Abstract
Autism is a neurodevelopmental disability characterized by deficits in verbal communications, impairments in social interactions, and repetitive behaviors. Several studies have indicated strong involvement of multigenic components in the etiology of autism. Linkage analyses and candidate gene search approaches so far have not identified any reliable susceptibility genes. We are using a proteomic approach to identify protein abnormalities due to aberrant gene expression in autopsied autism brains. In four of eight autism brains, we have found an increase in polarity (more acidic) of glyoxalase I (Glo1) by two-dimensional gel electrophoresis. To identify the molecular change resulting in the shift of Glo1 polarity, we undertook sequencing of GLO1 gene. Direct sequencing of GLO1 gene/mRNA in these brains, has identified a single nucleotide polymorphism (SNP), C419A. The SNP causes an Ala111Glu change in the protein sequence. Population genetics of GLO1 C419A SNP studied in autism (71 samples) and normal and neurological controls (49 samples) showed significantly higher frequency for the A419 (allele frequency 0.6 in autism and 0.4 in controls, one-tailed Fisher's test P < 0.0079). Biochemical measurements have revealed a 38% decrease in Glo1 enzyme activity in autism brains (one-tailed t-test P < 0.026). Western blot analysis has also shown accumulation of advanced glycation end products (AGE's) in autism brains. These data suggest that homozygosity for A419 GLO1 resulting in Glu111 is a predisposing factor in the etiology of autism.
Keywords: autism, autopsied brain, susceptibility gene, proteomics, glyoxalase I, single nucleotide polymorphism
INTRODUCTION
Autism (OMIM 209850) is a highly heterogeneous, pervasive neurodevelopmental disability characterized by impairments in social interactions, deficits in verbal and non-verbal communication, and stereotyped and repetitive patterns of behavior [Lord et al., 2000]. The symptoms of autism are discernible in the first 3 years of an affected infant's life and manifest throughout the life span. Individuals with autism exhibit a wide spectrum of cognitive abilities, with about 30% displaying severe developmental delay and a significant proportion also experiencing epileptic seizures. The prevalence of this disability in the general population is over 1 in 1, 000 live births [Fombonne, 2002], although a recent estimate indicate three times higher prevalence rate [Yeargin-Allsopp et al., 2003]. Generally, boys are three to four times more commonly affected than girls. At present, autism is diagnosed through psychological evaluation of affected individuals or interviewing of parents/guardians; consequently, diagnosis is rarely possible before 3 years of life, and treatment is symptomatic. Availability of a reliable early biomarker for autism may help in molecular or biochemical classification of this highly heterogeneous disability and in initiation of early specific and targeted therapeutic intervention.
The causes of autism are still unknown; however, considerable evidence exists for the involvement of genetic factors. The evidence is based on the higher ratio of autism recurrence in siblings and a greater concordance in monozygotic twins [Maestrini et al., 2000], although lack of a 100% concordance rate in monozygotic twins also suggests involvement of environmental and other factors in the development or severity of autism [Szatmari, 1999]. Several laboratories around the world are engaged in linkage analysis and positional cloning approaches of genome-wide scans to identify autism susceptibility genes [International Molecular Genetic Study of Autism Consortium IMGSAC, 1998, 2001; Ashley-Koch et al., 1999; Barrett et al., 1999; Philippe et al., 1999; Buxbaum et al., 2001; Liu et al., 2001; Yonan et al., 2003]. These studies have narrowed a number of chromosomal loci, and strong involvement of genetic loci on chromosomes 2, 7, and 17 has been indicated. Possible oligogenic interaction of over 15 different genetic loci has been proposed in the etiology of autism [Pickles et al., 1995; Risch et al., 1999]. However, no susceptibility genes have been reliably identified. The candidate gene search approach for autism has resulted in the identification of genes, notably, WNT2 [Wassink et al., 2001], RELN (reelin) [Persico et al., 2001], and NLGN3 and NLGN4 (neuroligins3 and 4) [Jamain et al., 2003]. These studies have not been substantiated yet in other laboratories [Krebs et al., 2002; McCoy et al., 2002; Zhang et al., 2002]. Thus, so far no reliable susceptibility genes for autism has been identified. The oligogenic mode of genetic interplay as well as the failure of whole genome scanning of more than 140 families prompted Risch et al. [1999] to conclude that “positional cloning of susceptibility loci by linkage analysis may be a formidable task and that other approaches may be necessary in determining autism susceptibility genes.” As a powerful alternate approach, our laboratory is conducting proteomic analyses of autism brain with the ultimate goal of identifying the molecular defect. Analyses of autopsied autism brain by two-dimensional gel electrophoresis have identified altered mobility of a protein of molecular mass 24 kDa that was identified as glyoxlase I (Glo1). The altered mobility of Glo1 is the result of C419A SNP causing Ala111Glu in the protein sequence.
Glo1 is a cytosolic, ubiquitously expressed, zinc metalloenzyme enzyme involved in scavenging toxic α-oxoaldehydes such as methylglyoxal that are formed during cellular metabolic reactions [Thornalley, 1990]. The methylglyoxal rapidly reacts non-enzymatically with reduced glutathione to form hemithioacetal that is converted by Glo1 to S-D-lactoylglutathione. A second enzyme glyoxalase II catalyzes the conversion of S-D-lactoylglutathione to lactate regenerating the reduced glutathione. A dysfunction of the glyoxalase system will result in accumulation of the highly reactive methylglyoxal.
MATERIALS AND METHODS
Human autopsied autism and control brain tissues (Table I) were obtained from the Autism Tissue Program, Princeton, NJ; Brain and Tissue Bank for Developmental Disorders, Baltimore, MD and Miami, FL; Harvard Brain Tissue Resource Center, Boston, MA; National Disease Research Interchange, Philadelphia, PA; and National Neurological Research Specimen Bank, Los Angeles, CA. The data presented is from the tissues that matched closest. We have not succeeded in perfectly matching the tissue in every respect such as age, gender, and post-mortem interval. Lymphoid cells from individual autism patients or from multiplex families were obtained from the Autism Genetic Resource Interchange, Los Angeles, CA. A list of AGRE repository numbers, for these cell lines, is available from the corresponding author upon request. Phenotypic data about the subjects whose lymphoid cells were used, is available at the AGRE website at http://www.agre.org. Normal control lymphoid cells from six Utah pedigree families were obtained from Coriell Institute for Medical Research, Camden, NJ, while neurological controls comprising of cells from Batten disease and Fragile X syndrome patients were from our Institute Cell repository. Other than individuals within a family, all the autism as well as the control subjects are unrelated.
TABLE I.
Age, Ethnicity, Cause of Death, Autopsy Time, and Glo1 419 SNP for Brain Samples Used in the Current Study
| Subjects | Ethnicity | Gender | Death cause | Age (year) | Autopsy time (h) | Seizures | GLO1 C419A SNPc |
|---|---|---|---|---|---|---|---|
| Autism | |||||||
| UMB 144a | African-American | Male | Drowning | 10 | 22 | Negative | A/A |
| HSB 765 | Caucasian | Male | Suicide | 19 | 29 | Unknown | A/C |
| UMB 797 | Caucasian | Male | Drowning | 9 | 13 | Negative | C/C |
| BTB 2004 | Asian | Male | Drowning | 10 | 23 | Negative | A/A |
| BTB 3663 | African-American | Male | Neuroleptic | 27 | 30 | Unknown | A/A |
| BTB 3871 | Hispanic white | Male | Seizure | 5 | 25 | Positive | A/A |
| B 4498 | Caucasian | Male | Heart failure | 56 | 19.4 | Positive | A/C |
| B 5000 | Unknown | Male | Unknown | 27 | 8.3 | Unknown | A/C |
| Controls | |||||||
| 1014 | Caucasian | Male | Heart failure | 15 | 15 | Negative | A/A |
| 1076 | Caucasian | Male | Sudden collapse | 17 | 19 | Unknown | C/C |
| 1858 | African-American | Male | Unknown | 5 | 19 | Negative | C/C |
| 4480 | Unknown | Male | Unknown | 57 | 17.7 | Negative | Not determined |
| Neurological controls | |||||||
| Seizure | |||||||
| 820 | African-American | Male | Seizure | 15 | 26 | Positive | A/A |
| 1085 | Caucasian | Female | Hypoxic/ischemicencephalopathy | 6 | 9 | Positive | A/C |
| Down syndrome | |||||||
| RT-01-063 | Caucasian | Female | Cardiac arrest | 17 | 12 | Negative | C/C |
| Mental retardation | |||||||
| CNL 68b | Unknown | Female | Pneumonia | 33 | Unknown | Negative | A/C |
| CNL 672 | Unknown | Male | Liver carcinoma | 45 | Unknown | Positive | A/C |
| CNL 673 | Unknown | Male | Pneumonia | 40 | Unknown | Negative | C/C |
Another brother of this subject is also diagnosed with autism.
All CNL brains had the diagnosis of mental retardation of unknown etiology.
419SNP A/A and C/C encodes homozygous glutamyl and alanyl residues at position 111, respectively, while A/C encodes heterozygous glutamyl/alanyl form of the Glo1.
Brief Clinical History of Autism Brains
UMB 144 was diagnosed with mild autistic trait/pervasive developmental disorder, and also had a brother diagnosed with autism. Chromosome analysis and investigation for Fragile-X syndrome were normal. His verbal and performance IQ's were 57 and 72, respectively. His behavior was in the clinically significant range on the withdrawal, attention problems, hyperactivity, and adaptability.
HSB 765 was diagnosed with autism at age 3. The subject displayed limited eye contact with others, hand flapping and self-stimulation, and was always withdrawn.
UMB 797 was diagnosed with autism/attention deficit disorder who also had chronic migraine headache but no known seizure history. According to Autism Diagnostic Interview-Revised (ADI-R), the subject had impairments in reciprocal social interactions, deviant communication, and repetitive behaviors and stereotyped patterns with scores significantly above cutoffs for autism.
BTB 2004 was diagnosed with severe autism and hyperactivity without any history of seizures. The subject had inability to use oral language for communication, impairment in social interaction, inattentive behavior and displayed self-stimulating ritualistic behavior. He had Southern California Ordinal Scales of Cognitive Development (SCOSCD) rating 4-5 and childhood autism rating scale (CARS) scoring of 39.
B 4498 was diagnosed with autism by several psychologists. He lost language by age 2, was later declared legally blind and had history of seizures. The subject had difficulties with communication, social behaviors, and atypical interests. His scores on ADI-R met or exceeded cutoffs for autism in each of those areas.
Reports about the remaining autism brains are very limited to that provided by the pathologists.
Immobilized linear pH gradient strips (Immobiline strips) were purchased from Pharmacia Biotech, Piscataway, NJ. Premixed acrylamide/bis-acrylamide solutions (3.3% crosslinking) were purchased from Bio-Rad Laboratories, Hercules, CA. Trizol reagent, platinum Taq polymerase, and one-step RT-PCR kit were purchased from Invitrogen, Carlsbad, CA. Nitrocellulose membrane for Western blots was purchased from Schleicher and Schuell, Keene, NH. All other laboratory reagents were purchased from Sigma Chemical Co., St. Louis, MO. Trypsin digestion of silver-stained protein spots and identification by MALDI-TOF or LC/MS/MS were done at the Proteomic Research Services, Ann Arbor, MI.
2-D Gel Electrophoresis
Total brain proteins from frontal lobe gray matter were used. The first dimensional separation of extracts from brain tissue (250 μg protein) was performed at 20°C in 11 cm long, linear gradient pH 4-7 isoelectric focusing strips in a Bio-Rad Protean IEF Cell. The IPG strips were rehydrated overnight with protein-containing buffer comprising 9 M urea, 2% CHAPS, 2 mM tributylphosphine, and 0.2% ampholytes pH 4-7 and covered with mineral oil to prevent evaporation. Focusing was started at 250 V, and after 15 min, the current was increased linearly to 8, 000 V over 8 hr and maintained at this voltage for another 12 hr. The second dimensional separation was run after equilibration with SDS-denaturing buffer, on a 10-20% gradient SDS—PAGE, and protein spots were visualized by mass spectroscopy-compatible silver staining.
Protein Identification
The protein spot of interest was excised from the gel, rinsed with distilled water and subjected to robotic in-gel proteolytic digestion with trypsin. Molecular masses of peptides in the tryptic digest were identified by LC/MS/MS analysis on a Micromass Q-TOF2 mass spectrometer using a 75 μm C18 column at a flow rate of 200 nl/min. The observed m/z values were searched using the MASCOT software (www.matrixscience.com) against the NCBInr protein database.
Sequence Analysis of Glyoxalase I Gene
The sequence of GLO1 gene was determined either from the mRNA or directly from genomic DNA by direct sequencing. The total RNA in the brain tissues was prepared with Trizol reagent by the manufacturer's protocol and was stored frozen at -70°C until further processing. First-strand cDNA and further amplification was achieved by using one-step RT-PCR kit using the sense primer 5′GGGTGACTCCTCCGTTCC3′ and the antisense primer 5′CCAAGAGCCAAGAGCACAAT3′. The RT-PCR conditions were as follows: 45°C for 25 min, 94°C for 2 min, followed by 35 successive cycles of 94°C for 15 sec, 55°C for 30 sec, 68°C for 2 min, and additionally 68°C for 5 min.
For sequencing the genomic DNA, a 713 bp fragment was amplified by PCR using the sense primer 5′TCAGAGTGTGTGATTTCGTG3′ and the antisense primer 5′CATGGTGAGATGGTAAGTGT3′. The following PCR conditions were employed for amplification: 94°C for 4 min, followed by 35 successive cycles of 94°C for 30 sec, 55°C for 30 sec, 68°C for 60 sec, and additionally 68°C for 5 min. The amplified products were resolved on a 0.7% agarose gel, and the ∼1.5 kb fragment from mRNA or 0.7 kb fragment from genomic DNA was purified from the gel by electroelution and cleaned by ethanol precipitation using standard procedures.
DNA sequencing was carried out on a Beckman CEQ2000 sequencer equipped with capillary electrophoresis. The sequencing reaction mixture contained template DNA at 50 fmol and primers at 1 pmol in a volume of 20 μl along with the enzyme and fluorescent nucleotides supplied by the manufacturer. For each sample, reaction was done with one forward and one reverse primer. The following reaction conditions were used: 96°C for 2 min, followed by 50 cycles of 96°C for 10 sec, 55°C for 10 sec, and 60°C for 4 min. Fluorescence tagged oligos were purified by ethanol precipitation and dissolved in dime-thylformamide before loading into the sequencer.
The GLO1 C419A SNP was also detected by fragment analysis following restriction digestion. For fragment analysis, the genomic DNA is amplified with the same aforementioned sense and antisense primers. An aliquot of the reaction mixture following PCR amplification is digested with 0.2 U of SfaNI for 2 hr at 37°C. The SfaNI digest is then resolved by 1% agarose gel electrophoresis, and the fragments are visualized under UV light after staining with ethidium bromide.
Western Blot Analysis
Staining of protein bands following separation by 10-20% gradient SDS—PAGE and subsequent transfer onto nitrocellulose membrane were done as described earlier [Junaid et al., 2000]. Equal amount of brain gray matter proteins (50 μg) from all the brains were loaded onto the gel. The primary AGE antibody used was raised in rabbits against carboxymethylated-ribonuclease which is one of the markers for this modification on proteins. Secondary mouse antibody against rabbit IgG coupled to alkaline phosphatase was used, and the colored formazan developed after incubation with bromochloro-indoylphosphate and nitroblue tetrazolium visualized the immunocomplex.
Glyoxalase I Enzyme Activity
Glo1 enzyme activity was measured according to the technique of Kester and Norton [1975] with slight modifications. The reaction mixture contained 14.4 mM methylglyoxal, 0.72 mM reduced glutathione, 100 mM imidazole buffer pH 6.8, and 16 mM MgSO4 The reaction mixture was incubated at 22°C for 4 hr to ensure non-enzymatic hemithioacetal formation before adding tissue extract. The enzymatic production of S-lactoylglutathione (ε3.37 mM-1 cm-1) was measured at 240 nm at 15 sec interval for 2 min. Specific activity is expressed as μmol S-lactoylglutathione formed/hr/mg protein.
RESULTS AND DISCUSSION
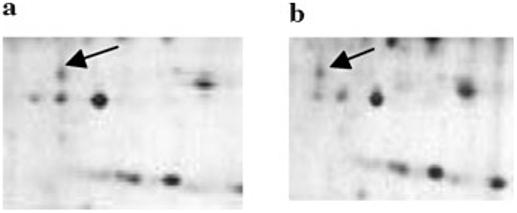
As an alternative to linkage analysis, positional cloning and candidate gene search approaches, we used a proteomics method to identify abnormal proteins in autopsied autism brain tissues [Junaid and Pullarkat, 2001], with the ultimate goal of identifying underlying genes and molecular defects responsible for the disorder [Sleat et al., 1997]. Brain tissues from well-characterized autism patients and approximately age-matched normal or neurological controls were used in the current study (Table I). Analyses of total brain gray matter proteins by proteomics revealed a shift in the charge of a protein spot of 24 kDa molecular mass in four out of eight autism brains (Fig. 1). In these brains, the 24 kDa protein spot is more acidic (pI 5.9) than in normal or neurological control brains (pI 6.8); indicating a gain of net negative charge. In contrast, only two of nine controls showed the charge shift in the 24 kDa protein.
Fig. 1.
Two dimensional gel electrophoresis. 2D gel electrophoresis of (a) control (C1076) and (b) autism brain (UMB 144) extracts. The total gray matter proteins from the control and autism extracts were resolved by IEF on pH 4-7 Immobiline strips, followed by 10-20% gradient SDS—PAGE and visualized by silver staining. Arrows indicate protein band that shift the mobility. The spot of interest in controls is on top of the middle spot (three spots in a row), while in autism, it is on top of the left spot.
Tryptic digestion of the silver-stained gel plugs followed by mass spectrometry (LC/MS/MS) of the resultant peptides identified these protein spots in autism and control brain as Glo1 (lactoylglutathione lyase, EC 4.4.1.5). The identity of Glo1 was established positively from 6 out of 17 peptides in the tryptic digest of the protein spot (data not shown). No other proteins were identified in the remaining peptides, although some of these peptides could be modified with as yet undetermined posttranslational modifications of Glo1. The matched amino acid residues represented 28% of the complete Glo1 sequence.
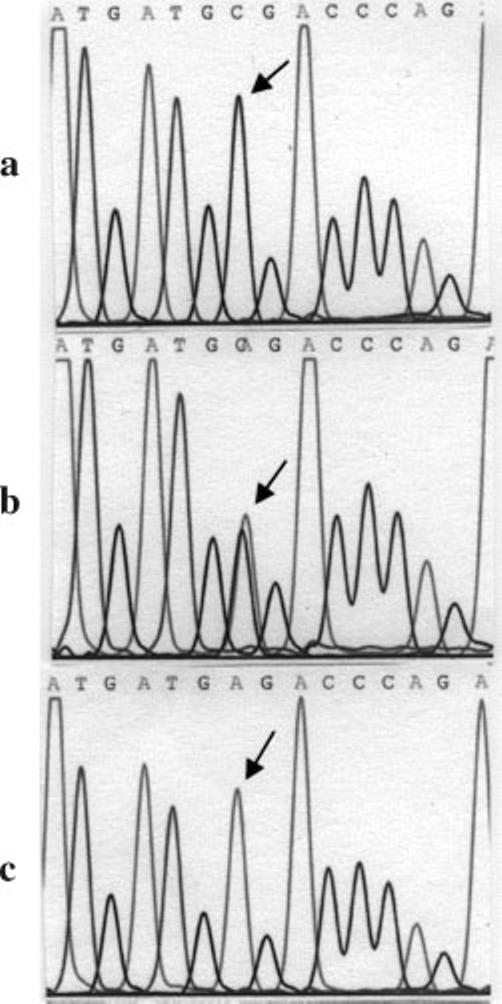
To identify the molecular defect resulting in the shift of net charge in Glo1, we undertook direct sequencing of mRNA or DNA. Sequencing of mRNA was preferred over that of DNA because it would provide the exact message transcribed rather than the genomic DNA sequence. However, in brains that had poor quality mRNA due to delayed autopsy intervals, direct sequencing was achieved from the DNA. For each sample, the sequencing reaction was performed with two different primers: one forward and one reverse, and the sequence was checked independently with both primers. Sequencing of cDNA prepared from total RNA or direct DNA sequencing of all autism and control brains (normal and neurological) showed a SNP C419A (Fig. 2 and Table I) in situations where Glo1 was more acidic (the SNP is numbered with respect to the cDNA sequence). This SNP is reported in the PubMed data base as rs2736654. The result is a GAG codon in place of a GCG codon in exon 4, which changes Ala111Glu (numbered with respect to the protein sequence). Four autism brains were homozygous for Glu-Glo1 (both alleles code for Glu111), whereas three others were heterozygous for Glu/Ala-Glo1 (one allele each codes for Ala111 and Glu111). One autism brain (UMB 797) was homozygous for Ala-Glo1 (both alleles code for Ala111). Two of three normal controls were homozygous for Ala-Glo1, whereas the third was homozygous for Glu-Glo1. Among the neurological controls, one case with Down syndrome and one case with mental retardation of unknown etiology were homozygous for Ala-Glo1, whereas two other mental retardation cases and one seizure case were heterozygous for Ala/Glu-Glo1. One case with seizure disorder was homozygous for Glu-Glo1.
Fig. 2.
DNA sequencing electropherogram. Direct sequencing of genomic DNA for identification of GLO1 C419A SNP in lymphoid cells. DNA sequencing electropherogram shown when both alleles at position 419 code for C (a), each allele coding for A and C (b), or both alleles coding for A(c). The position of SNP is indicated by an arrow.
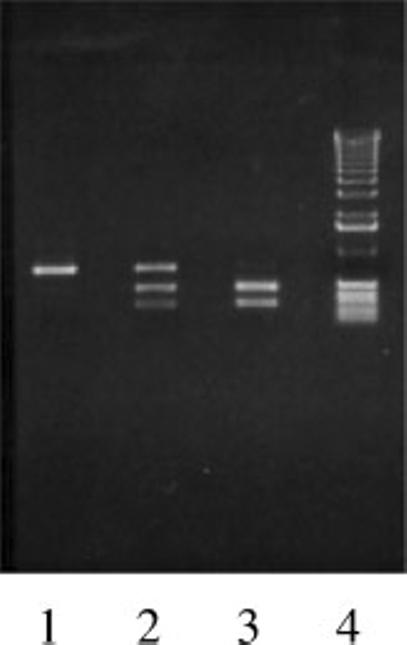
To gain insights into the involvement of Glu-Glo1 in autism and the mode of inheritance, we determined the SNP in DNA samples obtained from multiplex families with autism. For determining the SNP we used direct sequencing as well as restriction fragment length polymorphism (RFLP) analysis following digestion with SfaNI. The RFLP analysis involves restriction digestion with SfaNI of the 713 bp PCR product amplified from genomic DNA. When C419 exists, a restriction site for SfaNI is present which is lost because of A419 SNP. Thus, SfaNI will cleave a PCR product spanning the GLO1 C419A SNP depending upon whether or not C419 is present (Fig. 3). The DNA samples were selected randomly from the available Autism Genetic Resource Exchange (AGRE) catalog available at http://www.agre.org [Geschwind et al., 2001]. Analysis of the SNP data from brain and lymphoid cells showed that in 38% (27/71) of the autism subjects both alleles coded for Glu-Glo1 (Table II). 45% (32/71) of the subjects were heterozygotes. We also analyzed the GLO1 C419A SNP in 50 controls comprised of 10 brains, lymphoid cells from 6 different Utah pedigree families (33) and neurological controls, including patients with Batten disease (5) and Fragile X syndrome (2). Of 50 controls, only 9 (18%) were homozygous for Glu-Glo1. Genotype frequency distributions for both autistic and control samples are in agreement with Hardy—Weinberg equilibrium. The allele frequency of A419 in autism and control samples is0.6056 and 0.44, respectively, a highly significant difference (one-tailed Fisher's Test P < 0.0079). Numerous reports of GLO1 allele frequency (for Glu-Glo1) in different populations across the world and in the United States showed values between 0.306 and 0.49 [Goldman et al., 1991; Thornalley, 1991]. Our analysis of GLO1 (Glu-Glo1) allele frequency in controls also falls within this range. However, the GLO1 (Glu-Glo1) allele frequency in autism subjects in the present study is significantly higher, suggesting a predisposition of autism to GLO1 with homozygosity of A419. Additional support of GLO1 involvement in autism comes from its localization on chromosome 6p21.3-6p21.1 which is close to 6p24-6p22, a locus found to have linkage in an earlier genome-wide scan with the marker D6S260 [Philippe et al., 1999]. It is interesting that an earlier independent study by Philippe et al.[1999] involving DNA samples from Paris Autism Research International Sibpair Study showed association in the region close to GLO1. More recently, data from Yonan et al. [2003] using samples from the AGRE repository also indicated an increase in multipoint maximum LOD scores on chromosome 6, when narrow disease classification criteria were invoked. The two independent studies point towards involvement of the GLO1 gene in at least a subset of autism patients. There is also one report on the association of a depression locus to the GLO1 gene [Tanna et al., 1989]. The Glu111 variant in the present study that showed increased acidic mobility in the two-dimensional gel analysis is same as that reported by Goldman et al. [1991] and Thornalley [1991].
Fig. 3.
Restriction fragment length polymorphism (RFLP) analysis for GLO1 C419A SNP determination. The 713 bp PCR amplified fragment from genomic DNA samples from autism subject (lane 1) or controls (lane 2) and (lane 3) were digested with SfaNI for 2 hr at 37°C. The digested samples were resolved by 1% agarose gel electrophoresis and visualized under UV light following staining with ethidium bromide. Sample in lane 1 is homozygous A419, lane 2 is heterozygous A419/C419, and lane 3 is homozygous C419. Lane 4 is 1 kb DNA ladder. The digested fragments in lane 2 and 3 are 453 and 260 bp long.
TABLE II.
Genotype and Allele Frequency for GLO1 C419A SNP in Autism and Control Samples
| Autism |
Controls |
|||
|---|---|---|---|---|
| Genotype | Number | Frequency | Number | Frequency |
| GAG* | 27 | 0.380 | 9 | 0.18 |
| GAG/GCG | 32 | 0.451 | 26 | 0.52 |
| GCG | 12 | 0.169 | 15 | 0.30 |
| A allele frequency** | 86 | 0.61 | 44 | 0.44 |
| C allele frequency | 56 | 0.39 | 56 | 0.56 |
χ2=6.51 with 2-degrees of freedom, P < 0.039.
One-tailed Fisher's test P < 0.0079.
It has been suggested that marked differences in pairwise concordance between monozygotic and dizygotic twins and the rapid falloff in the recurrence rate of autism with decreasing genetic relatedness could indicate multiple susceptibility genes in autism [Lamb et al., 2002]. Several investigators have suggested that because of oligogenic interaction of genes, simple Mendelian inheritance may not explain sufficiently the transmission of autism from parents to children [Risch et al., 1999]. Analyses of inheritance in 11 multiplex families has shown that inheritance of the GLO1 C419A SNP follows the conventional Mendelian mode of transmission from parents to affected children (data not shown).
In order to substantiate the results, we have evaluated another SNP in the GLO1 sequence at position 459 (numbered with respect of the cDNA sequence) for association with autism. Only 4 autism samples out of 71 analyzed had the homozygous nucleotide T, while rest of the samples had T/A at position 459. Irrespective of the SNP, the codon will result in Gly124 (numbered with respect to the protein sequence), however, the fact that majority of autism samples had only one SNP give credence to our contention that the SNP A419 is involved in the autism etiology.
Molecular changes resulting from the substitution of Ala111Glu in Glo1 due to the SNP may be caused by conformational changes of the protein. Glo1 is a dimeric zinc containing metalloenzyme. The zinc atom is held by electro-static forces with amino acid residues that also include Glu99, and functions in enediolate intermediate formation and stabilization during catalysis [Ridderstrom et al., 1998]. This residue is close to the Glu111 that is replaced by the SNP. The 3D structure of Glo1 with Glu111 has shown that the active Glo1 functions as a dimer with the two zinc atoms, two active sites as well as two glutathione-binding sites formed at the interface [Cameron et al., 1997]. Although, these studies have not shown direct interaction of Glu111 in zinc binding, any chemical interactions that change the 3D structure or disrupt electrostatic bonds are expected to affect enzyme activity. The 3D structure of Glo1 with Ala111 is not available for comparison.
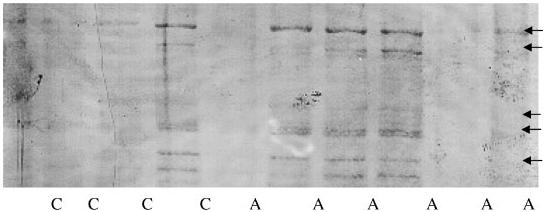
Enzyme activity measurements in brain extracts (Table III) have shown a 38% reduction in Glo1 specific activity in autism subjects when compared to normal controls (one-tailed t-test P < 0.026). This study was done with a very limited number of autopsied brain tissues, and needs to be replicated in a larger set of matched tissues. Glo1 activities in all neurological controls were higher than normal controls probably as a consequence of the primary neurological condition, hence, these were omitted while calculating the control average value. There is no evidence of Glo1 activity differing between gender and the age-range of patients and controls assessed in the present study. Reduced Glo1 activity will result in the accumulation of AGE's due to inability of the cells to detoxify the highly reactive methylglyoxal that is formed during cellular metabolism of sugars, amino acids, and fatty acids [Ray and Ray, 1983; Reichard et al., 1986; Lyles and Chalmers, 1992]. Indeed, Western blot analysis of brain samples with antibody against carboxymethylated-ribonuclease has confirmed accumulation of AGE in autism brains (Fig. 4). This observation is in agreement with the notion that autism brains accumulate AGE due to elevated methylglyoxal resulting from decreased Glo1 activity. Glo1 is expressed in fetal tissues three times higher than those in adult tissues indicating its importance during fetal growth. Due to reduced enzyme activity a specific methylglyoxal-derived AGE may have altered function that is deleterious at crucial events during neurogenesis in an individual with autism.
TABLE III.
Glo1 Enzyme Activity in Control and Autism Brain Extracts
| Brains | Glo1 activity* μmol/hr/mg protein |
|---|---|
| Controls (n=4) | 47.12 ± 5.72 |
| Autism (n=8) | 29.30 ± 4.65 |
Glo1 activities in neurological controls were omitted from the controls, as these were significantly higher than controls, and would have given bias to the data in favor of controls.
Values are mean SEM and n= number of brains (one-tailed t-test P < 0.026).
Fig. 4.
AGE's in control and autism brain. Western blot analysis of control and autism brain tissue showing accumulation of advanced glycation end products in autism brains. Total cellular protein (50 μg) in brain gray matter were separated by SDS—PAGE on a 10-20% gradient gel, transferred to nitrocellulose membrane and probed with an antibody for advanced glycation end products (against AGE-ribonuclease). Arrows indicate accumulated AGE's.
Prediction by protein phosphorylation software, NetPhos v2.0 available at http://www.cbs.dtu.dk/services/NetPhos/ [Blom et al., 1999], has shown increased probability of phosphorylation of Ser114 and Tyr115 residues in Glu-Glo1. The probabilities of phosphorylation of other Ser or Tyr residues in the protein are not changed due to Glu-Glo1. Ranganathan et al. [1993] have predicted four Ser and Thr phosphorylation sites in Glo1. Increased phosphorylation has been shown to modulate Glo1 activity, resulting in accumulation of specific methylglyoxal-derived AGE [Van Herreweghe et al., 2002]. Thus, the increased acidic mobility and reduced enzyme activity of Glu-Glo1 variant could be the net result of additional acidic charge from Glu111 as well as increased phosphorylation of Ser/Tyr residues.
Our approach of detecting relevant polymorphic proteins via proteomics was quite successful in brains from autistic patients. Increased acidity of Glo1 in 50% of autism brains analyzed is due to the substitution of Ala with Glu and the possible additional phosphorylation. Most genes show multiple SNP's; however, only a few SNPs actually result in amino acid substitutions. In the case of GLO1, the polymorphism results in Ala111Glu, and this isoform has decreased enzyme activity. Reduced Glo1 activity would be expected to result in the accumulation of methylglyoxal [Shamsi et al., 2000; Miyata et al., 2001]. Methylglyoxal is highly reactive and is known to react with biomolecules, notably proteins, forming AGE. These AGE's may have altered or compromised function at crucial stages during neurogenesis. Methylglyoxal has been shown to target several proteins involved in the regulation of cell growth and differentiation [Godbout et al., 2002; Sakamoto et al., 2002; Speer et al., 2003]. Overexpression of Glo1 in cells treated with phorbol esters [Gillespie, 1981] and cancer tissues [Thornalley, 1995] have also led to the suggestion that Glo1 activity is related to cell growth. Since autism in an early developmental disorder, molecules that affect cell growth can be possible sites of biological defects. Courchesne et al. [2003] have recently shown that autistic children have abnormal growth of the brain in the first year of life.
The present data suggest that homozygosity of the Glu-Glo1 variant could be one of the susceptibility factors in the etiology of autism. Because autism is a heterogeneous disorder, it is not expected that all autism tissue will have one particular molecular defect. Searches for other molecular defects need to be continued. Complex disorders result from a cumulative effect of a number of genetic, dietary, and environmental factors.
ACKNOWLEDGMENTS
We are thankful to Lucille McLendon and Michael Fenko for help with cell culture and DNA sequencing. We are thankful for the resources provided by the Autism Genetic Resource Exchange (AGRE) and the participating AGRE families, and the NAAR.
Footnotes
Part of this study was presented as a poster at the 2nd International Meeting for Autism Research, Orlando, FL 2002.
Grant sponsors: National Institutes of Health; National Alliance for Autism Research (NAAR); New York State Office of Mental Retardation and Developmental Disabilities; Grant number: NS40691.
REFERENCES
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA. Genetic studies of autistic disorder and chromosome 7. Genomics. 1999;61:227–236. doi: 10.1006/geno.1999.5968. [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, Folstein SE, Garcia M, Gardiner MB, Gilman S, Haines JL, Hopkins K, Landa R, Meyer NH, Mullane JA, Nishimura DY, Palmer P, Piven J, Purdy J, Santangelo SL, Searby C, Sheffield V, Singleton J, Slager S, Struchen T, Svenson S, Vieland V, Wang K, Winklosky B. An autosomal genomic screen for autism: Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet. 2001;68:1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AD, Olin B, Ridderstrom M, Mannervik B, Alwyn-Jones T. Crystal structure of human glyoxalase I—Evidence for gene duplication and 3D domain swapping. EMBO J. 1997;16:3386–3395. doi: 10.1093/emboj/16.12.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N. Evidence of brain over-growth in the first year of life in autism. JAMA. 2003;290:337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiological trends in rates of autism. Mol Psychiatr. 2002;7:S4–S6. doi: 10.1038/sj.mp.4001162. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, AGRE Steering Committee The autism genetic resource exchange: A resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie E. The tumor promoting phorbol diester, 12-O-tetradecanoylphorbol-13-acetate (TPA) increases glyoxalase I and decreases glyoxalase II activity in human polymorphonuclear leukocytes. Biochem Biophys Res Commun. 1981;98:463–470. doi: 10.1016/0006-291x(81)90862-7. [DOI] [PubMed] [Google Scholar]
- Godbout JP, Pesavento J, Hartman ME, Manson SR, Freund GG. Methylglyoxal enhances cisplatin-induced cytotoxicity by activating protein kinase Cdelta. J Biol Chem. 2002;277:2554–2561. doi: 10.1074/jbc.M100385200. [DOI] [PubMed] [Google Scholar]
- Goldman D, O’Brien SJ, Lucas-Derse S, Dean M. Linkage mapping of human polymorphic proteins identified by two-dimensional electrophoresis. Genomics. 1991;11:875–884. doi: 10.1016/0888-7543(91)90010-c. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet. 2001;10:973–982. doi: 10.1093/hmg/10.9.973. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg C, Soderstrom H, Giros B, Leboyer M, Gillberg IC, Bourgeron T, Paris Autism Research International Sibpair Study Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junaid MA, Pullarkat RK. Proteomic approach for the elucidation of biological defects in autism. J Autism Dev Disord. 2001;31:557–560. doi: 10.1023/a:1013242910574. [DOI] [PubMed] [Google Scholar]
- Junaid MA, Wu G, Pullarkat RK. Purification and characterization of bovine brain lysosomal pepstatin-insensitive proteinase, the gene product deficient in the human late-infantile neuronal ceroid lipofuscinosis. J Neurochem. 2000;74:287–294. doi: 10.1046/j.1471-4159.2000.0740287.x. [DOI] [PubMed] [Google Scholar]
- Kester MV, Norton SJ. The isolation and characterization of mouse liver glyoxalase I. Biochim Biophys Acta. 1975;391:212–221. doi: 10.1016/0005-2744(75)90168-0. [DOI] [PubMed] [Google Scholar]
- Krebs MO, Betancur C, Leroy S, Bourdel MC, Gillberg C, Leboyer M, Paris Autism Research International Sibpair (PARIS) Study Absence of association between a polymorphic GGC repeat in the 5′ untranslated region of the reelin gene and autism. Mol Psychiatr. 2002;7:801–804. doi: 10.1038/sj.mp.4001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Parr JR, Bailey AJ, Monaco AP. Autism: In search of susceptibility genes. Neuromolecular Med. 2002;2:11–28. doi: 10.1385/NMM:2:1:11. [DOI] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC, The Autism Genetic Resource Exchange Consortium A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69:327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Lyles GA, Chalmers J. The metabolism of aminoacetone to methylglyoxal by semicarbazide-sensitive amino oxidase in human umbilical artery. Biochem Pharmacol. 1992;43:1409–1414. doi: 10.1016/0006-2952(92)90196-p. [DOI] [PubMed] [Google Scholar]
- Maestrini E, Paul A, Monaco AP, Bailey A. Identifying autism susceptibility genes. Neuron. 2000;28:19–24. doi: 10.1016/s0896-6273(00)00081-7. [DOI] [PubMed] [Google Scholar]
- McCoy PA, Shao Y, Wolpert CM, Donnelly SL, Ashley-Koch A, Abel HL, Ravan SA, Abramson RK, Wright HH, DeLong GR, Cuccaro ML, Gilbert JR, Pericak-Vance MA. No association between the WNT2 gene and autistic disorder. Am J Med Genet. 2002;114:106–109. doi: 10.1002/ajmg.10182. [DOI] [PubMed] [Google Scholar]
- Miyata T, van Ypersele de Strihou C, Imasawa T, Yoshino A, Ueda Y, Ogura H, Kominami K, Onogi H, Inagi R, Nangaku M, Kurokawa K. Glyoxalase I deficiency is associated with an unusual level of advanced glycation end products in a hemodialysis patient. Kidney Int. 2001;60:2351–2359. doi: 10.1046/j.1523-1755.2001.00051.x. [DOI] [PubMed] [Google Scholar]
- Persico AM, D'Agruma L, Maiorano N, Totaro A, Militerni R, Bravaccio C, Wassink TH, Schneider C, Melmed R, Trillo S, Montecchi F, Palermo M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Conciatori M, Marino R, Quattrocchi CC, Baldi A, Zelante L, Gasparini P, Keller F, Collaborative Linkage Study of Autism Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatr. 2001;6:150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, LeCouteur A, Sim CH, Rutter M. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: A twin and family history study of autism. Am J Hum Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Walsh ES, Godwin AK, Tew KD. Cloning and characterization of human colon glyoxalase-I. J Biol Chem. 1993;268:5661–5667. [PubMed] [Google Scholar]
- Ray S, Ray M. Formation of methylglyoxal from aminoacetone by amine oxidase from goat plasma. J Biol Chem. 1983;258:3461–3462. [PubMed] [Google Scholar]
- Reichard GA, Jr, Skutches CL, Hoeldtke RD, Owen OE. Acetone metabolism in humans during diabetic ketoacidosis. Diabetes. 1986;35:668–674. doi: 10.2337/diab.35.6.668. [DOI] [PubMed] [Google Scholar]
- Ridderstrom M, Cameron AD, Jones TA, Mannervik B. Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I. J Biol Chem. 1998;273:21623–21628. doi: 10.1074/jbc.273.34.21623. [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, Nguyen L, Yang J, Harper C, Thorpe D, Vermeer S, Young H, Hebert J, Lin A, Ferguson J, Chiotti C, Wiese-Slater S, Rogers T, Salmon B, Nicholas P, Petersen PB, Pingree C, McMahon W, Wong DL, Cavalli-Sforza LL, Kraemer HC, Myers RM. A genomic screen of autism: Evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Mashima T, Yamamoto K, Tsuruo T. Modulation of heat-shock protein 27 (Hsp27) anti-apoptotic activity by methylglyoxal modification. J Biol Chem. 2002;277:45770–45775. doi: 10.1074/jbc.M207485200. [DOI] [PubMed] [Google Scholar]
- Shamsi FA, Sharkey E, Creighton D, Nagaraj RH. Maillard reactions in lens proteins: Methylglyoxal-mediated modifications in the rat lens. Exp Eye Res. 2000;70:369–380. doi: 10.1006/exer.1999.0800. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Donnelly RJ, Lackland H, Liu CG, Sohar I, Pullarkat RK, Lobel P. Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science. 1997;277:1802–1805. doi: 10.1126/science.277.5333.1802. [DOI] [PubMed] [Google Scholar]
- Speer O, Morkunaite-Haimi S, Liobikas J, Franck M, Hensbo L, Linder MD, Kinnunen PK, Wallimann T, Eriksson O. Rapid suppression of mitochondrial permeability transition by methylglyoxal. Role of reversible arginine modification. J Biol Chem. 2003;278:34757–34763. doi: 10.1074/jbc.M301990200. [DOI] [PubMed] [Google Scholar]
- Szatmari P. Heterogeneity and the genetics of autism. J Psychiatr Neurosci. 1999;24:159–165. [PMC free article] [PubMed] [Google Scholar]
- Tanna VL, Wilson AF, Winokur G, Elston RC. Linkage analysis of pure depressive disease. J Psychiatr Res. 1989;23:99–107. doi: 10.1016/0022-3956(89)90001-0. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. The glyoxalase system: New developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ. Population genetics of human glyoxalases. Heredity. 1991;67:139–142. doi: 10.1038/hdy.1991.73. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. Advances in glyoxalase research. Glyoxalase expression in malignancy, anti-proliferative effects of methylglyoxal, glyoxalase I inhibitor diesters and S-D-lactoylglutathione, and methylglyoxalmodified protein binding and endocytosis by the advanced glycation endproducts receptor. Crit Rev Oncol Hematol. 1995;20:99–128. doi: 10.1016/1040-8428(94)00149-n. [DOI] [PubMed] [Google Scholar]
- Van Herreweghe F, Mao J, Chaplen FWR, Grooten J, Gevaert K, Vandekerckhove J, Vancompernolle K. Tumor necrosis factor-induced modulation of glyoxalase I activities through phosphorylation by PKA results in cell death and is accompanied by the formation of a specific methylglyoxal-derived AGE. Proc Natl Acad Sci. 2002;99:949–954. doi: 10.1073/pnas.012432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Vieland VJ, Huang J, Swiderski RE, Pietila J, Braun T, Beck G, Folstein SE, Haines JL, Sheffield VC. Evidence supporting WNT2 as an autism susceptibility gene. Am J Med Genet. 2001;105:406–413. doi: 10.1002/ajmg.1401. [DOI] [PubMed] [Google Scholar]
- Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C. Prevalence of autism in a US metropolitan area. JAMA. 2003;289:49–55. doi: 10.1001/jama.289.1.49. [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcon M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, Grunn A, Juo SH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu X, Zhang C, Mundo E, Macciardi F, Grayson DR, Guidotti AR, Holden JJ. Reelin gene alleles and susceptibility to autism spectrum disorders. Mol Psychiatr. 2002;7:1012–1017. doi: 10.1038/sj.mp.4001124. [DOI] [PubMed] [Google Scholar]