22-fold). IE19 also showed no effect on IE86-mediated activation of the UL54 promoter.
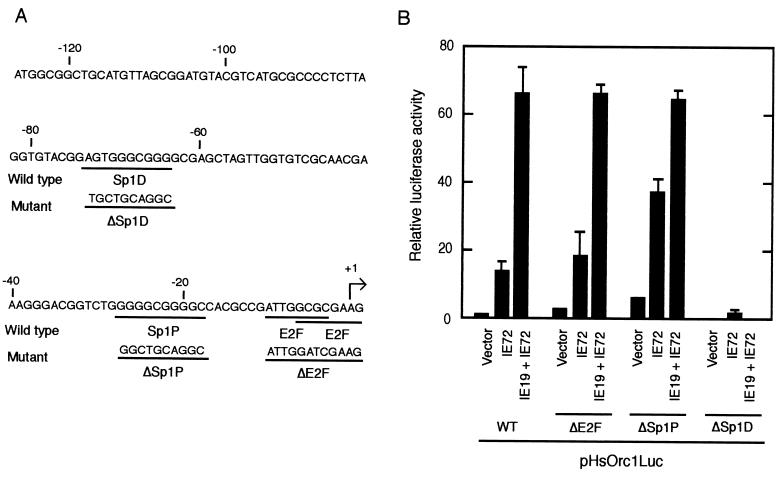
It has been reported elsewhere that the regulatory elements responsive to IE72 are the Sp1- and E2F-binding sites (37, 39). The HsOrc1 promoter region inserted in the pHsOrc1Luc reporter contains overlapping E2F sites at the transcription initiation site and two Sp1 sites located at nt −22 (Sp1P) and −66 (Sp1D) (Fig. 8A). To determine the importance of these cis-acting elements in the activation of the HsOrc1 promoter, several mutations were introduced into the promoter region of pHsOrc1Luc (Fig. 8A). It was shown elsewhere that the E2F sites at the transcription initiation site are responsible for the suppression of the HsOrc1 promoter (45). Consistent with this, our assay revealed that the basal activity of the HsOrc1 promoter was elevated about threefold by the mutations at these E2F sites (ΔE2F) (Fig. 8B). Expression of IE72 activated the E2F-mutated promoter to the same extent as it did the wild-type promoter. Similarly, the mutation of the Sp1P site (ΔSp1P) did not reduce the IE72-induced activation but rather up-regulated it. These results indicate that the E2F and Sp1P sites were not required for IE72-induced activation. In contrast, the mutation of the Sp1D site (ΔSp1D) totally abolished the promoter activity, indicating that the Sp1D site is an essential element of the HsOrc1 promoter.
FIG. 8.
Mutational analysis of HsOrc1 promoter. (A) Nucleotide sequence of HsOrc1 promoter region is shown. The major transcription initiation site is numbered as +1, and the upstream promoter region is numbered relative to the initiation site. The putative Sp1-binding sequences, Sp1P and Sp1D, and the E2F-binding sequences are underlined. Nucleotide sequences of the mutated promoters ΔSp1P, ΔSp1D, and ΔE2F are shown under the wild-type sequence. (B) Activity of the mutated HsOrc1 promoter was examined using the luciferase reporter plasmid. The expression plasmid of IE72 and/or IE19 (1.5 μg each) was transfected with pHsOrc1Luc or its mutated reporter plasmid (1 μg) in a reporter assay similar to that described in the legend to Fig. 7.
DISCUSSION
IE72, IE86, IE55, and IE18 have been identified as alternative splicing products of the HCMV MIE gene (Fig. 1A). In this study, a novel IE protein, IE19, was identified. IE19 is a polypeptide of 172 aa which appeared at 38 kDa on an SDS-polyacrylamide gel. IE19 has the amino-terminal region (aa 1 to 87) common to all MIE proteins and a carboxyl-terminal region (aa 88 to 174) identical to the corresponding region of IE72. Because IE55 and IE18 are splicing variants of exon 5 of the MIE gene, IE19 is the first splicing variant of exon 4 to be identified. IE19 was expressed at a low level in the immediate-early, the early, and the late period of HCMV infection in HEL cells. Our data suggested that IE19 is a transcriptional coactivator that cooperates with IE72.
IE72 activates several genes required for cellular DNA synthesis, including those of DNA polymerase and dihydrofolate reductase (18, 40, 64). In addition to genes of the replication machinery, expression of IE72 activated the promoter of the HsOrc1 gene, which encodes a replication origin-binding factor, HsOrc1. IE72 increased the basal activity of the HsOrc1 promoter by about 16-fold, and coexpression of IE19 and IE72 increased it by 115-fold. Because IE19 activated the promoter only fivefold, it has a transcriptional coactivator function cooperating with IE72. Our site-directed mutation analysis showed that mutation of the Sp1D site located at nt −66 abolished the promoter activity. This indicates that the Sp1D sequence is the essential promoter element of the HsOrc1 gene. In contrast, mutations of the proximal Sp1 site (Sp1P) and E2F sites or deletion of the region upstream of the SpiD site did not affect the level of activation induced by IE19 and IE72. Because IE72 is believed to regulate target gene promoters through Sp1 and/or E2F sites, these results suggest that IE72 interacts with the basic transcriptional machinery formed at the Sp1D site and enhances subsequent transcription. It was recently shown elsewhere that IE72 and IE86 interact with TAFIIs on a simple TATA promoter (37). TAFIIs and TBP compose the basic transcription factor TFIID, which binds to the TATA element. TAFIIs also form a TBP-free TAFII-containing complex and support transcription of TATA-less promoters (68). Therefore, IE72 may activate transcription by interacting with TAFIIs on the TATA-less promoter of the HsOrc1 gene. IE72 associates with a TAF, hTAFII130, primarily through a domain from aa 215 to 378. A region from aa 379 to 491 also mediates modest interaction between IE72 and hTAFII130. Among these regions, IE19 covers the carboxyl-terminal part of the latter domain from aa 405 to 491. This suggests that IE19 may also associate with hTAFII130. As a transcriptional coactivator, IE19 showed promoter selectivity. IE19 did not enhance IE72-mediated activation of the UL54 promoter, which is a typical TATA promoter with the upstream IE72-responsive Sp1 site (Fig. 7). IE72 bridges the Sp1 protein at the site and the basal transcription factor TFIID on the TATA element in the UL54 promoter (34, 38). In contrast, the putative IE72-responsive Sp1 site (Sp1D) in the HsOrc1 gene is the essential promoter element, where the basic transcriptional complex would be assembled. This functional difference in Sp1 sites may be related to the promoter selectivity of IE19. It is reported elsewhere that IE72 is a protein kinase that phosphorylates E2F and the Rb-related proteins p130 and p107 (46). The kinase domain of IE72, which is encoded in the 5′ region of exon 4, is spliced out in the IE19 mRNA.
Both the dhfr and the HsOrc1 genes have E2F-binding sequences at the transcription initiation site and are repressed by E2F. However, IE72 activates the dhfr promoter, but not the HsOrc1 promoter, through the E2F site (Fig. 8) (40, 64). This suggests that these E2F sites have a functional difference. The HsOrc1 promoter has an Sp1 site (Sp1P) adjacent to its E2F site, while the dhfr gene has four Sp1 sites in the promoter region but none near its E2F site (8). Therefore, the function of the E2F site of the HsOrc1 promoter may be affected by the adjacent Sp1 site. A recent study showed that the regulation of cdc2 or B -myb genes in the cell cycle differs from that of dhfr (22). Like the HsOrc1 promoter, the E2F-responsive promoter of cdc2 or B -myb genes also has an Sp1 site close to its E2F site (31, 63). Our reporter assay showed that mutation of the E2F site enhanced moderately the basal (2.6-fold) and IE72-activated (1.3-fold) promoter activity of HsOrc1 (Fig. 8B) and confirmed that the HsOrc1 promoter is under negative regulation through the E2F site in growing cells (45). In addition, the assay revealed that the mutation of the Sp1P site also resulted in enhancement of the basal (6-fold) and IE72-activated (2.6-fold) promoter activity. This suggests that the Sp1P site functions in the negative regulation of the HsOrc1 promoter. We showed that HsOrc1 is actually activated by HCMV infection but did not demonstrate that this activation fully depends upon IE72 and/or IE19. Therefore, other viral transcriptional regulators may be involved in the activation of HsOrc1.
HsOrc1 is a component of the prereplicative complex that is assembled with the replication licensing factor Mcm in G1 at the replication initiation sites of the cellular genome (4, 35, 61). Recently, it was suggested that cellular replicative complexes constitute a huge replication factory in nuclei which is similar to the replication compartment (11, 27). Therefore, it would be interesting to elucidate whether HsOrc also plays a role in the formation of the replication compartment or more directly in the replication of the HCMV genome (1, 49, 51, 69).
Acknowledgments
We thank K. Ohtani for pHsOrc1Luc and pHsOrc1Luc(−E2F) plasmids and the HsOrc1 cDNA, B. Roizman for HEL cells, and M. Yanokura of the Laboratory of Biological Information for technical assistance in DNA sequencing.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Science, Sports and Technology of Japan.
REFERENCES
- 1.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford, C. A., and W. J. Britt. 1990. Cytomegalovirus. Raven Press, Ltd., New York, N.Y.
- 3.Baracchini, E., E. Glezzer, K. Fish, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1992. An isoform variant of the cytomegalovirus immediate-early autorepressor functions as a transcriptional activator. Virology 188:518-529. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiple complex. Nature (London) 357:128-134. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi, and B. Stillman. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83:563-568. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. [DOI] [PubMed] [Google Scholar]
- 7.Caswell, R., C. Hagemeier, C.-J. Chou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y.-C., S. Illenye, and N. H. Heintz. 2001. Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol. Cell. Biol. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou, C. J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 12.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMarchi, J. M., C. A. Schmidt, and A. S. Kaplan. 1980. Patterns of transcription of human cytomegalovirus in permissively infected cells. J. Virol. 35:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depto, A. S., and R. M. Stenberg. 1989. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J. Virol. 63:1232-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari, B. A., E. Poma, T. F. Kowalik, S.-M. Huong, and E.-S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator protein in eukaryotes. Science 270:1667-1671. [DOI] [PubMed] [Google Scholar]
- 17.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helin, K., and E. Harlow. 1993. The retinoblastoma protein as a transcriptional repressor. Trends Cell Biol. 3:43-46. [DOI] [PubMed] [Google Scholar]
- 20.Hermiston, T. W., C. L. Malone, P. R. Witte, and M. F. Stinski. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J. Virol. 61:3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, M. 1991. Cytomegalovirus: biology and infection. Plenum Publishing Corp., New York, N.Y.
- 22.Hurford, R., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 23.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal., and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, B. K., D. A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerry, J. A., M. A. Priddy, and R. M. Stenberg. 1994. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J. Virol. 68:4167-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerry, J. A., A. Sehgal, S. W. Barlow, V. J. Cavanaugh, K. Fish, J. A. Nelson, and R. M. Stenberg. 1995. Isolation and characterization of a low-abundance splice variant from the human cytomegalovirus major immediate-early gene region. J. Virol. 69:3868-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerry, J. A., M. A. Priddy, T. Y. Jervey, C. P. Kohler, T. L. Staley, C. D. Vanson, T. R. Jones, A. C. Iskenderian, D. G. Anders, and R. M. Stenberg. 1996. Multiple regulatory elements influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J. Virol. 70:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, E. W.-F., and R. J. Watson. 1993. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 12:2705-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Thangue, N. B. 1994. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem. Sci. 19:108-114. [DOI] [PubMed] [Google Scholar]
- 34.Li, J., T. Yamamoto, K. Ohtsubo, M. Shirakata, and K. Hirai. 1999. Major product pp43 of human cytomegalovirus UL112-113 gene is a transcriptional coactivator with two functionally distinct domains. Virology 260:89-97. [DOI] [PubMed] [Google Scholar]
- 35.Liang, C., M. Weinreich, and B. Stillman. 1995. ORC and Cdc6p interact and determine the frequency of initiation of the DNA replication in the genome. Cell 81:667-676. [DOI] [PubMed] [Google Scholar]
- 36.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luu, P., and O. Flores. 1997. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J. Virol. 71:6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonough, S., S. Staprans, and D. Spector. 1985. Analysis of the major transcripts encoded by the long repeat of human cytomegalovirus strain AD169. J. Virol. 53:711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mocarski, E. 1996. Cytomegalovirus and their replication, p. 2447-2479. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 43.Mocarski, E. S., G. Kemble, J. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1 491 aa is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevins, J. R. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258:424-429. [DOI] [PubMed] [Google Scholar]
- 45.Ohtani, K., J. DeGregori, G. Leone, D. R. Herendeen, T. H. Kelly, and J. R. Nevins. 1996. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 16:6977-6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pajovic, S., E. L. Wong, A. R. Black, and J. C. Azizkhan. 1997. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol. Cell. Biol. 17:6459-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pazzagli, M., J. H. Devine, D. O. Peterson, and T. O. Baldwin. 1992. Use of bacterial and firefly luciferase as reporter genes in DEAE-dextran-mediated transfection of mammalian cells. Anal. Biochem. 204:315-323. [DOI] [PubMed] [Google Scholar]
- 49.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 50.Rodems, S. M., C. L. Clark, and D. H. Spector. 1998. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection. J. Virol. 72:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenberg, R. M., D. R. Thomsen, and M. F. Stinski. 1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 49:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stenberg, R. M., P. R. Witte, and M. F. Stinski. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 56:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stenberg, R. M., and M. F. Stinski. 1985. Autoregulation of the human cytomegalovirus major immediate-early gene. J. Virol. 56:676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stenberg. R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stinski, M. F., D. R. Thomsen, R. M. Stenberg, and L. C. Goldstein. 1983. Organization and expression of the immediate early genes of human cytomegalovirus. J. Virol. 46:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 62.Thomsen, D. R., R. M. Stenberg, W. F. Goins, and M. F. Stinski. 1984. Promoter regulatory region of the major immediate early gene of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 81:659-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tommasi, S., and G. P. Pfeifer. 1995. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 15:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wade, M., T. F. Kowalik, M. Mudryj, E.-S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wara-Aswapati, N., Z. Yang, W. R. Waterman, Y. Koyama, S. Tetradis, B. K. Choy, A. C. Webb, and P. E. Auron. 1999. Cytomegalovirus IE2 protein stimulates interleukin 1β gene transcription via tethering to Spi-1/PU.1. Mol. Cell. Biol. 19:6803-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wathen, M. W., D. R. Thomsen, and M. F. Stinski. 1981. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J. Virol. 38:446-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wathen, M. W., and M. F. Stinski. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J. Virol. 41:462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature (London) 393:187-191. [DOI] [PubMed] [Google Scholar]
- 69.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature (London) 349:429-431. [DOI] [PubMed] [Google Scholar]
- 70.Wu, J., J. O'Neill, and M. S. Barbosa. 1998. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J. Virol. 72:236-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoo, Y. D., C.-J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S.-J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J. Virol. 70:7062-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E.-S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoter. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]