Abstract
Previously we have shown that the African swine fever virus (ASFV) NL gene deletion mutant E70ΔNL is attenuated in pigs. Our recent observations that NL gene deletion mutants of two additional pathogenic ASFV isolates, Malawi Lil-20/1 and Pr4, remained highly virulent in swine (100% mortality) suggested that these isolates encoded an additional virulence determinant(s) that was absent from E70. To map this putative virulence determinant, in vivo marker rescue experiments were performed by inoculating swine with infection-transfection lysates containing E70 NL deletion mutant virus (E70ΔNL) and cosmid DNA clones from the Malawi NL gene deletion mutant (MalΔNL). A cosmid clone representing the left-hand 38-kb region (map units 0.05 to 0.26) of the MalΔNL genome was capable of restoring full virulence to E70ΔNL. Southern blot analysis of recovered virulent viruses confirmed that they were recombinant E70ΔNL genomes containing a 23- to 28-kb DNA fragment of the Malawi genome. These recombinants exhibited an unaltered MalΔNL disease and virulence phenotype when inoculated into swine. Additional in vivo marker rescue experiments identified a 20-kb fragment, encoding members of multigene families (MGF) 360 and 530, as being capable of fully restoring virulence to E70ΔNL. Comparative nucleotide sequence analysis of the left variable region of the E70ΔNL and Malawi Lil-20/1 genomes identified an 8-kb deletion in the E70ΔNL isolate which resulted in the deletion and/or truncation of three MGF 360 genes and four MGF 530 genes. A recombinant MalΔNL deletion mutant lacking three members of each MGF gene family was constructed and evaluated for virulence in swine. The mutant virus replicated normally in macrophage cell culture but was avirulent in swine. Together, these results indicate that a region within the left variable region of the ASFV genome containing the MGF 360 and 530 genes represents a previously unrecognized virulence determinant for domestic swine.
African swine fever virus (ASFV) is the sole member of the newly named Asfarviridae (19), whose genomic organization and cytoplasmic replication strategy show similarities with the Poxviridae (14, 28, 45, 54). ASFV is the only known DNA arbovirus (8, 14, 20). In nature, this virus cycles between two highly adapted hosts, Ornithodoros ticks and wild pig populations (warthogs and bushpigs) in sub-Saharan Africa (47, 48, 59, 63). In the warthog host, infection is subclinical, characterized by low-titer viremias (49, 60). Persistent infection of ticks, domestic and wild pigs, as well as warthogs occurs following infection (10, 16, 17, 32, 33).
In domestic pigs, ASF occurs in several disease forms, ranging from highly lethal to subclinical infections, depending on contributing viral and host factors (13, 41). ASFV infects cells of the mononuclear-phagocytic system, including highly differentiated fixed-tissue macrophages and reticular cells; affected tissues show extensive damage after infection with highly virulent viral strains (13, 34, 35, 39, 42). ASFV strains of lesser virulence also infect these cell types, but tissue involvement and resulting tissue damage are much less severe (31, 40, 41). The abilities of ASFV to replicate and induce marked cytopathology in these cell types in vivo appear to be critical factors in ASFV virulence. The viral factors responsible for the different outcomes of infection with strains of high virulence and of lesser virulence are largely unknown.
Variations in genome size and restriction fragment patterns are observed among different ASFV isolates. Like poxviruses, the diversity within the ASFV genome is localized primarily in the terminal genomic regions (6, 7, 18, 56, 62). With poxviruses, genes contained within the terminal variable regions are often nonessential in vitro, instead performing functions related to viral host range (26, 37, 53). ASFV variable regions comprise the left 35-kb and the right 15-kb ends of the genome and contain at least five multigene families (MGFs): MGF 100, MGF 110, MGF 300, MGF 360, and MGF 530 (3, 4, 15, 27, 61, 64). Variations in these regions, including gene deletion events, are observed during ASFV adaptation to monkey cell lines (6, 58). Recently, MGF 360 and MGF 530 genes in the left variable region of the ASFV genome have been shown to function in macrophage host range (65). ASFV variable-region genes, including the multigene families, are thought to be associated with important host range functions in the pig host.
Previously, we described an ASFV gene, NL, with similarity to the neurovirulence gene ICP34.5 of herpes simplex virus (11, 12, 38), a myeloid differentiation primary response gene, MyD116 (36), and a growth arrest DNA damage gene, GADD34 (24). The NL gene was highly conserved among pathogenic ASFV isolates, encoding either a long (23-NL, 184 amino acids) or short (NL-S, 70 to 72 amino acids) form (57, 66). Deletion of NL from the virulent European isolate E70 resulted in marked attenuation of the virus in the swine host. Pig infections with the null mutant were characterized by a lack of clinical disease apart from a transient fever response, reduced viremia titers (1,000-fold), and no mortality (66). However, gene deletion mutants of two additional pathogenic African ASFVs, Malawi Lil-20/1 (Mal) and Pretoriuskop/96/4 (Pr4), remained highly virulent, with mortality rates approaching 100% in domestic pigs. Apart from a modest reduction in mean viremia titers for MalΔNL-infected animals, MalΔNL exhibited virulence characteristics similar to its revertant, Mal-NLR (2, 30, 33).
These data indicated that NL gene function is not an absolute requirement for ASFV virulence and that NL's contribution to viral virulence is strain (isolate) dependent. The data suggested that both the MalΔNL and Pr4ΔNL isolates encode a virulence determinant capable of directly complementing the NL defect, which is either absent or inactive in the E70 isolate.
Here, using in vivo marker rescue, we have identified genes in the left variable region of the ASFV genome that are essential for swine virulence in the absence of NL gene function.
MATERIALS AND METHODS
Cell culture and viruses.
Primary porcine macrophage cell cultures were prepared from defibrinated swine blood as previously described (25). Briefly, heparin-treated swine blood was incubated at 37°C for 1 h to allow sedimentation of the erythrocyte fraction. Mononuclear leukocytes were separated by flotation over a Ficoll-Paque (Pharmacia, Piscataway, N.J.) density gradient (1.079 specific gravity). The monocyte/macrophage cell fraction was cultured in plastic Primaria (Falcon Becton Dickinson Labware, Franklin Lakes, N.J.) tissue culture flasks containing RPMI 1640 medium with 30% L929 supernatant and 20% fetal bovine serum (FBS) for 48 h (37°C in 5% CO2). Adherent cells were detached from the plastic with 10 mM EDTA in phosphate-buffered saline and then reseeded into Primaria T25 6-well or 96-well dishes at a density of 5 × 106 cells per ml for use in assays 24 h later.
Previously described ASFV NL gene deletion mutants MalΔNL and E70ΔNL were used in this study (2, 66).
In vivo marker rescue of pig virulence.
For in vivo marker rescue experiments, porcine macrophages (107 cells/25-cm2 flask) were infected with E70ΔNL at a multiplicity of infection (MOI) of 10 and transfected with 5 μg of cosmid DNA or plasmid DNA in 20 μl of Lipofectin (Gibco-BRL, Gaithersburg, Md.) in 2 ml of serum-free medium (Opti-MEM; Gibco-BRL). After 5 h, RPMI 1640 medium containing 20% FBS (2 ml) was added to the transfected cells. Viral lysates were harvested when complete cytopathic effect was evident. Virus titers in the infection-transfection lysates were determined.
Yorkshire pigs (30 to 35 kg) were inoculated intramuscularly with 107 50% tissue culture infective doses (TCID50) of E70ΔNL infection-transfection lysates. Clinical disease, i.e., fever (defined as a rectal temperature greater than or equal to 40°C), anorexia, lethargy, shivering, cyanosis, and recumbency, was monitored daily. Blood samples were collected every other day for up to 30 days postinfection. Virus isolation and titration were done in primary swine macrophage cultures. Virus titers were calculated using the method of Spearman-Karber and expressed as TCID50 (22). To select rescued virulent viruses, viremic blood (2 ml) was taken from pigs infected with infection-transfection lysates at 7 days postinfection and inoculated into a second group of naïve pigs. Virulent recombinant viruses were isolated from the second pig group at 7 days postinfection.
Viral DNA isolation and cosmid and plasmid cloning.
MalΔNL-infected macrophage cell cultures were harvested, cytoplasmic virions were purified, and genomic DNA was prepared as previously described (62). An overlapping cosmid library in pHC79 representing 35 to 45 kb of the MalΔNL genome was constructed as previously described (66). Cosmid clones were amplified and screened by dot blot hybridization using probes representing various ASFV genomic regions. DNAs from selected cosmid clones were purified, and the nucleotide sequence of the vector insert borders was determined to define the exact genomic regions represented by each cosmid. PCR-amplified products were cloned into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.) and introduced into Escherichia coli INVαF′ according to the manufacturer's protocols. Southern blot, radiolabeling, and hybridization analyses were performed using standard methods (52).
Nucleotide sequencing and analysis.
Viral DNA was extracted from the cytoplasm of infected cells as described above. Random DNA fragments were obtained by incomplete enzymatic digestion with RsaI and AluI endonucleases. DNA fragments of 1.5 to 2.5 kb were isolated after separation on agarose gels, cloned into the dephosphorylated SmaI site of pUC19 plasmids, and grown in E. coli DH10B cells (Gibco-BRL). Plasmids containing sequences for the left variable region of the E70ΔNL genome were identified by in situ colony filter hybridization using 32P-labeled probes derived from the MalΔNL cosmid C4 (Fig. 1) and the E70 cosmid G7 (65). Plasmids were purified by the boiling method (52). DNA templates were sequenced from both ends with M13 forward and reverse primers using dideoxy chain terminator sequencing chemistries and the Applied Biosystems Prism 377 automated DNA sequencer (PE Biosystems, Foster City, Calif.). ABI sequencing analysis software (version 3.3) was used for lane tracking and trace extraction.
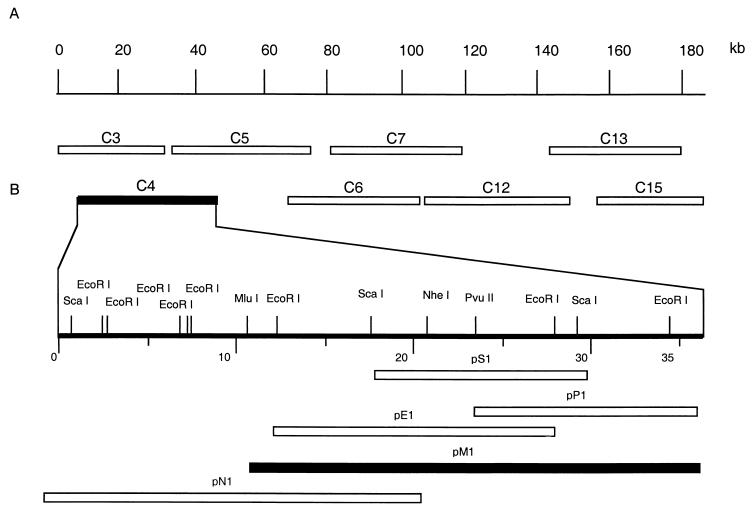
FIG. 1.
(A) Diagram showing the locations of the eight MalΔNL cosmid clones used for in vivo marker experiments. (B) Locations of plasmid subclones of cosmid C4 (pS1, pP1, pE1, pM1, and pN1) are indicated. Solid boxes denote clones capable of restoring virulence to E70ΔNL.
Bases were called from chromatogram traces with Phred (21), which also produced a quality file containing a predicted probability of error at each base position. DNA sequences were assembled with Phrap (23) using the quality files and default settings to produce a consensus sequence which was manually edited with Consed (29). Gap closure was achieved by primer walking of genomic DNA and sequencing of PCR products. The final consensus sequence represented on average eightfold redundancy at each base position. Open reading frames (ORFs) greater than or equal to 60 codons with a methionine start codon were identified by using Staden (55) and MacVector programs (Oxford Molecular Group, Madison, Wis.). Alignment of the consensus sequence to the ASFV sequences in the GenBank database was achieved using Sim2 alignment programs (5). Homology searches were conducted using Blast (5) and FastA (46) with the GenBank database.
Construction of MalΔSVD.
The MalΔSVD recombinant virus containing deletions or truncations of three MGF 360 and three MGF 530 genes (see Fig. 4) was constructed by homologous recombination between the parental virus, MalΔNL, and an engineered recombination transfer vector in primary swine macrophages as previously described (43, 66). DNA fragments mapping from bp 23600 to 24626 (left flank) and from bp 32290 to 33428 (right flank) were amplified by PCR using MalΔNL genomic DNA as a template.
FIG. 4.
Characterization of MalΔSVD recombinants. (A) Diagram of the swine virulence determinant gene region in the parental MalΔNL isolate and the swine virulence determinant deletion mutant virus MalΔSVD. E denotes EcoRI restriction endonuclease site. (B) Southern blot analysis of parental virus (lane 1) and four independently isolated MalΔSVD viruses (lanes 2, 3, 4, and 5). Purified viral DNA was digested with EcoRI, electrophoresed, blotted, and hybridized with 32P-labeled L3IL gene probe (panel I) or 32P-labeled probe generated from the recombination transfer vector (panel II). Molecular size markers are in kilobase pairs at the left of panel ll. Panel III, PCR analysis of MalΔSVD (lanes 2 and 4) and parental MalΔNL (lanes 3 and 5) for L3IL gene sequences (lanes 4 and 5). Positive-control PCR for the MalΔSVD viral DNAs using primers for a region of the genome immediately flanking the swine virulence determinant region are included (lanes 2 and 3). Molecular size markers are shown in lane 1.
Primer sets, each of which introduced a BamHI site adjacent to the region to be deleted and a BglII site at the other end for the left and right flank, were as follows: left flank forward primer, 5′-AGATCTTCGTGAAGGTCGAACACGTTG-3′; reverse primer, 5′-GGATCCAGCTGCTGCTATTCTTAGGGCTTC-3′; right flank forward primer, 5′-GGATCCACGAGACGTTTCAATGAAGGGG-3′; and right flank reverse primer, 5′-AGATCTAATGGCTGCTTTTTCGAGGTG-3′ (introduced restriction sites are in boldface).
Amplified fragments were cloned and verified by nucleotide sequence analysis. The flanking fragments were then digested with BamHI and BglII and sequentially cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.) to give pTM6. The reporter cassette, p72GAL (2), was inserted into BamHI-digested pTM6 to yield the p72GALΔM6 transfer vector, which removed or truncated three members from each of the multigene families MGF 360 and MGF 530. Porcine macrophage cell cultures were infected with MalΔNL and transfected with p72GALΔM6 as described previously (43). Four recombinant viruses representing independent primary plaques were purified to homogeneity and verified as products of a double-crossover recombination event by Southern hybridization and PCR analysis (43).
Nucleotide sequence accession numbers.
The E70ΔNL sequences were assigned GenBank accession no. AF327841, and the MalΔNL sequences were assigned GenBank accession no. AF327842.
RESULTS
Left variable region of the MalΔNL genome encodes a swine virulence determinant.
In vivo marker rescue experiments were used to identify ASFV (MalΔNL) sequences capable of rescuing E70ΔNL virulence in domestic pigs. Porcine macrophage cell cultures were infected with avirulent E70ΔNL virus, followed by transfection with eight overlapping clones (Fig. 1A), representing the complete virulent MalΔNL genome. Recombinant E70ΔNL viruses with enhanced virulence were selected and purified by serial passage in domestic pigs.
In initial animal experiments, infection-transfection lysates derived from each of the eight selected cosmids were screened for their ability to rescue virulence. Only pigs infected with an infection-transfection lysate containing cosmid C4 presented with more severe clinical disease (febrile response for 7 days and increased blood viremia titers of 5.5 log10 TCID50/ml) than did the control group. In swine virulence assays, pigs were infected with the purified cosmid C4-derived recombinant virus, E70C4 (isolated after a second in vivo passage). Infected pigs presented with fever at 3 to 4 days postinfection, were viremic (7.8 log10 TCID50/ml of blood), and died between 5 and 7 days postinfection (Table 1). These results indicated that cosmid C4 was capable of completely restoring virulence to E70ΔNL.
TABLE 1.
Survival and viremia following infection of swine with virulence-rescued E70ΔNL recombinant virusesa
| Virus | No. of survivors/total | Mean time to death (days) ± SEM | Viremia
|
||
|---|---|---|---|---|---|
| Mean time to onset (days) ± SEM | Mean titer (log10 TCID50/ml) ± SEM | Max titer (log10 TCID50/ml) ± SEM | |||
| E70ΔNL | 4/4 | 9 ± 3.0 | 3.4 ± 1.0 | 3.8 ± 0.8 | |
| E70C4 | 0/2 | 5 ± 1.5 | 3 ± 1.0 | 7.8 ± 0.2 | 8.0 ± 0.2 |
| E70 M1 | 0/6 | 6.3 ± 0.2 | 3 ± 0.0 | 7.6 ± 0.2 | 8.1 ± 0.2 |
Rescued E70ΔNL recombinants selected in the in vivo marker rescue experiments are designated E70C4 for the cosmid C4- derived isolate and E70 M1 for the virus isolate selected using the plasmid subclone pM1.
The genetic determinant(s) in C4 responsible for the restoration of virulence was fine mapped using subclones derived from C4 (Fig. 1B) in further in vivo marker rescue experiments. Animals infected with infection-transfection lysates containing the pM1 subclone exhibited severe clinical disease, whereas all other C4 subclones had no effect on virulence. In swine virulence assays, pigs infected with the purified pM1-rescued viral isolate E70 M1 presented with fever at 3 to 4 days postinfection, were viremic (7.6 log10 TCID50/ml of blood), and died 6 to 7 days postinfection (Table 1). The pM1 subclone, which contains a 28-kb fragment from the left variable region of MalΔNL, was both necessary and sufficient for complete virulence rescue.
Results from pig virulence assays show that virulence was fully restored in both the E70C4 and E70 M1 recombinants. Virulence assay results obtained here are consistent with previously published swine virulence experiments using E70NLR (revertant of E70ΔNL) and MalΔNL virus (2, 66).
Genetic characterization of virulent E70C4 and E70 M1 recombinants.
The genomic structures of the virulent E70C4 and E70 M1 viruses obtained from in vivo marker experiments were analyzed to define MalΔNL genomic sequences present in the virulent recombinants. Genomic DNA was extracted and examined by restriction endonuclease and Southern blot hybridization analysis (Fig. 2C). The restriction patterns for the E70C4 and E70 M1 rescued viruses were similar to that of the parental E70ΔNL isolate with the exception of bands corresponding to MalΔNL fragments and new fragments generated by recombination. For example in Fig. 2C, four new EcoRI fragments (16, 11, 5, and 4.6 kb) present in the E70C4 recombinant virus (Fig. 2CI) replaced the 24-kb, 5.1-kb, and 4.8-kb fragments in the parental E70ΔNL genome. The 16-kb and 5-kb fragments correspond to the 16-kb and 5-kb fragments in MalΔNL, while the 11-kb and 4.6-kb fragments are novel and presumably contain recombination junctions.
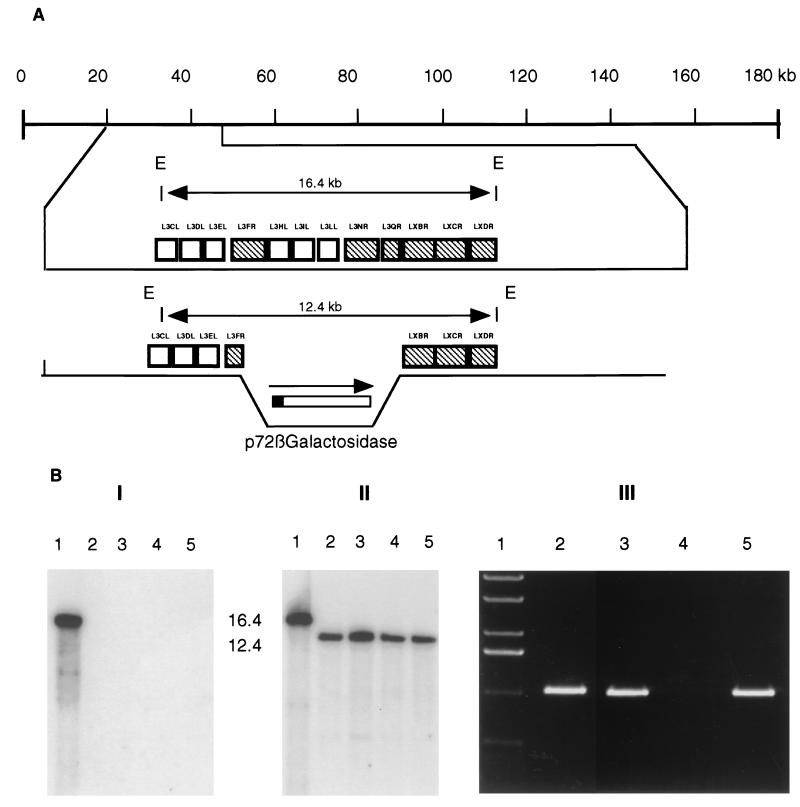
FIG. 2.
(A) (l) Diagram showing the genomic location of cosmid C4 on the MalΔNL genome. (II and III) EcoRI restriction maps of cosmid C4 (II) and the corresponding region on virulence-rescued E70ΔNL (E70C4) following in vivo marker rescue with cosmid C4 (III). (IV) MalΔNL DNA sequence which is present in the rescued virulent virus is indicated (dashed lines). (B) Schematic representation of the MalΔNL fragment and MGF ORFs (MGF 360 genes open boxes and MGF 530 genes are hatched boxes) present in E70 M1 rescued virus. (C) Purified DNAs from the parental virus, E70ΔNL (lane 1); rescued viruses (E70C4 in panels l and lland lanes 2 and 3; E70Ml in panel lll and lanes 2 and 3); and MalΔNL (lane 4) were digested with EcoRI (panels I and II) or BglII (panel III), electrophoresed, blotted and hybridized with 32P-labeled C4 DNA probe (panels Il and IIl). DNA fragments derived from MalΔNL are indicated.
Southern hybridization analysis of genomic DNAs digested with several restriction endonucleases and probed with 32P-labeled radioactive DNA probes generated from DNA fragments located in the left variable region of the MalΔNL virus indicated that E70C4 and E70 M1 contain approximately 22 to 28 kb and 20 to 22 kb of MalΔNL genomic sequence, respectively (Fig. 2Aiii and 2B). The hybridization patterns obtained for E70C4 and E70 M1 were indicative of a homogenous viral population in both instances. Recombination junctions in E70 M1 DNA were determined by nucleotide sequence analysis of PCR products spanning the predicted recombination site. E70 M1 contained a genomic fragment of 20 kb mapping from 0.11 to 0.22 map units on the MalΔNL genome (Fig. 2B).
Swine virulence determinant genomic region is deleted in E70ΔNL.
As shown above, replacement of a segment of the E70ΔNL left variable region with a 20-kb fragment from the Malawi Lil-20/1 left variable region was sufficient to rescue E70ΔNL virulence, indicating that differences in this region were responsible for the virulence phenotype.
The left variable regions of the E70ΔNL and Malawi Lil-20/1 genomes were sequenced in their entirety and compared. An 8-kb deletion was observed in the left variable region of E70ΔNL, resulting in the deletion or truncation of three MGF 360 genes (L3HL, L3IL, and L3LL) and four MGF 530 genes (L3FR, L3NR, L3QR, and LXBR) (Fig. 3). These seven MGF genes are present in all seven of the pathogenic ASFVs sequenced to date (Kutish et al., unpublished data).
FIG. 3.
Comparison of the left variable regions of the MalΔNL and E70ΔNL genomes. (A) ORFs corresponding to MGF 360 family members (hatched boxes) and MGF 530 genes (hatched boxes). The relative position and extent of the left variable region deletion of E70ΔNL is shown. (B) Comparative analysis of ORFs encoded in the left variable region of MalΔNL and E70ΔNL. Amino acid (AA) lengths and amino acid identities (ID) are indicated.
Analysis of the MGF 360 and MGF 530 family members present in both E70ΔNL and MalΔNL shows a high degree of conservation (80 to 90% amino acid identity) (Fig. 3B). E70ΔNL does however encode the Ba71v MGF 530 gene A528R, which is absent in MalΔNL. Analysis of the left variable region from seven additional pathogenic ASFVs has identified isolates which contain either the A528R homolog or LXDR homolog and others which contain both genes, illustrating the variability in the number of MGF 530 genes present in given ASFV isolates (Kutish et al., unpublished data).
Deletion of swine virulence determinant attenuates MalΔNL in swine.
Insertion of a 20-kb fragment from the left variable region of MalΔNL into E70ΔNL restored viral virulence. The nucleotide sequence of the E70ΔNL left variable region identified an 8-kb deletion within this region. This specific deletion in E70ΔNL suggests a role for genes in this location in ASFV virulence.
To confirm that genes encoded by the 8-kb region were responsible for attenuation of E70ΔNL, a recombinant MalΔNL virus with a 7.6-kb deletion in the left variable region, MalΔSVD, was constructed by homologous recombination between the parental virus, MalΔNL, and a recombination transfer vector, p72GALΔMGF. Three MGF 360 genes (L3HL, L3IL, and L3LL) and three MGF 530 genes (L3FR, L3NR, and L3QR) (Fig. 3) (64) were either deleted or truncated in p72GALΔM6 (see Materials and Methods). The introduced deletion replaced 7.6 kb from the left variable region of the MalΔNL genome with a 3.6-kb p72βgal reporter cassette (3) (Fig. 4A). The resultant virus was designed to contain the same number of functional MGF 360 and MGF 530 genes in the left variable region as E70ΔNL.
Four recombinant viruses, representing individual primary plaques, were purified to homogeneity. Genomic DNAs from MalΔNL and the recombinant viruses were analyzed by Southern hybridization to confirm genomic structure (Fig. 4B). Viral DNAs were digested with EcoRI, gel electrophoresed, Southern blotted, and hybridized with 32P-labeled DNA ORF L3IL or the transfer vector, p72GALΔM6 (Fig. 4B, panels I and II, respectively). As expected, the L3IL probe only hybridized to the 16.4-kb EcoRI DNA band in the parental lane. The p72GALΔM6 probe hybridized to the 16.4-kb EcoRI DNA in the parental virus and to a 12.4-kb fragment in the recombinant viruses. The 12.4-kb fragment resulted from replacement of the 7.6-kb MalΔNL fragment by the 3.6-kb p72βgal reporter cassette. The purity of the virus stocks was examined using primers specific for a deleted ORF, L3IL (Fig. 4B, panel III). A primer set specific for the right arm of the transfer vector was used for the positive control.
All characterized mutant virus isolates had the predicted restriction endonuclease patterns (Fig. 4A) and were free from contaminating parental viruses when examined by PCR analysis (Fig. 4B, panel III). One isolate, designated MalΔSVD, was selected for further study.
MalΔSVD is attenuated in swine.
Macrophage growth characteristics of MalΔSVD were compared to the parental MalΔNL by infecting primary macrophage cell cultures (MOI = 0.01) and then determining virus yield at various times postinfection. MalΔSVD growth kinetics and virus yields were statistically indistinguishable from those of MalΔNL (Fig. 5). Thus, deletion of this MGF gene-containing region from MalΔNL did not affect viral replication in swine macrophage cell cultures.
FIG. 5.
Growth characteristics of MalΔNL and MalΔSVD in swine macrophage cell cultures. Primary swine macrophages were infected with MalΔNL and MalΔSVD (MOI = 0.1). At indicated times, duplicate samples were collected and titrated for total virus yield. Titers are expressed as log10 TCID50. Data represent means and standard errors of two independent experiments.
To examine the effect of this deletion on virulence, pigs were inoculated intramuscularly with 3 log10 TCID50 of parental MalΔNL and MalΔSVD. Data from this experiment (Table 2) showed that all MalΔSVD-infected pigs survived infection, whereas all MalΔNL-infected animals died. A delay in the onset of clinical disease with a transient fever response (2 to 4 days) and lethargy (1 to 2 days) was observed for MalΔSVD-infected pigs compared to the MalΔNL group. Maximum viremia titers in the MalΔSVD-infected animals were significantly reduced by 1,000-fold over those of MalΔNL-infected animals. The MalΔSVD virulence phenotype was similar to that described previously for E70ΔNL (66).
TABLE 2.
Survival, fever response, and viremia in swine following infection with MalΔSVD
| Virus | No. of survivors/total | Mean time to death (days) ± SEM | Fever
|
Viremia
|
||
|---|---|---|---|---|---|---|
| Mean time to onset (days) ± SEM | Mean duration (days) ± SEM | Mean time to onset (days) ± SEM | Mean max titer (log10 TCID50/ml) ± SEM | |||
| MalΔNL | 0/4 | 8.28 ± .5 | 2.5 ± 0.3 | 5.75 ± 0.2 | 2 ± 0.0 | 7.73 ± 0.3 |
| MalΔSVD | 4/4 | 12.5 ± 1.6 | 6 ± 0.0 | 7 ± 1.0 | 4.78 ± 0.5 | |
These findings indicate that MGF 360 and 530 genes encoded in the left variable region of the ASFV genome function as a swine virulence determinant complementing NL gene function.
DISCUSSION
Here we have shown that MGF 360 and MGF 530 genes in the left variable region of the ASFV genome encode a novel virulence determinant. ASFV isolates lacking NL together with MGF 530 genes 3FR, 3NR, and 3QR and the MGF 360 genes 3HL, 3IL, and 3LL are avirulent in swine. Using an in vivo marker rescue strategy, we have shown that genomic DNA (20 kb) from the left variable region of the MalΔNL genome was sufficient to rescue virulence in the attenuated E70ΔNL isolate. Nucleotide sequence analysis of the E70ΔNL left variable region identified an 8-kb deletion which resulted in the deletion or truncation of three MGF 360 genes and four MGF 530 genes in E70ΔNL. An MalΔNL recombinant virus, MalΔSVD, with an engineered deletion which removed or truncated three MGF 360 and three MGF 530 genes from this region was attenuated in pigs. These data indicate that in the absence of NL gene function, the swine virulence determinant is necessary for viral virulence in domestic pigs.
The swine virulence determinant locus contains ASFV MGF 530 and MGF 360 genes, indicating a role for these genes in swine virulence. MGF 530 and MGF 360 genes do not show similarity to other genes or motifs in the current databases. Individual MGF genes are conserved among ASFV isolates (80 to 98% amino acid similarity) (Fig. 4) (Kutish et al., personal communication). Transcriptional analysis of MGF 360 and MGF 530 genes showed that these ORFs are transcribed early during ASFV infection (51, 64; J. G. Neilan, unpublished), but translated proteins have not yet been identified or characterized. Alignments of predicted amino acid sequences of all MGF 360 and MGF 530 family members show three conserved regions at the amino terminus of these proteins (64). These conserved regions may indicate common ancestral relationships among the MGF genes or roles for these genes in common or related pathways. The functions of different multigene families in ASFV infection are unclear at present.
Recently the MGF 360 and 530 genes have been shown to have a significant role in ASFV macrophage host range (65). The swine virulence determinant and macrophage host range determinants share four MGF genes; however, these determinants differ phenotypically. The virulence phenotype associated with the swine virulence determinant is independent of the macrophage host range phenotype MHR.
MHR encompasses MGF 360 genes L3CL, L3DL, L3EL, L3HL, L3IL, and L3LL and the MGF 530 gene L3FR. Data indicate that MHR functions in the regulation of infected-cell death pathways. The MGF genes L3FR, L3HL, L3IL, and L3LL are common to both the swine virulence determinant and macrophage host range determinants. It is tempting to speculate that swine virulence determinant and macrophage host range determinant may have related functions in maintaining infected-cell survival. In addition, other studies have shown that macrophage cell cultures infected with ASFV NL null mutants exhibit a twofold decrease in cell viability at 24 h postinfection compared to wild-type-infected cultures (Afonso et al., unpublished data). This observation suggests a function for NL, albeit less dramatic than MHR, in the maintenance of infected-cell survival. It is possible that, in the absence of NL, the swine virulence determinant may function in the maintenance of infected-cell survival in specific ASFV target cells which are critical for a virulence phenotype in domestic pigs.
Two additional ASFV genes, 5HL and 4CL (also known as A224L) (1, 9, 44, 50), have also been shown to have roles in the maintenance of infected-cell survival. Thus, ASFV encodes multiple genes involved in aspects of maintaining infected-cell survival. It is possible that the functions of these genes, which may or may not be required for in vitro growth, may be critical for successful infection of specific lineages of target cells in vivo. ASFV infects cells within the mononuclear-phagocytic system. In vivo, these cell populations are heterogeneous with respect to their states of activation and maturation, and thus it is possible that while attenuated viruses exhibit normal replication in in vitro differentiated macrophage cell cultures, they may exhibit restricted growth in specific lineages of mononuclear cells in pigs, which are critical for virulence. ASFV virulence is associated with the ability of ASFV to replicate and spread within target tissues and not specifically with the ability to replicate in primary swine macrophage cultures in vitro (13, 34, 35, 39, 42).
The MGF 360 and MGF 530 genes are unique to ASFV, functioning in the maintenance of infected-cell survival and viral virulence. Studies to determine how these novel genes function in viral virulence and host range are ongoing.
Acknowledgments
We thank Aniko Zsak, Adriene Lakowitz, and the PIADC animal care staff for excellent technical assistance.
REFERENCES
- 1.Afonso, C. L., J. G. Neilan, G. F. Kutish, and D. L. Rock. 1996. An African swine fever virus bcl-2 homolog, 5-HL, suppresses apoptotic cell death. J. Virol. 70:4858-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., L. Zsak, C. Carrillo, M. V. Borca, and D. L. Rock. 1998. African swine fever virus NL gene is not required for virus virulence. J. Gen. Virol. 79:2543-2547. [DOI] [PubMed] [Google Scholar]
- 3.Almazán, F., J. M. Rodríguez, G. Andrés, R. Pérez, E. Viñuela, and J. F. Rodríguez. 1992. Transcriptional analysis of multigene family 110 of African swine fever virus. J. Virol. 66:6655-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almendral, J. M., F. Almazán, R. Blasco, and E. Viñuela. 1990. Multigene families in African swine fever virus: family 110. J. Virol. 64:2064-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco, R., M. Agüero, J. M. Almendral, and E. Viñuela. 1989. Variable and constant regions in African swine fever virus DNA. Virology 168:330-338. [DOI] [PubMed] [Google Scholar]
- 7.Blasco, R., I. de la Vega, F. Almazán, A. Agüero, and E. Viñuela. 1989. Genetic variation of African swine fever virus: variable regions near the ends of the viral DNA. Virology 173:251-257. [DOI] [PubMed] [Google Scholar]
- 8.Brown, F. 1986. The classification and nomenclature of viruses: summary of results of meetings of the International Committee on Taxonomy of Viruses in Sendai, September 1984. Intervirology 25:141-143. [DOI] [PubMed] [Google Scholar]
- 9.Brun, A., C. Rivas, M. Esteban, J. M. Escribano, and C. Alonso. 1996. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology 225:227-230. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo, C., M. V. Borca, C. L. Afonso, D. V. Onisk, and D. L. Rock. 1994. Long-term persistent infection of swine monocytes/macrophages with African swine fever virus. J. Virol. 68:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to g134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 12.Chou, J., and B. Roizman. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J. Virol. 64:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colgrove, G. S., E. O. Haelterman, and L. Coggins. 1969. Pathogenesis of African swine fever in young pigs. Am. J. Vet. Res. 30:1343-1359. [PubMed] [Google Scholar]
- 14.Costa, J. V. 1990. African swine fever virus, p. 247-270. In G. Darai (ed.), Molecular biology of iridoviruses. Kluwer Academic Publishers, Norwell, Mass.
- 15.De la Vega, I., E. Viñuela, and R. Blasco. 1990. Genetic variation and multigene families in African swine fever virus. Virology 179:234-246. [DOI] [PubMed] [Google Scholar]
- 16.DeKock, G., E. M. Robinson, and J. J. G. Keppel. 1994. Swine fever in South Africa. Onderstepoort J. Vet. Sci. Anim. Ind. 14:31-93. [Google Scholar]
- 17.DeTray, D. E. 1957. Persistence of viremia and immunity in African swine fever. Am. J. Vet. Res. 18:811-816. [PubMed] [Google Scholar]
- 18.Dixon, L. 1988. Molecular cloning and restriction enzyme mapping of an African swine fever virus isolate from Malawi. J. Gen. Virol. 69:1683-1694. [DOI] [PubMed] [Google Scholar]
- 19.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Vinuela, and P. J. Wilkinson. 2000. Family Asfarviridae, p. 159-165. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 20.Dixon, L. K., D. L. Rock, and E. Viñuela. 1995. African swine fever-like viruses. Arch. Virol. Suppl. 10:92-94. [Google Scholar]
- 21.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 22.Finney, D. J. 1984. Statistical methods in biological assays, 2nd ed., p. 524-533. Hafner Publishing Co., New York, N.Y.
- 23.Florea, L., G. Hartzell, Z. Zhang, G. Rubin, and W. Miller. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8:967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fornace, A. J., Jr., D. W. Nebert, M. C. Hollander, J. D. Luethy, M. Papathanasiou, J. Fargnoli, and N. J. Holbrook. 1989. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol. Cell. Biol. 9:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genovesi, E. V., F. Villinger, D. J. Gerstner, T. C. Whyard, and R. C. Knudsen. 1990. Effect of macrophage-specific colony-stimulating factor (CSF-1) on swine monocyte/macrophage susceptibility to in vitro infection by African swine fever virus. Vet. Microbiol. 25:153-176. [DOI] [PubMed] [Google Scholar]
- 26.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 27.González, A., V. Calvo, F. Almazan, J. M. Almendral, J. C. Ramirez, I. de la Vega, R. Blasco, and E. Viñuela. 1990. Multigene families in African swine fever virus: family 360. J. Virol. 64:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González, A., A. Talavera, J. M. Almendral, and E. Viñuela. 1986. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 14:6835-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 30.Haresnape, J. M., P. J. Wilkinson, and P. S. Mellor. 1988. Isolation of African swine fever virus from ticks of the Ornithodoros moubata complex (Ixodoidea: Argasidae) collected within the African swine fever enzootic area of Malawi. Epidemiol. Infect. 101:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess, W. R. 1982. African swine fever: a reassessment. Adv. Vet. Sci. Comp. Med. 25:39-69. [PubMed] [Google Scholar]
- 32.Heuschele, W. P., and L. Coggins. 1969. Epizootiology of African swine fever in warthogs. Bull. Epizootic Dis. Afr. 17:179-183. [PubMed] [Google Scholar]
- 33.Kleiboeker, S. B., G. F. Kutish, J. G. Neilan, Z. Lu, L. Zsak, and D. L. Rock. 1998. A conserved African swine fever virus right variable region gene, l11L, is nonessential for growth in vitro and virulence in domestic swine. J. Gen. Virol. 79:1189-1195. [DOI] [PubMed] [Google Scholar]
- 34.Konno, S., W. D. Taylor, and A. H. Dardiri. 1971. Acute African swine fever. Proliferative phase in lymphoreticular tissue and the reticuloendothelial system. Cornell Vet. 61:71-84. [PubMed] [Google Scholar]
- 35.Konno, S., W. D. Taylor, W. R. Hess, and W. P. Heuschele. 1971. Liver pathology in African swine fever. Cornell Vet. 61:125-150. [PubMed] [Google Scholar]
- 36.Lord, K. A., B. Hoffman-Liebermann, and D. A. Liebermann. 1990. Sequence of MyD116 cDNA, a novel myeloid differentiation primary response gene induced by IL6. Nucleic Acids Res. 18:2823.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massung, R. F., J. J. Esposito, L. Liu, J. Qi, T. R. Utterback, J. C. Knight, L. Aubin, T. E. Yuran, J. M. Parsons, V. N. Loparev, N. A. Selivanov, K. F. Cavallaro, A. R. Kerlavage, B. W. J. Mahy, and J. C. Venter. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature (London) 366:748-751. [DOI] [PubMed] [Google Scholar]
- 38.McGeoch, D. J., and B. C. Barnett. 1991. Neurovirulence factor. Nature (London) 353:609. [DOI] [PubMed] [Google Scholar]
- 39.Mebus, C. A. 1988. African swine fever. Adv. Virus Res. 35:251-269. [DOI] [PubMed] [Google Scholar]
- 40.Mebus, C. A., and A. H. Dardiri. 1980. Western hemisphere isolates of African swine fever virus: asymptomatic carriers and resistance to challenge inoculation. Am. J. Vet. Res. 41:1867-1869. [PubMed] [Google Scholar]
- 41.Mebus, C. A., J. W. McVicar, and A. H. Dardiri. 1981. Comparison of the pathology of high and low virulence African swine fever virus infections, p. 183-194. In P. J. Wilkinson (ed.), Proceedings of CEC/FAO expert consultation in African swine fever research, Sardinia, Italy, September 1981. Commission of the European Communities, Luxembourg, Belgium.
- 42.Moulton, J., and L. Coggins. 1968. Comparison of lesions in acute and chronic African swine fever. Cornell Vet. 58:364-388. [PubMed] [Google Scholar]
- 43.Neilan, J. G., Z. Lu, G. F. Kutish, L. Zsak, T. G. Burrage, M. V. Borca, C. Carrillo, and D. L. Rock. 1997. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitro and viral virulence. Virology 230:252-264. [DOI] [PubMed] [Google Scholar]
- 44.Nogal, M. L., G. Gonzalez de Buitrago, C. Rodriguez, B. Cubelos, A. L. Carrascosa, M. L. Salas, and Y. Revilla. 2001. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J. Virol 75:2535-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortin, J., L. Enjuanes, and E. Viñuela. 1979. Cross-links in African swine fever virus DNA. J. Virol. 31:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 47.Plowright, W., J. Parker, and M. A. Pierce. 1969. African swine fever virus in ticks (Ornithodoros moubata, Murray) collected from animal burrows in Tanzania. Nature (London) 221:1071-1073. [DOI] [PubMed] [Google Scholar]
- 48.Plowright, W., J. Parker, and M. A. Pierce. 1969. The epizootiology of African swine fever in Africa. Vet. Rec. 85:668-674. [PubMed] [Google Scholar]
- 49.Plowright, W., G. R. Thomson, and J. A. Neser. 1994. African swine fever, p. 568-599. In J. A. W. Coetzer, G. R. Thomson, and R. C. Tustin (ed.), Infectious diseases in livestock, with special reference to South Africa, vol. 1. Oxford University Press, Durban, South Africa. [Google Scholar]
- 50.Revilla, Y., A. Cebrian, E. Baixeras, C. Martinez, E. Vinuela, and M. L. Salas. 1997. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domain. Virology 228:400-404. [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez, J. M., R. J. Yáñez, R. Pan, J. F. Rodríguez, M. L. Salas, and E. Viñuela. 1994. Multigene families in African swine fever virus: family 505. J. Virol. 68:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 53.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 54.Sogo, J. M., J. M. Almendral, A. Talavera, and E. Viñuela. 1984. Terminal and internal inverted repetitions in African swine fever virus DNA. Virology 133:271-275. [DOI] [PubMed] [Google Scholar]
- 55.Staden, R. 1982. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 10:2951-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumption, K. J., G. H. Hutchings, P. J. Wilkinson, and L. K. Dixon. 1990. Variable regions on the genome of Malawi isolates of African swine fever virus. J. Gen. Virol. 71:2331-2340. [DOI] [PubMed] [Google Scholar]
- 57.Sussman, M. D., Z. Lu, G. Kutish, C. L. Afonso, P. Roberts, and D. L. Rock. 1992. Identification of an African swine fever virus gene with similarity to a myeloid differentiation primary response gene and a neurovirulence-associated gene of herpes simplex virus. J. Virol. 66:5586-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tabarés, E., I. Olivares, G. Santurde, M. J. Garcia, E. Martin, and M. E. Carnero. 1987. African swine fever virus DNA: deletions and additions during adaptation to growth in monkey kidney cells. Arch. Virol. 97:333-346. [DOI] [PubMed] [Google Scholar]
- 59.Thomson, G. R., M. Gainaru, A. Lewis, H. Biggs, E. Nevill, M. Van Der Pypekamp, L. Gerbes, J. Esterhuysen, R. Bengis, D. Bezuidenhout, and J. Condy. 1983. The relationship between ASFV, the warthog and Ornithodoros species in southern Africa, p. 85-100. In P. J. Wilkinson (ed.), Proceedings of CEC/FAO Research Seminar, Sardinia, Italy, September 1981, ASF, EUR 8466 EN. Commission of the European Communities, Luxembourg, Belgium.
- 60.Thomson, G. R., M. D. Gainaru, and A. F. V. Dellen. 1980. Experimental infection of warthog (Phacochoerus aethiopicus) with African swine fever virus. Onderstepoort J. Vet. Res. 47:19-22. [PubMed] [Google Scholar]
- 61.Vydelingum, S., S. A. Baylis, C. Bristow, G. L. Smith, and L. K. Dixon. 1993. Duplicated genes within the variable right end of the genome of a pathogenic isolate of African swine fever virus. J. Gen. Virol. 74:2125-2130. [DOI] [PubMed] [Google Scholar]
- 62.Wesley, R. D., and A. E. Tuthill. 1984. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev. Vet. Med. 2:53-62. [Google Scholar]
- 63.Wilkinson, P. J. 1989. African swine fever virus, p. 17-35. In M. B. Pensaert (ed.), Virus infections of porcines. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 64.Yozawa, T., G. F. Kutish, C. L. Afonso, Z. Lu, and D. L. Rock. 1994. Two novel multigene families, 530 and 300, in the terminal variable regions of African swine fever virus genome. Virology 202:997-1002. [DOI] [PubMed] [Google Scholar]
- 65.Zsak, L., Z. Lu, T. G. Burrage, J. G. Neilan, G. F. Kutish, D. M. Moore, and D. L. Rock. 2001. African swine fever virus multigene family 360 and 530 genes are novel macrophage host range determinants. J. Virol 75:3066-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zsak, L., Z. Lu, G. F. Kutish, J. G. Neilan, and D. L. Rock. 1996. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J. Virol. 70:8865-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]