Abstract
Eukaryotic viruses can maintain latency in dividing cells as extrachromosomal plasmids. It is therefore of vital importance for viruses to ensure nuclear retention and proper segregation of their viral DNA. The bovine papillomavirus (BPV) E2 enhancer protein plays a key role in these processes by tethering the viral DNA to the host cell chromosomes. Viral genomes that harbor phosphorylation mutations in the E2 gene are transformation defective, and for these mutant genomes, neither the viral DNA nor the E2 protein is detected on mitotic chromosomes, while other key functions of E2 in transcription and replication were wild type. Moreover, secondary mutations in both the E2 and E1 proteins lead to suppression of the phosphorylation mutant phenotype and resulted in reattachment of the viral DNA and the E2 protein onto mitotic chromosomes, suggesting that E1 also plays a role in viral genome partitioning. The E1 protein was cytologically always excluded from mitotic chromatin, either as a suppressor allele or as the wild type. In the absence of other viral proteins, an E2 protein containing alanine substitutions for phosphorylation substrates in the hinge region (E2-A4) was detected as wild-type on mitotic chromosomes. However, when wild-type E1 protein levels were increased in cells expressing either the A4 mutant E2 proteins or wild-type E2, the E2-A4 protein was much more sensitive to chromosomal dislocation than was the wild-type protein. In contrast, suppressor alleles of E1 were not capable of such abrogation of E2 binding (A4 or wild-type) to chromosomes. These results suggest that wild-type E1 can be a negative regulator of the chromosomal attachment of E2.
Some DNA viruses, such as papillomaviruses and lymphotropic herperviruses, maintain their genomes as stable episomal plasmids in the nuclei of infected cells. Papillomaviruses as a family usually infect the dividing basal epithelial cell layers and give rise to benign lesions (papillomas), while certain family members, such as bovine papillomavirus type 1 (BPV-1), can infect both epithelial and fibroblast cell types (7). Upon infection, the cells take up the virus, and the viral genome is transported to the nucleus, where it is kept as a multicopy plasmid. In this stage of infection the amplification of viral DNA and its stable maintenance have been modeled by viral amplification in transient DNA replication and stable transformation of cultured cells. The only players needed for replication and stable plasmid maintenance are the virally encoded E1 and E2 proteins and a plasmid containing the viral origin (17, 24, 25), making the BPV system a very useful model for the study of DNA replication and plasmid persistence in eukaryotic cells.
The virally encoded helicase, E1, is required for replication initiation and elongation (6, 7, 21, 28). E1 also binds specifically to the viral origin, and an assembly pathway targeting the initiator protein to the ori is regulated by the viral E2 protein (3, 4, 15, 19, 26, 27, 29). The multifunctional transcription factor E2 regulates gene expression from several viral promoters and enhances the functions of E1 by binding cooperatively with E1 to the viral origin (15, 20, 21, 29). A very stable heterotypic E2 dimer and E1 monomer complex can form without DNA, yet the physiological significance of this complex is unknown. Indeed, to date most studies have focused on the interactions between the two proteins, with an eye towards the ternary complex with DNA, which is stabilized by interactions between the amino-terminal activation domain of E2 and the carboxy-terminal helicase domain of E1. Two additional forms of the E2 protein, E2C and E8/E2, are N-terminal deletions of E2 that lack the transcriptional activation domain and act as repressors of E2-mediated transcription and replication (5, 10, 13).
In order to maintain the episomal viral genome in the nuclei of infected cells following mitosis, viruses like BPV and the large lymphotropic herpesviruses must ensure effective genome partitioning to the resulting daughter cells. If the viral genome is not somehow targeted to the nucleus, then following nuclear membrane reassembly, the genomes may be left behind in the cytoplasm and lost in the population of cells, either through degradation or dilution after cell division.
Several groups have shown that the E2 protein is a key player in the viral genome nuclear retention and segregation mechanism (2, 8, 11, 23). The cellular factor bound to mitotic chromosomes that serves as the receptor for viral attachment is not known. It is, however, clear that this factor, likely a protein, is conserved throughout many vertebrate species, as such E2 binding has been measured in hamster, mouse, and human cell lines (8, 11, 23; unpublished data). Studies have shown that the amino-terminal activation domain of E2 is by itself sufficient for chromosomal binding (2), though functional tethering of the viral plasmid requires both the activation and DNA-binding domains of E2. These data are consistent with the simple hypothesis that a reasonably abundant and evolutionarily conserved mitotic chromosomal protein binds the activation domain of E2 and the plasmid DNA hitchhikes onto chromosomes via binding to the DNA-binding domain of the viral protein.
Previously, we showed that the E2 protein is phosphorylated within the flexible hinge region and that at least one of the phosphorylation sites is critical for efficient viral transformation (11, 12). An E2 protein containing alanine substitutions at four serine residues in the hinge region (E2-A4), which serve as substrates for phosphorylation in wild-type E2, severely cripples viral transformation. Mutations of all four serines were required for this phenotype. The block in viral transformation is not due to insufficient DNA replication, since the E2-A4 as well as the E2-A3 mutant (E2-A3 has only three of the four serines substituted with alanine) resulted in even higher transient replication levels than that detected with the wild-type virus. The transformation defect of the E2-A4 mutant could be overcome by the substitution of A235 with aspartate, which is thought to mimic phosphorylation at serine residues. This result suggests that phosphorylation of the hinge region is critical for transformation.
Further insight into the role of the E2 protein in viral transformation was gained from a genetic screen for suppressors of the original A4 mutation. Isolation of BPV DNA from the few morphologically transformed foci that grew from an A4 transfection revealed that secondary mutations in both the E2 and E1 proteins could allow A4 transformation (11). In the context of these mutants and for wild-type viral genomes, a perfect correlation was uncovered between attachment of viral DNA and E2 protein to mitotic chromosomes and stable plasmid maintenance. With E2 mutants that could not be phosphorylated, neomycin-resistant colonies obtained by cotransformation with the drug resistance marker and viral plasmids quickly lost replicating viral DNA. These data thus genetically separated replication and transcription functions from a vital maintenance activity of E2.
How all of these data fit together was, however, not clear. For example, if the E2 protein by itself was sufficient for chromosomal binding, what role could E1 play in this process? The suppressor mutations that mapped to E1 established that phosphorylation of E2 was not obligatory for chromosomal attachment, and we speculated that E1 mutants could bypass this apparent requirement for modification (11). Moreover, recent work indicates that E2 phosphorylation leads to its degradation and that mutating phosphorylation sites increases E2 accumulation (16). In our laboratory, the mutant E2-A4 and wild-type alleles of E2 showed identical transactivation functions when tested in transient assays out of the context of the viral genome (12). This was perhaps due to the overexpression of E2 by the cytomegalovirus promoter in our vectors.
In the context of the viral genome, the apparent activities for transactivation by E2 as measured by the expression of a reporter cotransfected with the viral genomes showed a different phenotype for the wild type and E2-A4 mutant. The phenotypes of the viral genomes harboring the A4 allele of E2 have thus enhanced replication and transactivation activities (12), consistent with increased E2 activity and unstable plasmid maintenance. The rapid loss of viral DNA could not be ascribed to runaway replication for two reasons: (i) the E2-A3 mutant stably transforms cells with wild-type levels, and its transient replication was enhanced but identical to that of the genome harboring the E2-A4 allele and (ii) coselection with an integration drug marker resulted in equal numbers of drug-resistant colonies when comparing wild-type, E2-A3, and E2-A4 alleles. Thus, we concluded that a segregation function was affected.
In this work we show that wild-type E1 can block E2 binding to mitotic chromosomes and that the E2-A4 mutant expressed by itself is indeed as competent as the wild type for chromosome binding. The E2-A4 mutant, however, is more sensitive to the negative effect of wild-type E1, and the suppressor E1 mutants are not competent to block chromosomal attachment. These data are consistent with a model that has a pathway for the release of E2 from E1 in the tethering process.
MATERIALS AND METHODS
Cell culture.
C127, COS-7, and CMT4 cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal calf serum (FCS) supplemented with penicillin and streptomycin. Transfections through electroporation were carried out using a Gene Pulser (Bio-Rad) at 290 V and 960 μF, using 5 × 106 cells in 250 ml. The sample DNAs were cotransfected with 50 μg of sheared salmon sperm DNA as a carrier.
The cells were split into two to four plates posttransfection. When appropriate, neomycin-resistant (Neor) cells were selected with 1 mg of G418 (Gibco) per ml of medium 24 h posttransfection for 10 days, followed by maintenance in 0.5 mg of G418 per ml. Transfection with Lipofectamine (Gibco-BRL) was carried out using 1 μg of DNA, 50 μl of Lipofectamine, and 30 μl of PLUS reagent in serum-free medium. After 3 h the medium was replaced with serum-containing medium.
Plasmids.
The BPV genome plasmid pMLBPV has been described. The suppressor mutant genotypes were used in a repressor-minus environment. To knock out both E2 repressor forms E2C and E2/E8, the ATG codon for E2C was disrupted by a T to C change at nucleotide3092 and the splice donor AGGT was changed by a silent mutation to AGAT at nucleotide 1235.
Recombinant SV40 production.
SB11 DNA, a plasmid containing pBR322 and the simian virus 40 (SV40) genome without T antigen, and plasmid PAVA-RMV, recombinant SV40 DNA expressing the repressor-minus BPV E2 protein, were a generous gift from Dan DiMaio. The plasmids for expression of different E1 and E2 proteins were obtained by introducing a BamHI site immediately upstream of the ATG codon and immediately downstream of the stop codon of the respective gene. The fragments were cloned into the BamHI site of SB11. To release the SV40 fragment of SB11, the plasmid was digested with EcoRI and XmnI, and the purified SV40 DNA was religated overnight at 16°C with 1 U of ligase (NEB).
For transfection, 1 μg of religated recombinant SV40 DNA was mixed with 55 μl of Lipofectamine and incubated for 30 min. The DNA-Lipofectamine mix was brought up to 6 ml with serum-free medium and placed on top of CMT4 cells attached to the plastic dish and incubated for 3 h. After removing the medium, 10 ml of PAVA medium (DMEM, 10% FCS, HEPES [pH 7.2], Na2HCO3, 1 μM CdCl2, 100 mM ZnCl2) was added to the cells. CMT4 cells express SV40 T antigen under a metallothionine promoter, and therefore addition of the heavy metals induces T-antigen expression and enables viral amplification and packaging. After 3 days the cells were subjected to repeated freeze-thaw cycles. After removal of the cell debris by low-speed centrifugation, the supernatant was used for subsequent amplification. After three rounds of amplification, the virus titer was determined by immunofluorescence, employing an antibody directed against the viral capsid protein VP1.
The ratio of DAPI (4′,6′-diamidino-2-phenylindole)-stained cells to cells positive for VP1 gives a measure of the number of infected cells and infectious units per milliliter by a applying a standard Poisson equation (−ln [1 − number of infected cells/number of cells] = multiplicity of infection [MOI]). For infections, cells were plated on cover slips in six-well plates at a density of 1.7 × 105. The next day, different MOI of recombinant SV40 were allowed to adsorb in 1 ml of medium for 3 to 4 h at 37°C. After that, 2 ml more of medium was added to each well, and the cells were incubated for 24 to 48 h to allow protein expression.
Western blots.
To determine the amount of protein expressed or to check for correct protein molecular weight, ≈5 × 106 cells were taken up in 50 μl of loading buffer and boiled for 5 min. After a 30-s vortexing step to shear the genomic DNA, 5 to 20 μl of this extract was fractionated by electrophoresis through sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gels. After transfer to a polyvinylidene difluoride membrane, the proteins were detected with primary antibodies, followed by horseradish peroxide-conjugated secondary antibody and chemiluminescence reagents (Renaissance; NEN).
Protein immunolocalization.
Transformed C127 or cells infected with recombinant viruses were grown on cover slips in six-well plates (2 × 105) for 1 day. The experiments were performed as described previously (11). Briefly, the cells were fixed with 50% methanol-50% acetone for 5 min and rehydrated with phosphate-buffered saline (PBS) for 5 min. The slides were blocked with PBSTB (0.1% Triton X-100, 10%FCS, 3% bovine serum albumin [BSA] in PBS) for 30 min. Primary antibody was diluted in PBSTB to the appropriate dilution and allowed to bind for 1 h. After three 15-min washes with PBST, a goat anti-mouse immunoglobulin (Ig) secondary antibody coupled to either indocarbocyanine (Cy3) or fluorescein isothiocyanate was allowed to bind for 1 h in the dark. After three more washes with PBST, cells were counterstained with DAPI and mounted with 2% N-propylgallate (NPG; Sigma) in 70% glycerol. Fluorescence was detected by using either a Zeiss axioplan fluorescence microscope or a Leica TCSNT confocal laser scanning imaging system.
Coimmunoprecipitation.
Baculovirus-infected Sf9 cells were lysed in lysis buffer (100 mM Tris [pH 7.5], 100 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1 mM EDTA, 0.3% NP-40), and the extracts were incubated with anti-E1 antibody coupled to protein G-Sepharose for 30 min at 4°C. The E1 antibody was BPV104, a generous gift by A. Stenlund. After washing the BPV104 beads three times with a 10-fold excess of lysis buffer, the beads were incubated with 0.4% N-lauroylsarcosine in lysis buffer for 1 h at 4°C. N-Lauroylsarcosine is a detergent that disrupts protein-protein interactions, but under these conditions does not disrupt antibody-antigen interactions. Therefore, with an N-lauroylsarcosine elution, only the proteins that are bound to the antigen are eluted.
After detection of the E2 protein by Western blotting, the bands were quantitated using a Fluorchem 8000 (Alpha Innotech Corp.). E1 levels were also measured by quantitative chemiluminescence, and after normalization for the E1 protein levels, the value for the highest wild-type E1:E2 ratio for a given pair of interactions was set as 100% and the other values in one experiment were taken as a percentage of the maximum. The error bars for each point were calculated from three individual experiments.
RESULTS
E1 suppressor mutants restore E2-A4 colocalization onto mitotic chromosomes.
Previous work had established that E1 mutations could suppress the defective transformation phenotype of the E2-A4 allele and that this suppression was concomitant with viral DNA tethering to mitotic chromosomes. If the E2 protein is the key factor required for attachment of the viral genomes to the mitotic chromosomes, then the disrupted E2 localization in the A4 mutant should be reversed in the suppressor mutations. We therefore proceeded first to ask if, in cells stably transformed by the E2-A4/E1 suppressor genomes, the E2-A4 protein relocated to mitotic chromosomes.
In order to visualize the intact E2-A4 protein in such cells it was necessary to use viral genomes that do not encode the cross-reacting repressor forms of E2. These repressor forms, devoid of the activation domain, do not bind to chromosomes and obscure the immunocytological signal provided by the full-length E2 protein. Therefore, the cytological observation of E2 in stably transformed cells with wild-type BPV is impossible with current antibodies. When observing the intact E2 protein in these repressor-minus strains, we detected chromosomal association with the A3 genome in stable or transient assays and dislocation in the A4 variant in transient assays (11) (data not shown).
Interestingly, one or another of these repressor forms is required for stable morphological transformation and plasmid maintenance in a wild-type BPV-1 background (9), but in either the E2-A3 or E2-A4/E1 suppressor genotypes the repressor minus mutations do not show this defect (C. Lehman, C. Voitenleitner, and M. Botchan, unpublished observations). Cells stably transformed by the E2-A3 allele provided the only lines that showed a clear wild-type E1 signal, presumably because of the increased copy number and consequently higher levels of the E1 protein. The lines transformed with E2-A3, E2C−, and E8/E2− thus provide the best material to observe the compartmentalization of wild-type and suppressor forms of E1.
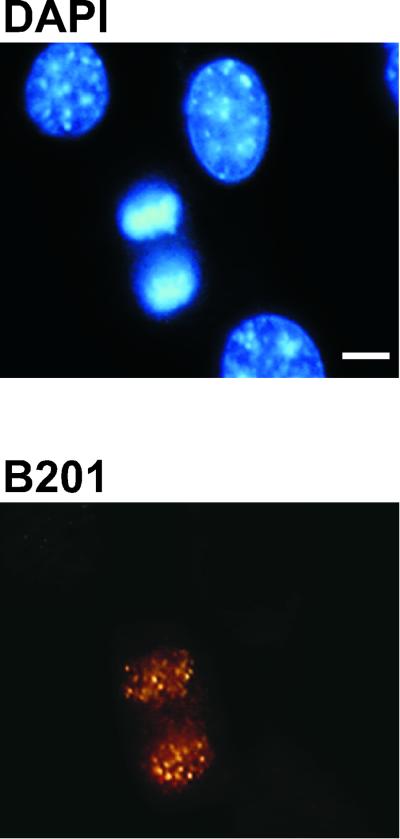
C127 cells were transfected with viral genomes harboring either the A3 mutation or several different A4 suppressor mutations. After drug-resistant colonies were selected and expanded, the transformed cells were maintained in G418-containing medium and stained with the E2 monoclonal antibody B201, and E2 localization in mitosis and interphase was monitored by immunofluorescence microscopy. For these experiments, a higher magnification was used than in our previous studies for E2 localization (11), and a punctate pattern rather than a diffuse E2 localization on mitotic plates could be uncovered (see, for example, Fig. 5). The use of high-magnification confocal microscopy also allowed us to take optical slices of the cell and more clearly distinguish the boundaries between condensed chromosomes and cytosol.
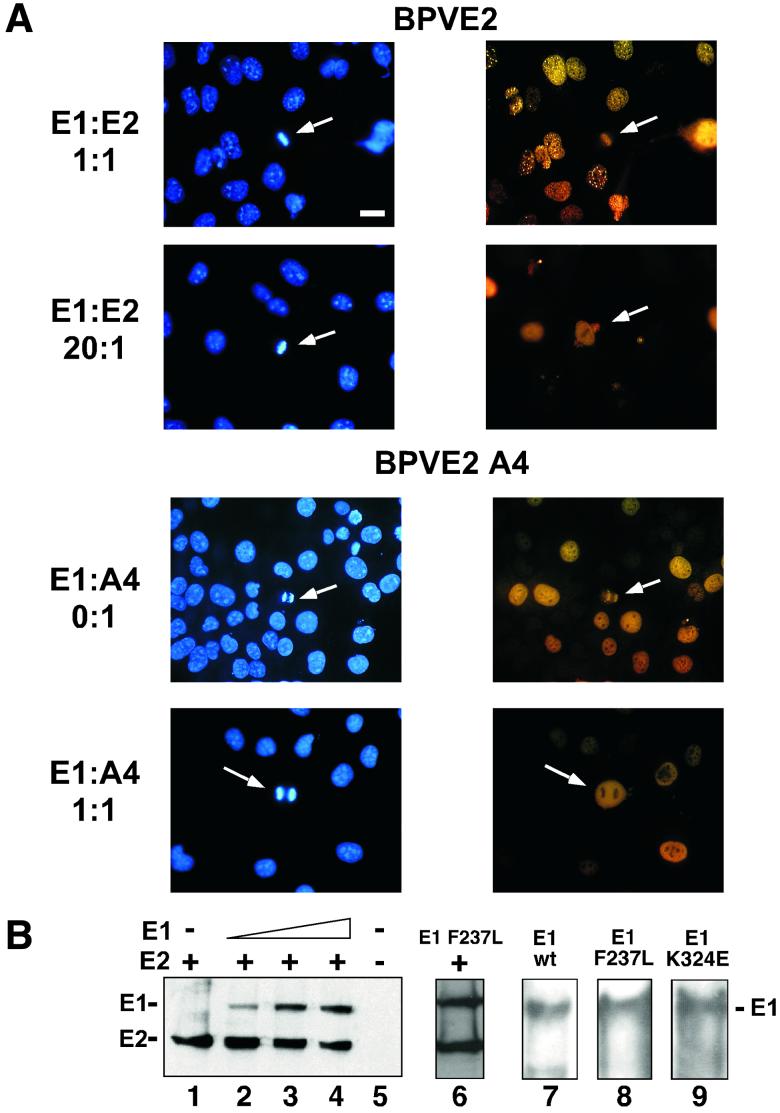
FIG. 5.
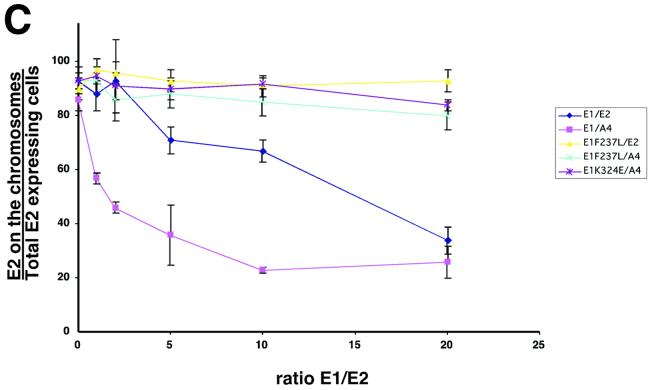
E1 expression is sufficient for E2 delocalization from mitotic chromosomes. (A) COS-7 cells were grown on cover slips and infected with a constant amount of PAVA-E2 or PAVA-A4 (1 MOI) and increasing amounts of PAVA-E1 (0 to 20 MOI). After 24 h, the cells were fixed, and E2 was detected by immunofluorescence. The panels show representative fields of E2 distribution for the outlined E1/E2 ratios. Right side, E2 immunostaining (yellow); left side, DAPI staining (blue). Bar, 20 μm. (B) Immunoblot analysis of PAVA-E2- and PAVA-E1-infected cell extracts. For detection, both the E1- and E2-specific monoclonal antibodies were used. Shown are extracts derived from COS-7 cell infections using constant amounts of PAVA-E2 (1 MOI) and increasing amounts of PAVA-E1 (0, 5, 10, and 20 MOI) (lanes 1 to 4, respectively), as well as mock-infected extracts (lane 5). At the levels of expression used for these and subsequent experiments, no differences in E2-A4 or wild-type levels were detected (data not shown). Lane 6 shows the level of the E1F237L mutant protein (10 MOI) and E2 (1 MOI). The points at which mutant and wild-type E1 accumulate to the same level as E2 are equivalent. Lanes 7 through 9 show the level of E1 that accumulated at an MOI of 10 for the wild-type, E1-F237L, and E1-K235E alleles in side-by-side infections from total cell extracts. The positions of the E1 and E2 protein bands are indicated on the left side of the panel. (C) Quantitative evaluation of the dislocation of the E2 proteins by wild-type and mutant E1 proteins. For each ratio of E1- and E2-encoding viruses, approximately 60 mitotic cells that were positive for E2 protein were examined for E2 localization. The percentage of mitotic cells that had chromosomally located E2 was plotted against the ratio of the two viruses. Shown are averages and the standard deviation for three experiments for each E1-E2 combination. ⧫, wild-type E1/wild-type E2; ▴, E1-F237L/wild-type E2; ▪, wild-type E1/E2-A4; ∗, E1-K324E/E2-A4; ×, E1-F237L/E2-A4).
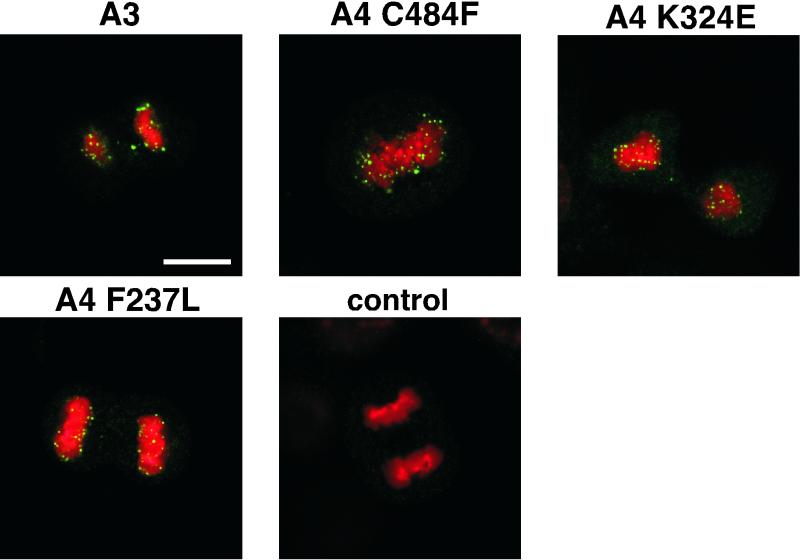
As shown in Fig. 1 the mutant E2 protein was, as anticipated, localized onto the mitotic chromosomes for three E1 suppressor mutants. In these E1 suppressor genomes, the E2-A4 was localized onto chromosomes in the same punctate manner as the A3 allele of E2. Thus, the E1 suppressor restores E2-A4 binding, as predicted. An E2 suppressor mutant, A4D255G (11), also resulted in E2's relocalizing back onto mitotic chromosomes (data not shown).
FIG. 1.
E2 protein is localized to mitotic chromosomes in suppressor mutant-transformed cells. Cells stably transformed with BPV-1 genomes of different genotypes were obtained via coselection with a Neor marker, and E2 proteins were detected by confocal microscopy using a Leica TCSNT confocal laser scanning imaging system. Green, E2 immunofluorescence with monoclonal antibody B201; red, DAPI staining of chromosomal DNA. For the confocal processing, we chose red for the DAPI signal. The upper left panel shows an anaphase mitotic cell from C127 cells transformed with the A3 mutant of BPV. The other panels show E1 suppressor mutants of the A4 genome of BPV at different stages of mitosis (E2-A4/E1 C484F, metaphase plate; E2-A4/E1 F237L, anaphase; and E2-A4/E1 324E, telophase). Mitotic figures from the nontransformed C127 cells did not stain for E2 protein (control). Bar, 10 μm. The E2-A4 mutant genome does not stably transform cells, and the localization of E2 for this allele from intact plasmids can only be assessed through transient assays and has been described previously (11); the protein in the genome context is not associated with mitotic chromosomes.
E1 does not colocalize with mitotic chromosomes.
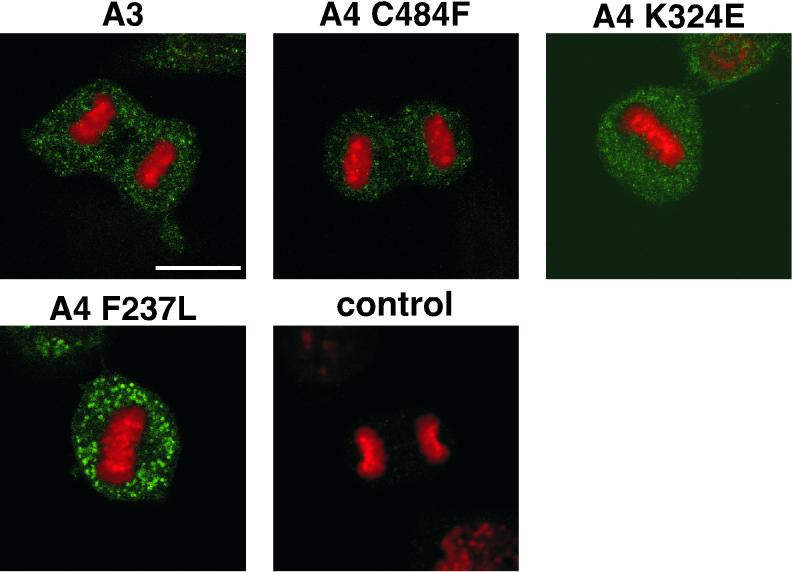
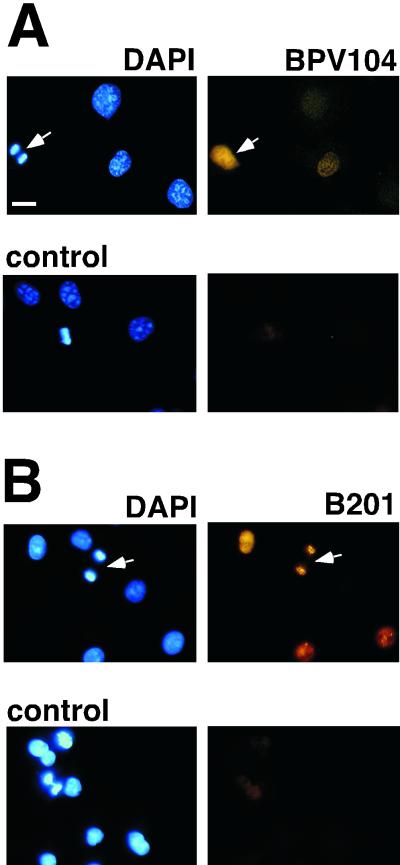
To explore the role of E1 in the localization of E2 in these transformed cells, we determined the cytological position of E1 itself in mitotic cells. If E1 were a cofactor involved in a complex with E2 on mitotic chromosomes, this might be evident upon immunostaining. Stably transformed cells harboring BPV genomes with the E2-A3 allele or the three E1 suppressor alleles within the E2-A4 allele were thus stained with a monoclonal antibody against the E1 protein (BPV104). E1 localization was examined following immunofluorescence microscopy. Figure 2 shows that in all cell lines examined, the E1 protein was not localized to mitotic chromosomes. The E1 protein was nuclear in interphase (data not shown) and was excluded from condensed chromosomes and localized outside of the chromatin during mitosis. These results suggest that E1 does not associate with mitotic chromosomes and does not play a role for maintenance of E2 attachment to the mitotic chromosome, but provide an insight into the roles that E1 might play in the initiation of this process.
FIG. 2.
Wild-type and mutant E1 proteins do not associate with mitotic chromosomes. The transformed cell lines described in Fig. 1 were used here. Wild-type and mutant E1 proteins were detected by confocal microscopy using monoclonal antibody BPV104, followed by Cy3-conjugated Ig secondary incubation (green). Red, DAPI staining of chromosomal DNA. Bar, 10 μm. We could not detect E1 by these cytological methods in wild-type BPV-1-transformed cells. Thus, the E2-A3 variant, which transforms stably and shows no segregation defects, served as the wild-type control. Control, nontransformed C127 anaphase cell.
E2-A4 mutant behaves differently outside the viral context.
To examine the chromosome binding of the E1 and E2 proteins in the absence of other virally encoded proteins and the influence of E1 on the chromosomal binding of E2, we used recombinant SV40 viruses that encoded either the E1 or E2 protein alone. These so-called PAVA viruses express the desired protein in place of large T antigen and can be replicated and packaged in simian cells expressing the SV40 large T antigen (22).
Figure 3A shows that when COS-7 cells were infected with a PAVA virus encoding E1, the E1 protein localized in mitotic cells as it did when E1 was expressed in the context of the entire genome: E1 was found in the nucleus during interphase and was not associated with mitotic chromosomes when the cells entered mitosis (Fig. 3A). A distinct chromosomal sparing of E1 stain was observed in mitotic cells.
FIG. 3.
PAVA-E2 and PAVA-E1 expressed protein is detected in COS-7 cells by immunofluorescence. COS-7 cells were grown on cover slips and incubated with 5 MOI of virus for 24 h. After fixation, E2 and E1 were detected by immunofluorescence with a Zeiss axioplan fluorescence microscope using monoclonal antibodies B201 and BPV104, respectively, and the DNA was stained with DAPI. As a negative control, mock-infected cells were incubated with B201 or BPV104, followed by the Cy3-conjugated secondary antibody. Bar, 20 μm.
As reported previously (23), we found that when the wild-type E2 protein was expressed alone, it was attached to mitotic chromosomes, as shown in Fig. 3B. Mock-infected cells showed no E1 or E2 signal. We infected COS-7 cells with a PAVA virus encoding E2-A4. This experiment was important even though others have reported that the hinge region of E2 can be deleted for mediation of tethering or chromosomal binding of E2 (1, 23). For example, perhaps only in the context of an intact protein would phosphorylation of the hinge region be of some significance to chromosomal binding per se. Comparing the intensities of the chromosomal signals for the wild-type and E2-A4 proteins observed at a variety of MOIs, we found no difference in the signal (Fig. 4). From these data we conclude that the behavior of the E2-A4 protein in binding to mitotic chromosomes in cells transformed by BPV-1 must be affected by other factors. Given the well-known interaction between E1 and E2 in forming complexes and the genetic interactions for tethering uncovered by the suppressor mutations in E1, we sought to measure the effects of E1 on E2 chromosomal binding.
FIG. 4.
E2-A4 protein by itself colocalizes with mitotic chromosomes. COS-7 cells were infected with the recombinant SV40 virus PAVAE2-A4, and after 24 h the E2-A4 protein was detected as described for Fig. 3. An infected anaphase cell is shown by fluorescence microscopy, using a Zeiss axioplan fluorescence microscope. Lower panel, E2-A4 immunofluorescence (yellow); upper panel, DAPI staining (blue). Bar, 10 μm.
E1 dislocates E2 from mitotic chromosomes.
The data presented above, taken together with our previous studies (11), suggest that either E1 transiently helps E2 to bind more efficiently to chromosomes (and the suppressor E1 allele might do so more efficiently) or that E1 interferes with E2 binding (and conversely that the suppressor alleles are deficient for this effect). We coinfected COS-7 cells with constant MOIs of the E2 PAVA virus and increasing levels of the E1 PAVA virus, as shown in Fig. 5. At 24 h postinfection, E2 protein localization was monitored by immunofluorescence microscopy.
In the case of the wild-type E1 and E2 proteins, E2 was routinely localized onto chromosomes at an E1/E2 viral ratio of 1:1 (Fig. 5A, first row of panels). At higher levels of the E1-encoding virus (20:1 ratio), the E2 protein was dramatically dislocated from the mitotic chromosomes (Fig. 5A, second row). Similarly, when the E2-A4-encoding virus was used, the A4 protein also dissociated from mitotic chromosomes (Fig. 5A, third and fourth rows). However, in this combination, only a 1:1 ratio of E1 to E2-A4 virus was sufficient for a significant increase in the number of cells with dislocated E2-A4 protein. In both cases of dislocation, a distinct chromosomal sparing for stain was detected on top of a diffuse but bright E2 staining of the entire cellular compartment.
Conspicuously, when the E2 protein binds to mitotic chromosomes by itself, essentially no staining other than chromatin association is observed. This reciprocal pattern (cytosolic, or condensed chromosome) was paralleled in the patterns reported previously for the A4 versus A3 protein in the context of the viral genome. Cytologically we did not detect a change in E2 brightness upon increasing E1 coinfections. Immunoblot analysis of PAVA virus-infected extracts showed that within the range of E1 virus levels used in these experiments, the E2 protein expression levels did not change significantly (Fig. 5B), suggesting that the dislocation of E2 from mitotic chromatin is not due to decreased E2 protein levels.
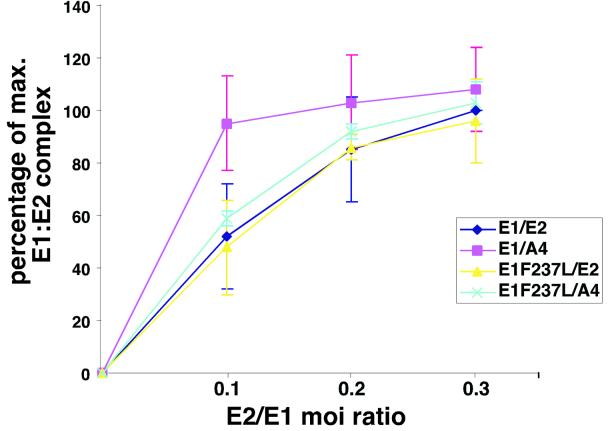
In order to quantitate the findings in Fig. 5B, we set up a series of infections with wild-type E2- or E2-A4-encoding PAVA viruses and increasing levels of wild-type E1- or E1 suppressor mutant-encoding PAVA viruses. For each ratio of viruses, the localization of the E2 protein in mitotic cells that expressed the E2 protein was scored and numbers were tabulated. For each point, at least 60 mitotic cells were scored, and an average for three separate experiments is presented. Figure 5C shows a graph depicting the percentage of cells with E2 localization onto the mitotic chromosomes versus the ratio of E1- and E2-encoding viruses. In the case of both E1 and E2 wild-type proteins, decreasing amounts of the E2 protein were found on mitotic chromosomes as the E1 virus levels were increased. However, for the wild-type case, a significant drop in the mitotic chromosome localization of E2 only occurred when the E1/E2 ratio was at 5:1, and at a 20:1 ratio of E1 to E2 virus levels, 40% of the cells examined still showed E2 protein localized on the mitotic chromosomes. In contrast, the A4 mutant was dislocated significantly at a ratio of 1:1 (Fig. 5C). As E1 PAVA virus levels increased, the percentage of E2-A4 bound to mitotic chromosomes decreased to 20%. We note that the endpoints for titrating E2 off chromosomes likely represent a pseudosaturation. We assume a Poisson distribution of infection multiplications per cell, and a significant fraction of cells will presumably not have the excess E1 virus.
In contrast to the wild-type E1, with the E1 suppressor mutant, increasing levels of virus encoding these proteins had no significant effect on either E2 or E2-A4 protein localization (Fig. 5C). From these data we conclude that the wild-type E1 protein interferes with E2 binding to mitotic chromosomes and that the E2-A4 mutant is more sensitive to this interference than wild-type E2. Moreover, the suppressor mutations in E1 abolish this blocking effect.
One notion that might help to explain the negative effects of E1 on E2 tethering function would be the formation of an E1-E2 complex that competes with E2 binding to a chromosomally bound factor. To partially explain the mutant phenotypes, we might posit that normally a phosphorylated E2 would weaken this complex and increase the pool size of free E2 for tethering. One direct approach to this speculation is to biochemically monitor E1-E2 complex formation between the E2-A4 protein and various E1 proteins and compare this to wild-type interactions. To do so in late G2 or mitotic stages would be ideal, but data relevant for such predicted differences in affinity are more reliably obtained from interactions between overexpressed forms in recombinant systems.
We set up a series of infections in Sf9 cells infected with baculoviruses encoding the E1 protein or the E1F237L mutant and increasing amounts of viruses encoding the E2 protein or the E2-A4 protein. Total soluble protein was captured by beads with an E1 antibody (BPV104). For each E1/E2 ratio point, we examined the amount of coimmunoprecipitated E2 protein with Western blotting utilizing an E2 monoclonal reagent. Figure 6 shows a graph depicting the percentage of E1-E2 complex as formed for each E1/E2 ratio at constant levels of E1. In the case of wild-type E1 and E2 protein, the percentage of E1-E2 complex increased, reaching a saturation point, which was determined by the set amount of E1 protein present in the extracts. For this figure, we assumed that each E1 molecule can bind one dimeric E2 molecule, so increasing the E2 protein concentration beyond a certain point as expected had no effect on the amount of E1-E2 complex once E1 is saturated.
FIG. 6.
Coimmunoprecipitation of E2 proteins with E1. Sf9 cells were infected with 1 MOI of baculoviruses encoding either E1 or mutant E1F237L proteins and increasing MOIs of baculoviruses encoding either wild-type E2 or E2-A4 protein. At 48 h postinfection, the cells were lysed, and the extracts were incubated with monoclonal antibody BPV104 coupled to protein G-Sepharose. After three wash cycles, the E2 protein was eluted with 0.4% N-lauroylsarcosine and detected by Western blotting. After quantitation of the bands, the values were normalized for equal E1 protein concentration and plotted against the E1/E2 MOI ratio, with the value of the highest wild-type E1/E2 ratio set at 100%.
At each multiplicity of infection for the wild-type E2 and E2-A4 virus, the levels of total soluble E2 protein detected were equivalent. When a virus encoding the E2-A4 protein was used, an equivalent level of saturation was reached, but at a lower E1/E2 ratio than in the wild-type case (Fig. 6). One likely explanation for this effect is that the E2-A4 protein has a higher affinity for E1 than does wild-type E2 and saturation is therefore reached at a lower level of E2 protein. Therefore, the two- to threefold difference in saturation point for the E1-E2 complex shown in Fig. 6 likely represents a modest increase in binding affinity for the E2-A4 protein to E1 relative to the wild-type E2. In the case of the mutant suppressor E1 protein, the graphs for both the E2 protein and the E2-A4 protein resemble that of the wild-type case. Thus, for the suppressor forms of E1, no measurable differences in affinities with a phosphorylated or hypophosphorylated form of E2 were apparent.
E1 and E2 levels are comparable in transformed C127 cells.
Our experiments suggested that the ratio of E1 to E2 is critical for maintenance of the viral genome in transformed cells, perhaps with some optimal transformation occurring when E1 and E2 levels are regulated so that there is not an excess of E1 to interfere with the critical late step in the cell cycle mediated by E2. Extrapolating from the data in Fig. 5, we might suggest that when E2 levels fall much below the levels of E1 in a wild-type E1 background, tethering will become problematic (especially in the A4 case).
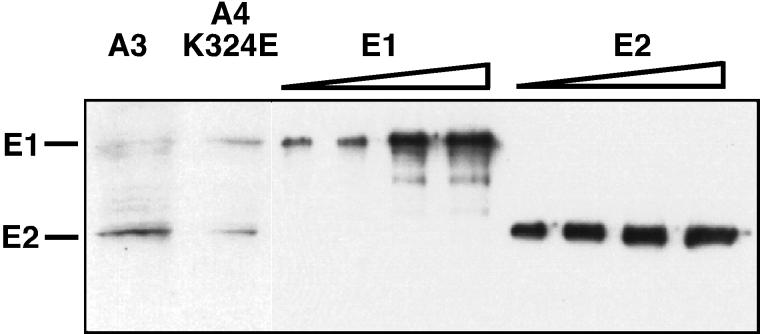
To determine the levels of the E1 and E2 proteins in transformed cell lines, which surprisingly have never been measured side by side, we performed immunoblot analysis on two transformed C127 cell lines: one transformed with the A3 genome (11), and the other transformed with an E1 suppressor mutant genome (K324E). To correct for different sensitivities between the E1 and the E2 antibody, increasing levels of Escherichia coli purified E1 and E2 were used as standards and processed in parallel to the extracts, and the signals were detected by quantitative immunoblot analysis (Fig. 7, lanes 3 to 10) (see Materials and Methods). The standards reveal that at low E1 and E2 levels, the signal for the E2 protein was about two to three times higher than the signal for the corresponding amount of E1 protein (compare Fig. 7, lanes 3 and 7 or lanes 4 and 8). From these results (Fig. 7, lanes 1 and 2), we can conclude that in the A3 case, the levels of the E1 and E2 proteins are about equal, whereas in the case of the E1 suppressor mutant, there seems to be only a slightly elevated E1 protein level. Unfortunately, in BPV-1 wild-type-transformed cells, the low levels of both proteins precluded an accurate assessment of the ratios. These experiments do show that in stably transformed cells of different genotypes, the ratio of E1 to E2 does not vary dramatically.
FIG. 7.
E1 and E2 are expressed at comparable levels in stably transformed cells. Shown is an immunoblot analysis of nuclear extracts derived from C127 cells that were transformed with the A3 or the E1-suppressor mutant K324E. The location of the E1 E2 proteins is indicated on the left. Lane1: A3-transformed cells; lane 2: E1 suppressor mutant K324E transformed cells; lanes 3-6: increasing amounts (1, 3, 5, 10 ng) of E. coli produced E1; lanes 7-10: increasing amounts (1, 3, 5, 10 ng) of E. coli produced E2.
DISCUSSION
BPV-1 establishes its genome in transformed cells as a multicopy nuclear plasmid with the ability to amplify its genome, and as such it seems likely that the key segregation function mediated by E2 is nuclear retention. Upon nuclear membrane breakdown, viral plasmids untethered to mitotic chromosomes may be lost after reassembly of the new nuclear membranes in late telophase. This model would be consistent with the very rapid and catastrophic loss of plasmids, as detected as sectored colonies by fluorescent in situ hybridization in earlier studies of the A4 mutant genome. It seems possible that the BPV-1 minichromosomes are too large to efficiently reenter the nucleus after cytosolic dispersion.
These issues clearly bring into focus the problem of how effectively and by what mechanisms these viruses deliver the initial minichromosomes to the nucleus in the first place. These questions aside, as E2 is not known to be associated with the virion, the A4 mutations in E2 do not diminish any direct replication or transcriptional activity of E2. Thus, the genetics clearly separated the tethering pathway from other known activities of the proteins. The central purpose of the present study was to understand more directly the nature of the mutations and the role played by the suppressor alleles of E1.
The key data and conclusions presented here are that phosphorylation of E2 must only indirectly affect the tethering activity of the regulatory protein. When expressed from recombinant PAVA viruses, no differences in chromosomal binding by the proteins were found even at limiting E2 levels. The other important finding is that E1 serves, when its levels increase significantly above those of E2, as a negative regulator of such tethering activities. It seems rather obvious but perhaps inescapable to posit that the E1-E2 complex forms in the cell and that this complex blocks the E2 activation domain from finding a chromosomally bound factor to interact with. What role would phosphorylation play in such a regulatory pathway? We suspect that there is no simple model as yet that can explain all of our data, though several points indicate that the tethering process may require a pathway rather then a simple binding step.
The disruption of the major phosphorylation sites in E2 at residues 298 and 301 by themselves lead to an increase in E2 trans-acting activity in transient assays (12) and increased E2 protein accumulation and a longer half-life for mutant E2 (16). A simple model might then include an overexpression of E2 in the A4 mutant genomic context, leading subsequently to an overexpression of E1. Such an imbalance of E1 could then in turn “squelch” E2 from its tethering functions. In previous studies, we found that the E2-A3 genomes stably transformed cells with a higher copy number than the wild type, perhaps because of increased E2 activity. However, no further increase in accumulated E2 levels could ever be detected in comparing the E2-A3 to the E2-A4 alleles in transient assays (12), yet the A4 genomes are severely transformation defective. Furthermore, the simplest model does not at all account for our finding that the E2-A4 protein is much more sensitive to E1 squelching than is the wild-type allele when the factors are expressed from recombinant vectors and chromosomal binding is measured (Fig. 5).
Another idea that must be considered for an understanding of the mutant phenotypes is that phosphorylation of E2 may disrupt E1-E2 complexes and that the A4 alleles of E2 allow a more stable complex and accumulate throughout interphase. Data from a previous study (14) indicated that E1 would only associate with E2 that was not modified by phosphorylation. Our data are consistent with a somewhat increased affinity (two- to threefold) between the unmodified and modified forms. This small difference may indeed be of significance when one considers the abundance and affinity that E2 might have for the unknown cellular chromosomal factor that E2 must interact with. Perhaps this factor is not as abundant as E1 and has a comparable binding affinity. In such a scenario, very small effects on E1-E2 interactions may be relevant. In the absence of any data relevant to receptor concentrations and affinities, such speculation is of course premature.
We must, however, point out that this concept—phosphorylation by itself destabilizes inactive complexes—does not simply explain why the suppressor alleles of E1 are not capable of squelching either the wild-type E2 or the A4 mutant form. In our experiments, both forms of E1 accumulated in mitotic cells to the same level, as judged from fluorescent signals, and no indication of differences in levels of proteins was measured by Western blotting (data not shown). Thus, the E1 suppressors have lost some activity that does not manifest in standard E1 experiments. For example, its ability to form a complex with the mutant and wild-type E2 forms of the protein is identical, a point that by itself would be somewhat problematic with a simple requirement for phosphorylation of E2 in the tethering pathway. In a final scenario, some combination of E1 levels, E2 modification, and perhaps other factors may be playing a role in this pathway.
The squelching effects of excess E1 on the mitotic tethering functions of E2 may also be relevant to the requirements for the E2C or E8:E2C repressor protein in the wild-type genome. In repressor-minus BPV-1 plasmids, transient replication is higher than wild type, yet stable transformation is not measurable. Moreover, in the E2-A3 background, the repressor forms are not required for stable transformation. A speculation that would explain these findings might be that for the wild-type case, the repressors are needed to keep E1 levels below a threshold to keep E1 from blocking E2's late step in the cell cycle for plasmid maintenance. In the case of the A3 allele, an increased level of E2 due to increased stability may in this case throw the balance between E1 and E2 back towards a manageable ratio.
We would posit that a single phosphorylation site in the E2-A3 allele at residue 235 in the hinge region can, when modified, serve to weaken the E1-E2 complex just enough to allow tethering. In the E2-A4 genome, this site for modification is not available, and E1 levels may be intolerable for stable maintenance. Continuing along this line of thought, the E1 suppressor mutations, through some unknown effect, alleviate the squelching behavior.
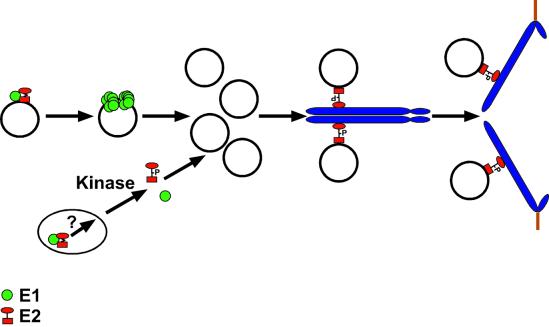
Figure 8 summarizes our current data and ideas pertaining to the roles of E2 and E1 in plasmid replication and tethering. To understand the E2-A4 allele phenotypes, an excess of E1 may squelch E2 chromosomal binding, but, as argued above, E2 modification by itself does not reasonably account for all these data. Clearly E1 may be modified (or unmodified) in post-S-phase periods, and such modifications as the sumoylation of BPV-1 E1 (18) do indeed affect E1 localization in the cell. Such loss (or gain) of modification or the association with other factors may release E2 from inactive E1-E2 complexes in the nucleus.
FIG. 8.
Model for the function of the E1 and E2 proteins in viral replication and nuclear retention of the viral genome during mitosis. E1 (green circles) and E2 (red circle/square) interaction facilitates the origin-specific binding of E1 to the viral DNA. It is believed that E2 must be removed in order for E1 to form double hexamers and perform its unwinding functions on viral origins for replication of the viral genome to occur (19). We propose that a second pool of E1-E2 complexes is present in the nucleus, especially when E1 levels are higher, as in the mutants. An unknown pathway that includes phosphorylation modification of the hinge region of E2 helps to disassemble such complexes. E2 can then bind to the pool of viral DNA that accumulates after S phase and, through interaction of the transactivation domain with a chromosomal receptor, associate with mitotic chromosomes. Phosphorylation of E2 proteins peaks in G2/M phase (15a). However, we do not know if chromosomally bound E2 is indeed phosphorylated. As the cellular chromosomes separate in mitosis, the tethered viral DNA is also segregated, ensuring that each daughter cell contains a nuclear copy of the viral genome following cell division.
A major goal is to uncover the nature of the chromosomal protein to which E2 binds for tethering. Perhaps insight into the behavior of this activity will provide further clues to the pathway by which stable BPV-1 plasmids are segregated.
Acknowledgments
We thank Anette Chan for help with the confocal microscopy. We gratefully acknowledge Eileen Beall and Eric Abbate for critical reading of the manuscript.
This study was supported by grants from the National Institutes of Health (CA-42414 and P30 ES01 896) and by a postdoctoral fellowship from the DFG (VO 800/1-1).
REFERENCES
- 1.Allikas, A., D. Ord, R. Kurg, S. Kivi, and M. Ustav. 2001. Roles of the hinge region and the DNA binding domain of the bovine papillomavirus type 1 E2 protein in initiation of DNA replication. Virus Res. 75:95-106. [DOI] [PubMed] [Google Scholar]
- 2.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 3.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G., and A. Stenlund. 2000. Two patches of amino acids on the E2 DNA binding domain define the surface for interaction with E1. J. Virol. 74:1506-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe, J., P. Vaillancourt, A. Stenlund, and M. Botchan. 1989. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J. Virol. 63:1743-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 7.Howley, P. M. 1995. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lipincott-Raven, Philadelphia, Pa.
- 8.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which Is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert, P. F., B. C. Monk, and P. M. Howley. 1990. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 64:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69-78. [DOI] [PubMed] [Google Scholar]
- 11.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehman, C. W., D. S. King, and M. R. Botchan. 1997. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol. 71:3652-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim, D. A., M. Gossen, C. W. Lehman, and M. R. Botchan. 1998. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J. Virol. 72:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusky, M., and E. Fontane. 1991. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequences within the carboxyl terminus of E1. Proc. Natl. Acad. Sci. USA 88:6363-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 15a.Nottoli, T. 1992. Regulation of bovine papillomavirus transformation and gene expression. Ph.D. thesis, University of California, Berkeley.
- 16.Penrose, K. J., and A. A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 74:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 18.Rangasamy, D., K. Woytek, S. A. Khan, and V. G. Wilson. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999-38004. [DOI] [PubMed] [Google Scholar]
- 19.Sanders, C. M., and A. Stenlund. 2000. Transcription factor-dependent loading of the E1 initiator reveals modular assembly of the papillomavirus origin melting complex. J. Biol. Chem. 275:3522-3534. [DOI] [PubMed] [Google Scholar]
- 20.Sedman, J., and A. Stenlund. 1995. Cooperative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo, Y. S., F. Muller, M. Lusky, E. Gibbs, H. Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Settleman, J., and D. DiMaio. 1988. Efficient transactivation and morphologic transformation by bovine papillomavirus genes expressed from a bovine papillomavirus/simian virus 40 recombinant virus. Proc. Natl. Acad. Sci. USA 85:9007-9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, V. G., and J. Ludes-Meyers. 1991. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J. Virol. 65:5314-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 28.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, L., I. Mohr, R. Li, T. Nottoli, S. Sun, and M. Botchan. 1991. Transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction Cold Spring Harbor Symp. Quant. Biol. 56:335-346. [DOI] [PubMed] [Google Scholar]