Abstract
The M184V mutation in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) causes resistance to lamivudine, but it also increases the sensitivity of the virus to zidovudine (3′-azido-3′-deoxythymidine; AZT). This sensitization to AZT is seen both in the presence and the absence of the mutations that confer resistance to AZT. AZT resistance is due to enhanced excision of AZT 5′-monophosphate (AZTMP) from the end of the primer by the RT of the resistant virus. Published data suggest that the excision reaction involves pyrophosphorolysis but that the likely in vivo pyrophosphate donor is not pyrophosphate but ATP. The mutations that lead to AZT resistance enhance ATP binding and, in so doing, enhance pyrophosphorolysis. The excision reaction is specific for AZT because HIV-1 RT, which can form a closed complex with a dideoxy-terminated primer and an incoming deoxynucleoside triphosphate (dNTP), does not form the closed complex with an AZTMP-terminated primer and an incoming dNTP. This means that an AZTMP-terminated primer has better access to the site where it can be excised. The M184V mutation alters the polymerase active site in a fashion that specifically interferes with ATP-mediated excision of AZTMP from the end of the primer strand. The M184V mutation does not affect the incorporation of AZT 5′-triphosphate (AZTTP), either in the presence or the absence of mutations that enhance AZTMP excision. However, in the presence of ATP, the M184V mutation does decrease the ability of HIV-1 RT to carry out AZTMP excision. Based on these results, and on the results of other excision experiments, we present a model to explain how the M184V mutation affects AZTMP excision.
Although combination therapy has been useful in the treatment of human immunodeficiency virus type 1 (HIV-1) infections, the development of drug resistance is a major problem. The drugs that have been approved for HIV-1 therapy target two viral enzymes, reverse transcriptase (RT) and protease (PR). There are two types of drugs that block reverse transcription, nucleoside analogs and nonnucleoside RT inhibitors. Because there are just three classes of approved anti-HIV-1 drugs, there is considerable cross-resistance within each of the three drug classes. This exacerbates the problem of drug resistance and places a premium on therapies that involve combinations of drugs that do not induce significant cross-resistance.
Nucleoside analogs lack the 3′ OH that is present in normal deoxynucleoside triphosphates (dNTPs). If a nucleoside analog is stably incorporated into viral DNA by HIV-1 RT, it acts as a chain terminator and blocks viral DNA synthesis. Viruses that are resistant to nucleoside analogs have specific mutations in RT. Because these drug-resistant RTs must be able to incorporate normal dNTPs reasonably efficiently, resistance must involve an increase in the discrimination between normal dNTPs and the analogs. This increased discrimination can arise either as a specific block to the incorporation of the nucleoside analog or by selective excision of the analog after it has been incorporated. For example, lamivudine (3TC) resistance most commonly involves the M184V mutation; this mutation interferes with the incorporation of 3TC 5′-triphosphate (3TCTP) (for a review, see reference 22). Conversely, the zidovudine (AZT) resistance mutations M41L, D67N, K70R, T215Y/F, and K219E/Q collaborate to enhance the selective excision of zidovudine 5′-monophosphate (AZTMP) after it has been incorporated (1, 7, 16-18).
Even better than drug combinations that do not lead to cross-resistance are those combinations in which the mutations that led to resistance to one drug cause hypersensitivity to another drug. This is the case for AZT and 3TC. The M184V mutation not only causes resistance to 3TC, it increases the sensitivity of the virus to AZT 5- to 10-fold (4, 14, 24, 25). This effect is seen both in the presence and in the absence of mutations that confer resistance to AZT, which suggests the possibility that the M184V mutation might somehow interfere with AZTMP excision from the end of the primer. Moreover, because the effect of M184V on AZT sensitivity is seen in the absence of any specific AZT resistance mutations, it would appear that wild-type HIV-1 RT carries out significant excision of AZTMP in vivo, an idea supported by in vitro data (7). The excision mechanism involves pyrophosphorolysis; however, there has been controversy about the nature of the pyrophosphate donor. Arion et al. (1) originally suggested that the in vivo pyrophosphate donor is pyrophosphate (PPi); however, other groups have suggested that the pyrophosphate donor is ATP (7, 16-18). There has also been disagreement as to the effects that the M184V mutation has on the rate of excision. Götte et al. (10) reported that the presence of the M184V mutation reduces the excision of AZTMP while Naeger et al. (19) reported that M184V does not impair AZTMP excision. Naeger et al. employed an assay that measured excision at a single position; this may explain why they did not see an effect on excision (see Results and Discussion). Lennerstrand et al. (15) reported that M184V reduces AZTMP excision when combined with the AZT resistance mutations M41L, D67N, K70R, and T215Y but not when combined with mutations M41L and T215Y.
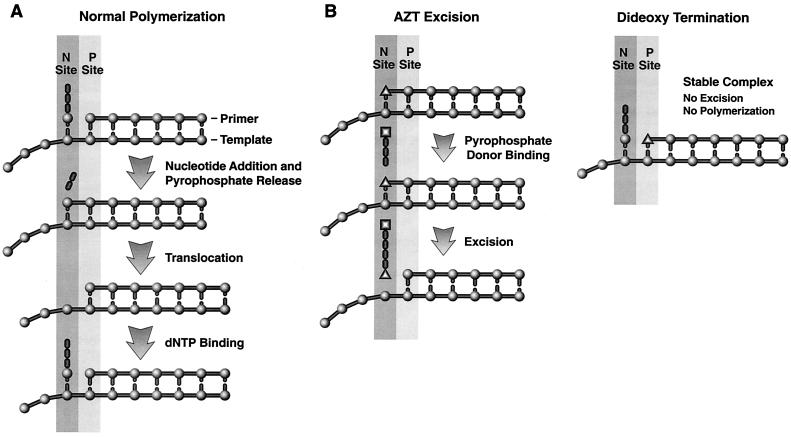
We have developed and tested a model for the selective excision of AZTMP (7). In the model, several of the mutations associated with AZT resistance serve to enhance the ability of HIV-1 RT to bind ATP but do not enhance the binding of PPi, suggesting that ATP is the pyrophosphate donor. This agrees with the excision results we obtained (7). So far as we can tell, there is no direct connection between the mutations that give rise to AZT resistance and the selective excision of AZTMP. Preferential excision of AZTMP appears to be an inherent property of the interactions between AZT and HIV-1 RT. The excision mechanism is closely related to the normal polymerization reaction run in reverse. Immediately after the incoming dNTP is joined to the end of the primer, the end of the primer lies in the site where the incoming dNTP is bound. We call this the N, or nucleotide binding, site (Fig. 1A). Before the next incoming dNTP can be bound and incorporated, RT must move relative to the nucleic acid substrate, a process called translocation. Translocation repositions the end of the primer into a site we have designated the P, or priming, site (Fig. 1A).
FIG. 1.
Polymerization and excision reactions carried out by HIV-1 RT. (A) Diagram of a simplified version of the polymerization reaction and the relationship of the nucleotide binding (N) and priming (P) sites. At the top, the incoming dNTP is bound at the N site; the end of the primer is at the P site. In the second step (second from the top), the α phosphate is joined to the end of the primer, releasing pyrophosphate. This leaves the end of the primer at the N site. Translocation (third from the top) moves the end of the primer to the P site. The next dNTP binds (bottom), and the cycle continues. (B) The drawings on the left show the steps of AZT excision; the drawing on the right shows the stable closed complex that forms when HIV-1 RT has incorporated a dideoxynucleotide, translocated the ddNMP block to the P site, and bound the incoming dNTP. This stable complex does not form with an AZTMP-terminated primer; the AZTMP-terminated primer resides preferentially at the N site (see text). Because the AZTMP-terminated primer preferentially resides in the N site, the binding of ATP, which acts as a pyrophosphate donor (middle of left drawing), can lead to excision, producing a dinucleotide tetraphosphate (bottom of left drawing).
For excision to occur, the end of the primer must be in the N site and a pyrophosphate donor must be bound to RT. It seems quite likely that the pyrophosphate moiety of the donor is in a position similar to the positions occupied by the β and γ phosphates of an incoming dNTP in the structure described by Huang et al. (11). There is relatively little excision of normal dNTPs incorporated by RT. Translocation rapidly moves the newly incorporated nucleotide away from the N site; additional rounds of polymerization embed it in the growing DNA chain. In a similar manner, if a dideoxynucleoside triphosphate (ddNTP) is incorporated, translocation moves it from the N to the P site and the next incoming dNTP can then bind. Because a dideoxynucleoside monophosphate (ddNMP) at the end of the primer strand lacks a 3′ OH, the incoming dNTP cannot be incorporated; however, the binding of the dNTP does cause the fingers to close on the incoming dNTP, forming a stable complex (18, 26). In this stable closed complex, the bound dNTP prevents the end of the primer from moving to the N site (Fig. 1B). There is sufficient dNTP present in vivo to generate this closed complex; this effectively blocks the excision of ddNTPs once they are incorporated and is the reason that a simple excision mechanism does not lead to resistance to ddNTPs. However, the azido group of AZT has sufficient steric bulk that it prevents a newly incorporated AZTMP from appropriately occupying the P site. Modeling experiments suggest that if an AZTMP-terminated primer is placed in the P site, there will be a steric conflict between the azido group and the aspartate at position 185 and one of the bound metals (7). This interferes with the ability of HIV-1 RT to form a closed complex with an AZT-terminated primer and an incoming dNTP (18, 26). More importantly, this means that even high levels of the incoming dNTP cannot block the access of an AZT-terminated primer to the N site, so AZTMP can be efficiently excised by ATP pyrophosphorolysis (7).
As has already been mentioned, the available data on the behavior of drug-resistant viruses imply that M184V sensitizes the virus to AZT, and when we developed this model for AZT excision, we considered why the M184V mutation in RT increased the sensitivity of HIV-1 to AZT. One possibility is that the mutation decreases the excision of AZTMP. We proposed, because the M184V mutation is at the position immediately adjacent to the site of steric conflict between the aspartic acid at position 185 and the azido group of AZTMP, that the M184V mutation could help relieve this steric hindrance, making it easier to move the AZTMP-terminated primer into the P site and form a stable closed complex. However, the effects of the M184V and M184I mutations on the structure of a binary complex of HIV-1 RT and double-stranded DNA suggest an alternate possibility. In the binary complex, with the end of the primer strand in the P site, one of the effects of substitution at M184 is to reposition the end of the primer strand (21). If there is repositioning when an AZTMP-terminated primer is in the N site, it could reduce the efficiency of the excision reaction (see Results and Discussion).
MATERIALS AND METHODS
Preparation of HIV-1 RT.
The open reading frames encoding wild-type HIV-1 RT and each of the RT mutants were cloned into a plasmid containing the HIV-1 PR open reading frame as previously described (6). The plasmid is based on the expression vector pT5m and was introduced into the Escherichia coli strain BL21(DE3)(pLysE). After induction with isopropyl β-d-thiogalactopyranoside, the plasmid expresses both the p66 form of HIV-1 RT (either wild-type or mutant) and HIV-1 PR. Approximately 50% of the overexpressed p66 RT is converted to the p51 form by HIV-1 PR, and p66/p51 heterodimers accumulate in E. coli. The p66/p51 heterodimers were purified by metal chelate chromatography (6).
Polymerase assays.
The polymerase assays were done as previously described (6). For each sample, 0.25 μg of single-stranded M13mp18 DNA (New England Biolabs) was hybridized to 0.5 μl of a solution of −47 sequencing primer (New England Biolabs) prepared by dissolving 0.1 optical density unit (OD260) in 0.1 ml of water. The template-primer was suspended in 100.0 μl of 25 mM Tris-Cl (pH 8.0); 75 mM KCl; 8.0 mM MgCl2; 100.0 μg of bovine serum albumin (BSA)/ml; 10.0 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS); 2.0 mM dithiothreitol (DTT); 10.0 μM (each) dATP, dTTP, and dGTP; 5.0 μM dCTP; 2.0 μM [α-32P]dCTP; and the indicated concentration of inhibitor. Extension was initiated by the addition of 1.0 μg of wild-type RT or RT variant. The mixture was incubated for 30 min at 37°C, and then the reaction was halted by the addition of 3 ml of ice-cold trichloroacetic acid. Precipitated DNA was collected by suction filtration through Whatman GF/C glass filters. The amount of incorporated radioactivity was determined by liquid scintillation counting.
Processivity and primer extension assays.
The processivity and primer extension assays have been previously described (6). In brief, for each sample to be assayed, 0.5 μl of 1.0 optical density −47 sequencing primer (New England Biolabs)/ml was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to single-stranded M13mp18 DNA (1.0 μl of a 0.25-μg/μl DNA stock for each sample to be assayed) by heating and slow cooling. The labeled template-primer was resuspended in 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg of BSA/ml, 10.0 mM CHAPS, and 2.0 mM DTT. One microgram of wild-type RT or RT variant was added to each tube and allowed to bind the labeled template-primer for 2 min. Extension was initiated by the addition of dNTPs to a final concentration of 10.0 μM each (for the primer extension assay) or 10.0 μM dNTPs plus 0.5 U of poly(rC) · oligo(dG)/ml (for the processivity assay). The addition of the poly(rC) · oligo(dG) trap limits the extension of the labeled primer by HIV-1 RT by binding the RT after it disassociates from the labeled template-primer.
Low dNTP extension assay.
The low dNTP extension assay has been described previously (6). Briefly, −47 sequencing primer (New England Biolabs) was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to single-stranded M13mp18 DNA (New England Biolabs) by heating and slow cooling. For each sample, 1.0 μg of wild-type RT or RT variant was added to the labeled template-primer in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, and 10.0 mM CHAPS. The reaction mixture was supplemented with 0.1 or 0.5 μM (each) dATP, dCTP, dGTP, and dTTP. The reactions were allowed to proceed at 37°C for 15, 30, or 60 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 6.0% polyacrylamide gel, and autoradiographed.
Strand displacement assay.
The construct PPT-PBS Litmus 28 contains the polypurine tract (PPT), a long terminal repeat (U3, R, and U5), and the primer binding site (PBS) of HIV-1 cloned into the vector Litmus 28 (New England Biolabs). Single-stranded PPT-PBS sense DNA was generated from this clone by using the M13 helper phage M13KO7. As previously described (6), an oligonucleotide complementary to the PBS was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to the single-stranded PPT-PBS sense DNA described above along with a 10-fold excess of four unlabeled DNA oligonucleotides by heating and slow cooling. The four unlabeled oligonucleotides will hybridize to regions of the HIV-1 long terminal repeat 3′ of the labeled PBS oligonucleotide and are separated from each other by three nucleotide gaps. There is a 10-nucleotide gap between the 3′ end of the labeled PBS primer and the 5′ end of the first unlabeled oligonucleotide. For each sample, 1.0 μg of wild-type RT or RT variant was added to the labeled template-primer in 25 mM Tris-Cl (pH 8.0), 35 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, 10.0 mM CHAPS, and 10.0 μM (each) dATP, dCTP, dGTP, and dTTP. The reactions were allowed to proceed at 37°C for 30 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 6.0% polyacrylamide gel, and autoradiographed. T4 DNA polymerase, which does not have strand displacement activity, was included as a control.
Primer block excision and extension assays.
The primer in these assays is complementary to the HIV-1 PBS sequence (5′-GTCCCTGTTCGGGCGCCA-3′). The primer was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to a fivefold excess of template oligonucleotide that is based on the sequence from the U5-PBS region of the HIV-1 genome (5′-AGTCAGTGTGGA CAATCTCTAGCAATGGCGCCCGAACAGGGACTTGAAAGCGAAAGTAAA-3′) by heating and slow cooling. The position in italics is normally an A in the pNL 4-3 sequence. It was changed to a C to alter a run of A residues. After the primer is annealed to the template, the underlined A residue will be the first base of the template strand after the double-stranded region. To block the primer, the template-primer was suspended in 25 mM Tris-Cl (pH 8.0), 35 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, 10.0 mM CHAPS, and a 10.0 μM concentration of either 3′-azido 3′-deoxythymidine 5′-triphosphate (AZTTP; Moravek Biochemicals) or 2′,3′ dideoxythymidine 5′-triphosphate (ddTTP; Boehringer Mannheim). Wild-type RT (1.0 μg) was added to the labeled template-primer, and the reactions were allowed to proceed at 37°C for 30 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol followed by an ethanol precipitation. The blocked template-primer was then resuspended in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 16.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, and 10.0 mM CHAPS. The level of MgCl2 was increased to 16.0 mM from 8.0 mM to ensure that the addition of ATP or sodium pyrophosphate did not bind all of the magnesium ions. The concentration of the template-primer is 0.15 nM. Depending upon the experiment (see the figure legends for details), the reaction buffer was supplemented with various amounts of dNTPs, nucleoside analogs (AZTTP or ddTTP), and a pyrophosphate donor (ATP or sodium pyrophosphate). Wild-type RT or variant RT (1.0 μg) was added to each reaction mixture with a final reaction volume of 50 μl; the approximate concentration of enzyme is 200 nM. The reactions were allowed to proceed for 10 min at 37°C and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 15.0% polyacrylamide gel, and autoradiographed. The total amount of template-primer (blocked and unextended plus deblocked and extended) and the amount of full-length product were determined by using a PhosphorImager.
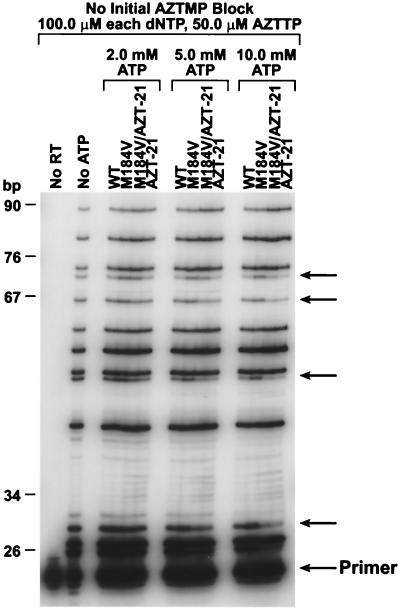
The assay for the M13-templated reaction (see Fig. 8) was similar to the assay described above except that the −47 M13 sequencing primer was 5′ labeled. The −47 primer was annealed to single-stranded M13mp18 DNA. Unlike the assays described above, the labeled −47 plus M13mp18 template-primer was not blocked by the addition of a nucleoside analog before the excision assay. The labeled template-primer was extended in the presence of 100.0 μM concentrations of each dNTP, 50.0 μM AZTTP, and the indicated amount of ATP. The samples were treated as described above.
FIG. 8.
Relative efficiency of AZTMP incorporation or excision at various positions. The abilities of the various HIV-1 RTs to be blocked by AZTMP incorporation at various positions on an M13mp18 template were compared at various concentrations of ATP (see Materials and Methods). The primer was labeled with 32P, and the reaction products were fractionated by electrophoresis on a 6% polyacrylamide gel. On the left is a scale showing the sizes of the DNA products. Arrows on the right indicate positions at which the excision efficiency differs for the RT mutants.
RESULTS AND DISCUSSION
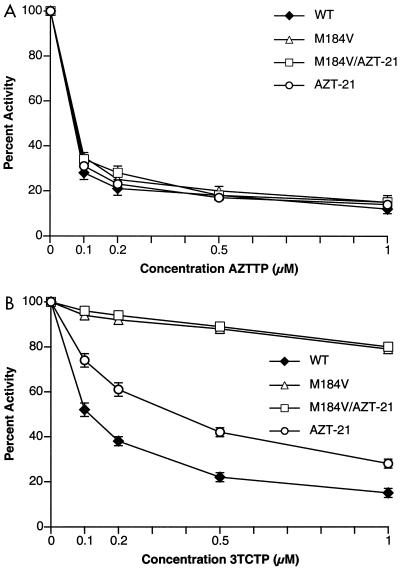
Experiments testing the AZT susceptibility of HIV-1 replicating in cultured cells showed that the presence of the M184V mutation enhanced the susceptibility of the virus to AZT. This effect of the M184V mutation is seen both in the presence and the absence of mutations that confer resistance to AZT (specifically M41L, D67N, K70R, T215F/Y, and K219E/Q). These mutations enhance the excision of AZTMP after it has been incorporated by HIV-1 RT. We proposed that the M184V mutation specifically interferes with the excision of AZTMP (7). There is an alternative possibility: the M184V mutation could interfere with the incorporation of AZTTP. To rule out this possibility, the abilities of AZTTP to block the polymerase activity of purified wild-type HIV-1 RT, the M184V mutant, the AZT-resistant mutant RT carrying M41L, D67N, K70R, T215Y, and K219Q (hereafter referred to as AZT-21), and a mutant RT carrying both the M184V mutation and the AZT-21 mutations were compared. To make the comparisons as accurate as possible, all four enzymes were purified at the same time and were stored under identical conditions. All were equally susceptible to inhibition by AZTTP (Fig. 2A), eliminating the possibility that the M184V mutation somehow enhances the incorporation of AZTMP during the polymerization reaction. However, as expected, the M184V mutation does lead to increased resistance to 3TCTP (Fig. 2B).
FIG. 2.
Inhibition of polymerase activity by AZTTP and 3TCTP. The four HIV-1 RTs used in the subsequent experiments (wild-type [WT], M184V, AZT-21, and M184V/AZT-21) were tested for inhibition by AZTTP and 3TCTP. To simplify the comparisons, the activities of each of the enzymes were normalized to 100%. Various concentrations of AZTTP and 3TCTP were added to polymerization reactions containing a −47 sequencing primer annealed to an M13mp18 DNA template (see Materials and Methods). After 30 min, the reactions were stopped by the addition of trichloroacetic acid and the newly synthesized DNA was collected on Whatman GF/C filters. Panel A shows the effects of adding AZTTP to the polymerization reactions; panel B shows the effects of adding 3TCTP.
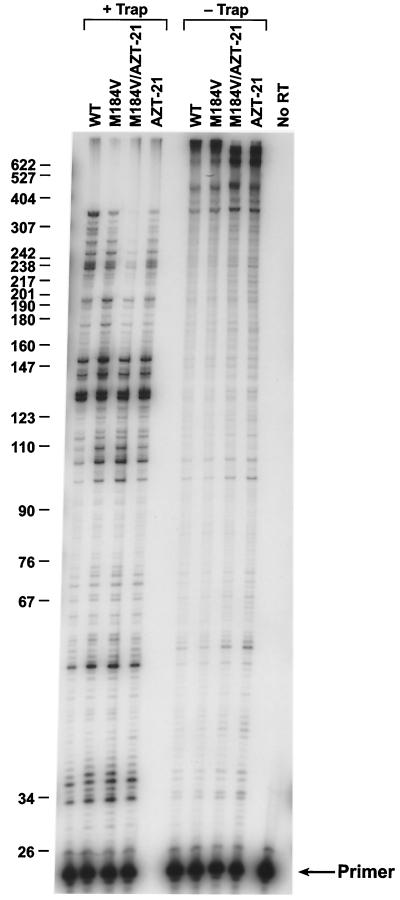
The excision reaction is closely related to the normal polymerization reaction; the excision reaction is, in essence, the reverse of the incorporation reaction with ATP used as the pyrophosphate donor (Fig. 1B). We asked whether the M184V mutation would significantly affect polymerization, either in the presence or the absence of the AZT-21 mutations. In a simple polymerase assay using the −47 sequencing primer annealed to single-stranded M13mp18 DNA (see Materials and Methods), the mutants were slightly less active than wild-type RT, which was designated as having 100% activity. M184V retained the most activity (90% of wild-type activity) while AZT-21 had 75% activity and M184V/AZT-21 had 71% activity. We previously showed that the M184V mutation had a modest but measurable negative effect on processivity (5), which is what was seen in the experiments described here (Fig. 3). This is in agreement with results from other groups (2, 19). The M184V mutant was slightly less able to polymerize at low dNTP concentrations than was wild-type RT (Fig. 4). Similar results were obtained with a strand displacement assay (data not shown). Taken together, these data suggest that the M184V mutation modestly impairs the ability of HIV-1 RT to polymerize. When compared to wild-type HIV-1 RT, we find that the AZT-21 RT mutant shows a modest decrease in processivity, primer extension, and strand displacement (Fig. 3 and data not shown) and an impaired ability to polymerize at low dNTP concentrations when compared to wild-type RT (Fig. 4). Arion et al. (1) and Caliendo et al. (9) have suggested that the mutations that confer AZT resistance enhance RT processivity; however, as they correctly point out, other mutations that affect the processivity of RT do not show this correlation with AZT resistance. Resistance to a nucleoside analog depends on an enhanced discrimination by the enzyme either at incorporation or through excision. In an excision-based resistance mechanism, the relative excision of the analog must be increased. A more-rapid reaction (or greater processivity) will not necessarily lead to resistance; resistance requires a change in the ratio of incorporation to excision. During active polymerization, excision of a nucleoside analog is essentially an irreversible reaction; after excision, a normal dNTP is usually incorporated in place of the analog and polymerization proceeds. However, a long template provides many opportunities for the incorporation (and excision) of a nucleoside analog; after each such block, HIV-1 RT must stop polymerizing and excise the analog. In this sense, what matters in terms of the completion of the HIV-1 genome in the presence of a nucleoside analog is not the initial reaction rate of the enzyme, nor its processivity, but whether it will remain active long enough to carry out sufficient polymerization (and excision) reactions to complete the synthesis of the DNA genome. A faster reaction rate could help, particularly because the virion core may be subject to degradation or disruption inside the cell; however, an enhanced polymerization rate is only helpful if, in vivo, the whole polymerizing complex, including the enzyme, is more persistent than the wild type. Moreover, since we do not find AZT resistance to be directly connected to increased enzyme activity or processivity (7), we do not think that the effect(s) of the M184V mutation on AZT resistance is derived directly from changes in polymerase activity.
FIG. 3.
Processivity and primer extension of wild-type HIV-1 RT and the various RT mutants. As described in Materials and Methods, the presence of a poly(rC)·oligo(dG) trap binds the HIV-1 RT after it disassociates from the labeled template-primer. This limits the HIV-1 RT to one round of extension of the labeled primer and is a measure of processivity (on left side of figure). Without the trap, the HIV-1 RT can undergo multiple rounds of primer extension (on right side). The reactions were run for 10 min, and the reaction products were fractionated on a 6% polyacrylamide gel. The products were visualized by autoradiography. The scale on the left indicates the sizes of the products. WT, wild type.
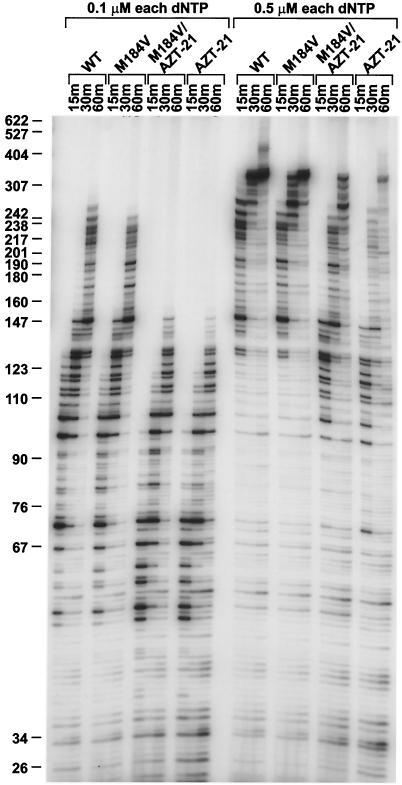
FIG. 4.
Low dNTP extension assay. The ability of the various enzymes to extend the −47 primer on an M13mp18 template was measured at a final concentration of either 0.1 or 0.5 μM each of the four dNTPs. The reactions were run as a time course with samples taken at 15, 30, and 60 min (see Materials and Methods). The reaction products were fractionated on a 6% polyacrylamide gel, and the DNA products were visualized by autoradiography. The scale at the left shows the sizes of the products. WT, wild type.
There is an alternative possibility: AZT resistance depends on a specific enhancement of the selective excision of AZTMP. Since a pyrophosphate-based excision mechanism is the inverse of polymerization, any changes that affect the excision rate should also affect the polymerization rate. One of the advantages of the models suggesting that ATP is the in vivo pyrophosphate donor is that this problem is resolved. In such models, the products of the polymerization reaction differ from the substrates for the excision reaction; this means that the mutations that cause AZT resistance can differentially affect excision and polymerization. The underlying mechanism is simple; the AZT resistance mutations enhance ATP (but not pyrophosphate) binding.
In an in vitro polymerization assay, particularly an assay that involves a relatively long template, the amount of full-length product will depend on the activity of the enzyme. As has already been discussed, we find that the drug resistance mutations modestly decrease the polymerase activity and processivity of HIV-1 RT. To minimize the effect of the overall polymerase activity on the excision assay, the excision assay is performed with a short template extension (24 bases). In the excision assay, there is no trap, so the extension assays (Fig. 4) are a better representation of the relative activities of the polymerases than are the processivity assays (Fig. 3). As has already been discussed, adding the M184V mutation either to the wild-type enzyme or to the AZT-resistant enzyme (AZT-21) has relatively little effect on the ability of the enzyme to polymerize.
These data suggest that any effects seen from the M184V mutation in the excision assay will be due to a specific effect on the excision reaction, rather than an overall effect on polymerization. What effect does the M184V mutation have on the excision reaction? The excision assays employ a labeled primer that is blocked at its 3′ end with AZTMP (or, where appropriate, with another chain terminator) and a pyrophosphate donor (PPi or ATP). The template was designed to provide multiple opportunities to incorporate and excise the chain terminator. Although there are multiple opportunities to incorporate (and excise) the chain terminator, there are far fewer positions where the chain terminator can be incorporated in the in vitro reaction (where the template extension is only 25 bases long) than in the viral genome. For this reason, the magnitude of the differences between the wild-type RT and AZT-21 is usually greater when measured in vivo than in in vitro reactions.
Excision reactions were run with wild-type RT, the M184V mutant, AZT-21, and M184V/AZT-21. If the level of dNTPs was low (10 μM) and the pyrophosphate donor was ATP, the M184V mutation had little, if any, effect, either in the presence or the absence of the AZT-21 mutations. However, as expected, the AZT-21 mutations did enhance excision (Fig. 5A). If the level of dNTPs was increased to 100 μM, a different pattern emerged (Fig. 5B). The M184V mutation decreased the efficiency of excision either in the presence or the absence of the AZT-21 mutations. However, all mutants with an AZT-21 genotype were more efficient in excision than wild-type RT (Fig. 5B).
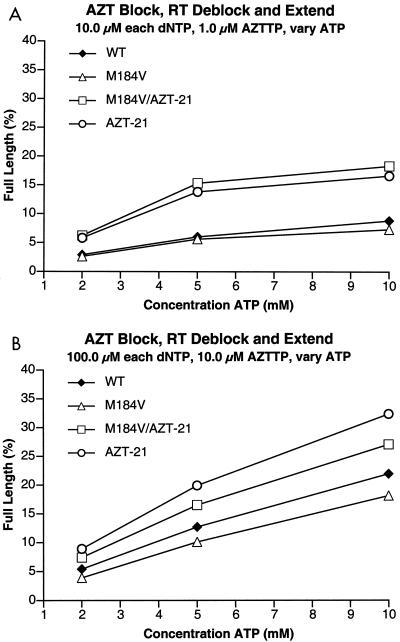
FIG. 5.
Comparison of the abilities of the various HIV-1 RTs (mutant and wild-type [WT]) to excise AZTMP from the end of the primer and extend the primer in the presence of AZTTP and various concentrations of ATP. The primer is fully blocked with AZTMP at the beginning of the reaction (see Materials and Methods). A short (25 nucleotides long) template extension provides an additional opportunity for the enzymes to incorporate AZTTP and excise AZTMP. The assay measures the percentage of the primer that is completely extended by each enzyme at various ATP concentrations. (A) Results obtained at low concentrations (10 μM) of each of the four dNTPs. (B) Results obtained with high concentrations (100 μM) of each of the four dNTPs.
The excision reaction was then run with a dideoxythymidine-blocked primer and ATP as the pyrophosphate donor. At a low dNTP concentration, both wild-type HIV-1 RT and the M184V RT excised dideoxythymidine 5′-monophosphate (ddTMP) poorly. However, RTs containing the AZT-21 mutations excised ddTMP relatively efficiently; the addition of the M184V mutation modestly reduced the ability of the AZT-21 mutant to excise ddTMP (Fig. 6A). Raising the dNTP concentration to 100 μM substantially reduced the excision of ddTMP. Neither wild-type RT nor the M184V mutant efficiently excised ddTMP at high dNTP concentrations. The AZT-21 mutant did excise a small amount of ddTMP at high dNTP concentrations; this was slightly reduced by adding the M184V mutation to the AZT-21 mutant (Fig. 6B).
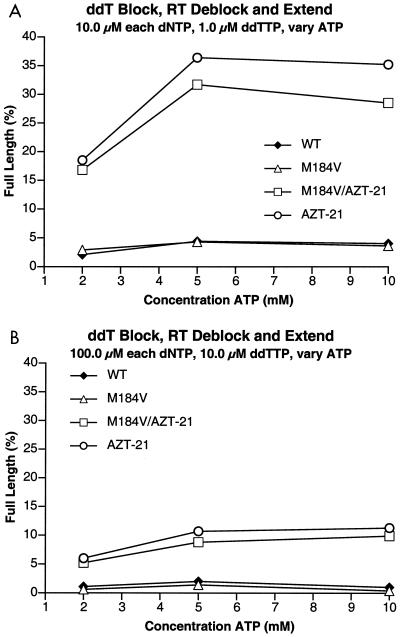
FIG. 6.
Comparison of the abilities of the various HIV-1 RTs (mutant and wild-type [WT]) to excise ddTMP and extend the primer in the presence of ddTTP and various concentrations of ATP. The assay conditions are similar to those described in the legend to Fig. 5, except that instead of AZTMP, the primer was terminated with ddTMP and, in the reaction mixtures, ddTTP was substituted for AZTTP. (A) Reactions run in the presence of 10 μM concentrations of each of the dNTPs. (B) Reactions run in the presence of 100 μM concentrations of each of the dNTPs.
This agrees with previous data; high levels of dNTPs substantially interfere with the excision of a ddNTP, but not AZTMP. ddNMP-terminated primers can move to the P site; binding a dNTP at the N site blocks excision (Fig. 1B). Although an AZTMP-terminated primer can bind normally at the N site, moving the end of the primer to the P site leads to steric conflicts between the azido group and the aspartic acid at position 185 and one of the active site metals. This steric conflict causes an AZT-terminated primer to preferentially reside at the N site, where it can be efficiently excised. Because the primer preferentially resides at the N site, high levels of dNTP do not block excision.
One possible explanation for the effect of the M184V mutation on AZT excision is that it relaxes the steric conflicts of the AZT-terminated primer when the end of the primer is at the P site. This should mean that the presence of an M184V mutation would make HIV-1 RT more susceptible to having the excision of AZTMP reduced by high levels of dNTPs. This is what was seen when AZTMP excision is measured in the presence of high levels of dNTPs (Fig. 5B). The presence of the M184V mutation reduces the excision of wild-type RT and of the AZT-21 mutant. This supports the model in which the M184V mutation relaxes the constraints on an AZTMP-terminated primer residing at the P site.
In terms of the active site of HIV-1 RT, the ability of M184V to relieve steric hindrance makes sense. The model predicts that the steric hindrance involves the aspartic acid at position 185 (the amino acid adjacent to the M184V mutation) and a metal partially chelated by D185. A simple form of the model would suggest that the change at position 184 leads to small changes in the amino acids in the immediate vicinity (including D185), which helps relieve the steric hindrance. If this were the explanation, then the presence of M184V would decrease excision if either ATP or pyrophosphate was used as the pyrophosphate donor.
To test this idea, the experiments were repeated with pyrophosphate instead of ATP. As previously noted, the AZT-21 mutant is less efficient than wild-type RT in excising the AZTMP block when pyrophosphate is the pyrophosphate donor (7). This supports the idea that, in vivo, the pyrophosphate donor is ATP, not pyrophosphate. Unlike the reactions with ATP as the pyrophosphate donor, there were only minor differences in the amount of excision at low (Fig. 7A) or high (Fig. 7B) concentrations of dNTPs. In either case, the mutants containing M184V were slightly better at excising AZTMP than were their counterparts without M184V. Since this was not the expected result, four new batches of enzyme were prepared (wild-type, M184V, AZT-21, and M184V/AZT-21). The excision experiments were repeated with both ATP and pyrophosphate, and the results were essentially identical to those already described (data not shown). These results suggest that the M184V mutation does not simply enhance the access of an AZT-terminated primer to the P site.
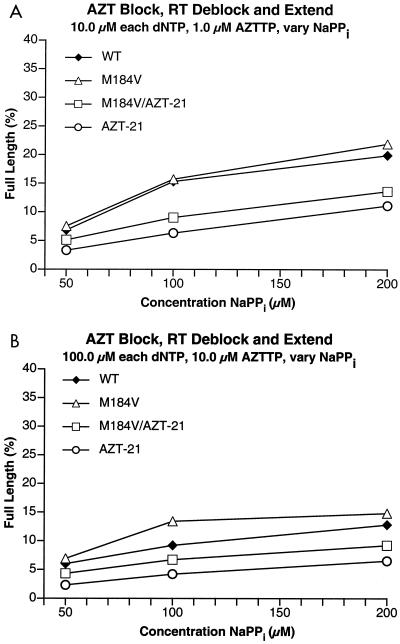
FIG. 7.
Comparison of the abilities of the various HIV-1 RTs (mutant and wild-type [WT]) to excise AZTMP from the end of the primer and extend it in the presence of AZTTP and various concentrations of sodium pyrophosphate (NaPPi). These reactions are quite similar to the reactions whose results are depicted in Fig. 5; however, in this case, the pyrophosphate donor is pyrophosphate. (A) Reactions run in the presence of 10 μM concentrations of each of the dNTPs. (B) Reactions run in the presence of 100 μM concentrations of each of the dNTPs.
How then can the effect of the M184V mutation on AZT excision be explained? There is an alternative possibility which is based on the crystal structures of the M184I and M184V mutations in complex with double-stranded DNA (21). In these binary complexes, the end of the primer is at the P site. However, the mutations at 184 alter the position of the DNA relative to the position of double-stranded DNA in a binary complex with wild-type HIV-1 RT. The end of the primer in the M184V/I structure is shifted toward the fingers subdomain relative to the end of the primer in the wild-type structure. This shift is away from the putative ATP binding site (7). Since the structure of the excision complex has not been solved, it is unclear whether there is (or is not) an alteration in the positioning of the nucleic acid when the last base at the end of the primer strand is in the N site of either the M184V or M184I enzymes. However, it is possible that there is a difference in the position of the nucleic acid and that this difference contributes directly to the difference in the efficiency of the excision reaction. In this version of the model, the M184 mutation causes a minor shift in the exact position where polymerization or excision occurs.
This reduction in the efficiency of the excision reaction could occur because the M184V mutation causes the end of the primer strand to move, but the ATP binding site does not move in concert. The site where ATP is bound is defined by amino acids that are relatively far from the active site (T215Y in particular). This could explain the apparent paradox that, despite the fact that the polymerization and excision reactions are closely related, the M184V mutation has a much greater effect on the excision of AZTMP than it does on polymerization. It could also explain why there are differential effects on the excision reactions with ATP and pyrophosphate. The pyrophosphate binding site is not the same as the ATP binding site. Interactions between the enzyme and the adenine base (and perhaps the ribose ring) of ATP could leave the pyrophosphate moiety too far away from the AZTMP-blocked end of the primer that has been shifted by the M184V mutation to excise efficiently. The binding of pyrophosphate itself is less constrained by the additional contacts that define the ATP binding site, so it is much easier for pyrophosphate to shift in concert with the active site. This would make the pyrophosphate excision reaction less susceptible to the effects of the M184V mutation than the ATP excision reaction. Altering the position of the primer could differentially affect the two related excision reactions.
If there is a repositioning of the nucleic acid, it could also play a role in relieving steric hindrance when the AZTMP-terminated primer is positioned at the P site. It is important to point out that the two ways the M184V mutation could reduce the efficiency of AZT excision (either by relieving the steric hindrance or by making it more difficult for the AZTMP to react with the γ phosphate of ATP) are not mutually exclusive; both could contribute to the reduced excision of AZTMP.
Whether the effect(s) of the M184V mutation on AZTMP excision involves only changes in the position(s) of the amino acids or whether the relative position of the nucleic acid is also important, it is likely that the differences in the position of the important components of the active site (both protein and nucleic acid) are modest, since all of the mutant enzymes are reasonably efficient polymerases. Any gross distortion of the polymerase active site would almost certainly interfere with polymerization. This suggests that the differences in AZTMP excision efficiency between the HIV-1 RTs that do, and do not, carry the M184V mutation might be small enough that the nucleic acid structure, which depends on nucleic acid sequence, might influence excision efficiency. A longer template was prepared from M13 DNA. A comparison of the pausing patterns of the various enzymes (Fig. 8) shows that there are sequence-specific differences in the ability of the different enzymes to excise AZTMP. This might account for the failure of Naeger et al. (19), who monitored AZTMP excision at a single site, to see an effect of the M184V mutation on AZTMP excision.
The results have implications for both the interpretation of in vivo resistance data and for the precise molecular effects of the M184V mutation on the polymerase active site. In terms of in vivo resistance, HIV-1 RT must normally excise significant amounts of AZTMP, at least in cultured cells. The M184V mutation sensitizes the wild-type enzyme to AZTMP by reducing excision. If excision did not occur in vivo, M184V (which decreases excision) could not enhance AZT sensitivity. This also suggests that, in some sense, AZT is an even better drug than it appears: AZT is a potent inhibitor of HIV-1 replication in spite of the preferential excision by wild-type HIV-1 RT.
A mechanism similar to what we described for M184V may also play a role with other combinations of mutations. Like M184V, the mutation Y181C is close to the active site of HIV-1 RT and has been shown to suppress the effects of the AZT resistance mutations (8, 12, 13, 20, 22). Other mutations that are known to suppress AZT resistance, e.g., K65R, L74V, and L100I (for reviews, see references 3, 22, and 23), may also alter the position of the template-primer and the RT active site. This repositioning may move the end of the primer away from the ATP pyrophosphate donor and render the excision reaction less effective.
Acknowledgments
We thank Pat Clark, Peter Frank, and Megan Bucheimer for preparing purified, wild-type, and mutant RT; Hilda Marusiodis for help in preparing the manuscript; and Anne Arthur for expert editorial advice.
Research in S.H.'s laboratory was supported by the National Cancer Institute and NIGMS. Research in E.A.'s laboratory was supported by grants AI 27690 and GM 55609 from the National Institutes of Health.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Back, N. K. T., M. Nijhuis, W. Keulen, C. A. B. Boucher, B. B. O. Essnik, A. B. P. van Kuilenburg, A. H. van Gennip, and B. Berkout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Balzarini, J. 1999. Suppression of resistance to drugs targeted to human immunodeficiency virus reverse transcriptase by combination therapy. Biochem. Pharmacol. 58:1-27. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, C. A., N. Cammack, P. Schipper, R. Schurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., and S. H. Hughes. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, P. L., H.-Q. Gao, P. K. Clark, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. YADD mutants of human immunodeficiency virus type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro. J. Virol. 75:6321-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrnes, V. W., E. A. Emini, W. A. Schleif, J. H. Condra, C. L. Schneider, W. J. Long, J. A. Wolfgang, D. J. Graham, L. Gotlib, A. J. Schlabach, B. S. Wolanski, O. M. Blahy, J. C. Quintero, A. Rhodes, E. Roth, D. L. Titus, and V. V. Sardana. 1994. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 38:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Götte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, H., R. Chopra, G. F. Verdin, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 RT: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 12.Larder, B. A. 1992. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 36:2664-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder, B. A. 1994. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J. Gen. Virol. 75:951-957. [DOI] [PubMed] [Google Scholar]
- 14.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 15.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, P. R., S. E. Matsurra, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, P. R., S. E. Matsurra, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, P. R., S. E. Matsurra, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naeger, L. K., N. A. Margot, and M. D. Miller. 2001. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal, and viral replication. Antivir. Ther. 6:115-126. [PubMed] [Google Scholar]
- 20.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, C.-K. Shih, M. Myers, and J. Griffin. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarafianos, S., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. 3TC (lamivudine) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 23.Sluis-Cremer, N., D. Arion, and M. A. Parniak. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 57:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian, H., J. M. Whitcomb, K. Linnoli, T. Wrin, G. Winslow, N. Parkin, D. Smith, Y. S. Lie, M. Bakthiari, D. Shugarts, R. T. Schooley, D. Kurizkes, and C. J. Petropoulos. 1998. Zidovudine/lamivudine co-resistance is preceded by a transient period of zidovudine hypersensitivity. Antivir. Ther. 3(Suppl. 1):22. [Google Scholar]
- 25.Tisdale, M., S. D. Kemp, N. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong, W., C.-D. Lu, S. K. Sharma, S. Matsuura, A. G. So, and W. A. Scott. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36:5749-5757. [DOI] [PubMed] [Google Scholar]