Abstract
Compared with the MHC of typical mammals, the chicken MHC is smaller and simpler, with only two class I genes found in the B12 haplotype. We make five points to show that there is a single-dominantly expressed class I molecule that can have a strong effect on MHC function. First, we find only one cDNA for two MHC haplotypes (B14 and B15) and cDNAs corresponding to two genes for the other six (B2, B4, B6, B12, B19, and B21). Second, we find, for the B4, B12, and B15 haplotypes, that one cDNA is at least 10-fold more abundant than the other. Third, we use 2D gel electrophoresis of class I molecules from pulse-labeled cells to show that there is only one heavy chain spot for the B4 and B15 haplotypes, and one major spot for the B12 haplotype. Fourth, we determine the peptide motifs for B4, B12, and B15 cells in detail, including pool sequences and individual peptides, and show that the motifs are consistent with the peptides binding to models of the class I molecule encoded by the abundant cDNA. Finally, having shown for three haplotypes that there is a single dominantly expressed class I molecule at the level of RNA, protein, and antigenic peptide, we show that the motifs can explain the striking MHC-determined resistance and susceptibility to Rous sarcoma virus. These results are consistent with the concept of a “minimal essential MHC” for chickens, in strong contrast to typical mammals.
Keywords: antigen presentation, avian, essential, evolution, minimal
Compared with typical mammals such as humans and mice, the chicken MHC is simpler and smaller, with the genes arranged differently (1-5). Moreover, the chicken MHC can determine decisive resistance and susceptibility to several infectious pathogens, as well as response to both live and killed commercial vaccines (6-9). The initial observations were made >40 y ago, when poultry breeders attempting to increase resistance to Marek's disease found large genetic effects attributable to a few loci. One major locus was the B blood group (10), later shown to contain the chicken MHC. Another well studied example is the MHC-determined response to Rous sarcoma virus (RSV) (8, 11, 12). Upon infection by RSV, transformation by the viral oncogene v-src leads to the appearance of tumors, which either regress or progress depending strongly on the MHC haplotype and on the presence of CD8-bearing cells.
Based on the fragmentary evidence for a single expressed classical class I molecule in chickens, we developed the hypothesis of a “minimal essential MHC” to explain the functional differences of the human and chicken MHCs in terms of differences at the structural level (3-5). In this view, the properties of the single class I molecule (and a single class II molecule) expressed by chickens can determine whether an individual chicken will respond to a particular pathogen (leading to strong MHC associations for that pathogen), whereas the multiple class I molecules expressed by humans confer response to most pathogens (leading to weaker associations).
However, a large and variable number of fragments was known to hybridize with class I and class II B probes in Southern blots of chicken genomic DNA (13). Moreover, two classical class I and two classical class II B genes were found to be present in the B-F/B-L region of the B locus, the region that corresponds to the classical MHC, in the B12 haplotype (3-5). We have reported that there is a single dominantly expressed class II B gene in common chicken MHC haplotypes (14). In this article, we examine classical class I genes at the levels of cDNA, protein, and antigenic peptide in several MHC haplotypes, and use this information to provide further evidence for the minimal essential MHC hypothesis.
Results
There Are Two Classical Class I cDNAs Found in Most Chicken MHC Haplotypes. At the start of this work, it had been reported that there were four classical class I genes in the chicken MHC (B12 haplotype), but studies of two haplotypes (B12 and B19) had reported cDNA clones for only one gene (2, 15).
To determine the number of classical class I genes expressed, we used a classical class I probe to exhaustively screen three cDNA libraries, each from a different tissue from a chicken line with a different MHC haplotype (B4, B12 and B19). For all three libraries, we found cDNA clones corresponding to two different genes (details in the legend to Fig. 1).
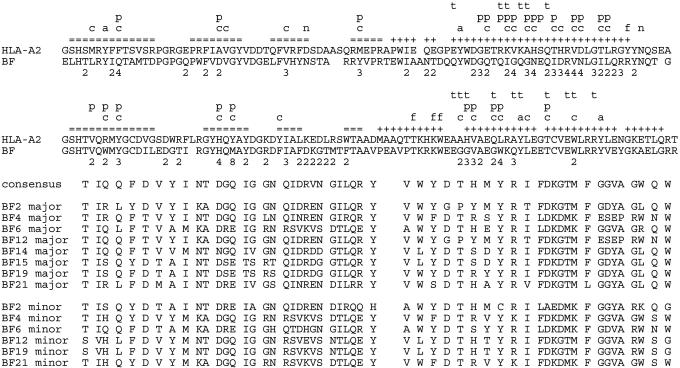
Fig. 1.
Amino acid sequences of the α1 and α2 domains derived from class I cDNAs of eight chicken MHC haplotypes amplified from purified B cells, T cells, or thrombocytes. (Upper) Comparison of the sequences of human HLA-A2 and a consensus sequence of the 14 chicken class I sequences reported in this article. Structural features above the HLA sequence are derived from the HLA structures and previous comparisons (15-22): +, α-helix; =, β-strand; a, pocket A; f, pocket F; c, other peptide contacts; t, T cell contact; n, glycosylation site; p, polymorphic residue in mammalian class I sequences. Numbers below the chicken class I sequence are the number of different residues found at the variable positions of the 14 chicken class I sequences reported in this article. (Lower) Comparison of all residues that differ between sequences for H.B2 line, B2 major (Z54321) and minor (Z54322); CC line, B4 major (Z54323) and minor (Z54324); GB2 line, B6 major (Z54330) and minor (Z54325); CB line, B12 major (Z54329) and minor (Z54314 with T4S); H.B14 line, B14 (Z54315 with I109T); H.B15 line, B15 (Z54316); H.B19 line, B19 major (Z54317) and minor (Z54318 with T4S); H.B21 line, B21 major (Z54319) and minor (Z54320). The sequences correspond to those isolated from cDNA libraries for CC spleen (B4 major, Z54362; B4 minor, Z54363), CB bursa (B12 major, Z54326; B12 minor, Z54359), H.B19 spleen (B19, M84766), and H.B19 intestine (B19 major, Z54360; B19 minor, Z54361).
To determine whether this finding was true for all of the available chicken lines, RT-PCR was performed on RNA from three cell types (T cells, B cells, and thrombocytes) from 10 lines carrying eight chicken MHC haplotypes, by using oligonucleotides based on conserved regions of the six sequences from cDNA clones (Fig. 1). We found sequences corresponding to two genes in six haplotypes (B2, B4, B6, B12, B19, and B21), but only one sequence in two other haplotypes (B14 and B15). These sequences have all of the features of classical class I molecules in chickens, based on the analysis of the B19 sequence (15). Interestingly, one of the sequences from B4 is identical to one from B21, and one from B12 is only one nucleotide different from one from B19.
There Is a Dominantly Expressed Class I Molecule at the Level of RNA in Three Common Chicken MHC Haplotypes. As mentioned above, the literature contained only reports of a single cDNA expressed in the B12 and B19 haplotypes. In the more thorough screening of B4, B12, and B19 libraries, we found cDNA sequences corresponding to two genes, but one was more abundant than the other (see legend to Fig. 1). For the haplotypes B12 and B19, the more abundant sequence corresponded to the previously reported sequence.
We quantitated this difference in the RT-PCR experiments. By counting the number of clones isolated from 32 independent amplifications of cDNA from three blood cell types, we found that overall one clone was present at approximately 10-fold the abundance of the other, as shown here for the three haplotypes B4, B12, and B15 (Table 1). We called the more and less abundant cDNAs the major and minor sequences, respectively.
Table 1.
Number of randomly selected clones designated major or minor sequence (as in Fig. 1) isolated from class I α chain sequences amplified from cDNA derived from purified peripheral blood B cells, T cells, and thrombocytes
| Sequence | B cells | T cells | Thrombocytes |
|---|---|---|---|
| B4 major | 10, 8 | 10 | ND |
| B4 minor | 0, 1 | 0 | ND |
| B12 major | 8, 7 | 10 | 9 |
| B12 minor | 2, 2 | 0 | 1 |
| B15 major | 10 | ND | 10 |
Two amplifications were performed for B cells from B4 and B12. ND, not determined.
For the B12 haplotype (5), the major sequence derives from the BF2 gene, and the minor sequence derives from BF1 gene. In agreement, some of the major but no minor sequences are identical with sequences amplified from cDNA by using oligonucleotides based on the so-called B-FIV (now BF2) gene of the B12 haplotype (16). Thus, despite having two class I genes, there is only one RNA expressed at a high level.
There Is a Dominantly Expressed Class I Molecule at the Level of Protein in Three Common Chicken MHC Haplotypes. We had previously found that there was a single N-terminal protein sequence found for class I molecules isolated from H.B19 spleen cell lysates using the mAb F21-21 to chicken β2-microglobulin (β2m) (23).
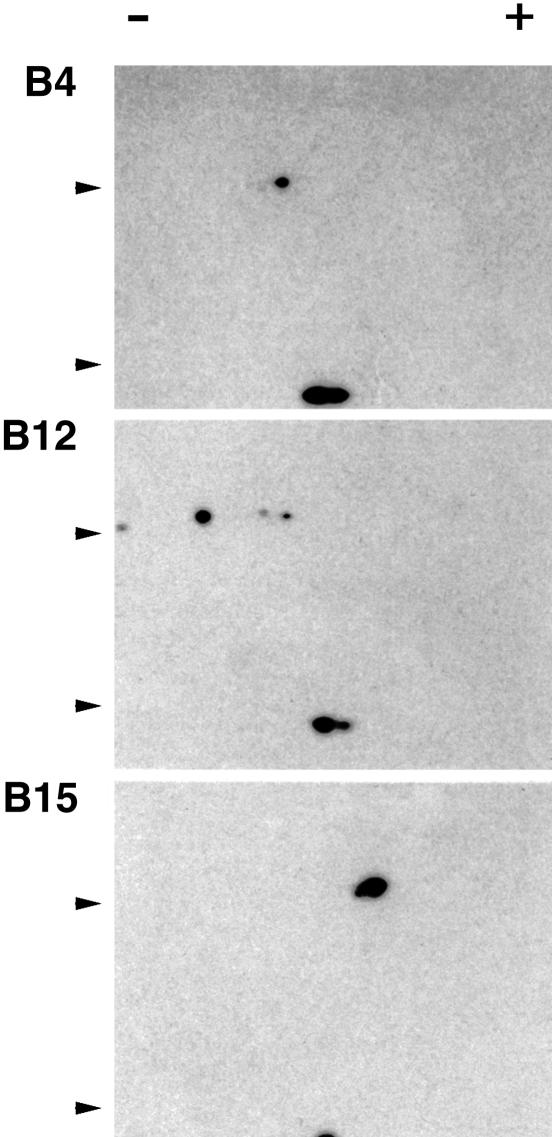
To see whether there was a dominantly expressed class I molecule expressed by other chicken MHC haplotypes, we performed 2D gel electrophoresis on F21-21 immunoprecipitates from pulse-labeled peripheral blood leukocytes (PBLs). Using an antibody to the invariant β2m subunit, we were able to capture all assembled class I molecules regardless of heavy chain sequence, and using pulse-labeled cells, we were able to analyze single protein species without confounding biosynthetic modifications. Under these conditions, there was a single spot for the heavy chain in the B4 and B15 haplotypes, and a single major spot for the B12 haplotype (Fig. 2).
Fig. 2.
Shown is 2D gel electrophoresis of class I heterodimers isolated from pulse-labeled PBL. Negative and positive signs indicate acidic and basic ends of the isoelectric focusing gel; arrows indicate markers of ≈18 and 45 kDa on SDS gel.
There Is a Dominantly Expressed Class I Molecule at the Level of Antigenic Peptide in Three Common Chicken MHC Haplotypes. At the start of this work, there were no reports of peptide motifs for MHC molecules outside of mammals (24). We used the mAb F21-21 to β2m to isolate all class I molecules from red blood cells, and peptides were eluted and separated by reverse phase chromatography. The total pool of peptides was sequenced by Edman degradation, and individual dominant peptides were sequenced by both Edman degradation and electrospray tandem mass spectrometry (MS/MS).
By pool sequencing, very clear motifs were found (Fig. 3A). The lengths of the peptides and the positions of anchor residues correspond to those expected for mammalian class I molecules: mostly octamers and nonamers, with anchors at peptide positions 2 and 8 for B15 (like most mammalian class I molecules), peptide positions 5 and 8 for B12 (like some mouse class I molecules), and peptide positions 2, 5, and 8 for B4. The discrimination of anchor positions is at least as great as mammalian class I molecules, with only one or two dominant amino acids found in each anchor position, but with some other amino acids found in low amounts at the C-terminal position. There are also strongly preferred residues at other positions, like peptide position 1 for B15, which would not have been expected to occupy deep pockets in the class I peptide-binding domain based on mammalian class I structures (17-22).
Fig. 3.
Peptide-binding motifs for three chicken strains. (A) Sequences of peptides bound to class I molecules isolated from three chicken strains determined from peptide pools showing anchor and strong and weak signals. Basic residues in blue, acidic residues in red, all others in black. (B) Sequences of individual peptides, with potential source proteins: peptide 1, chicken tubulinβ6 (P09207; peptide found in draft chicken genome sequence in contig 296.1 on chromosome 1); peptide 2, chicken DEAD-box helicase (NP990158, contig 7.29 on 1); peptide 3, FUS proto-onc gene (P35637 representative of peptide sequence in many species); peptide 4, elongation factor α chain (Q6DYT7 representative; 52.85 on 21); peptide 5, chicken β2m (P21611; 9.294 on 6); peptide 6, vimentin (Q9MZA9 representative; 2.352 on 2); peptide 7, chicken hemoglobin β chain (P02112; 1785.2 on 1); peptide 8, chicken ubiquitin (Q9PST8; 37.143 on 15); peptide 9, chicken MHC class I α chain (Q31412; contig 368.19 on 16); peptide 10, chicken calcium binding protein (Q90940; 71.39 on 10); peptide 11, chicken hsp70 (P08106, similar to 7.547 on 1); peptide 12, chicken 60S ribosomal protein L24 with terminal W (P83731, 34.64 on 1); peptide 13, proteasome subunit β type (Q59GN1 representative, 64.178 on 3). (C) Peptide anchor residues in large letters (or question marks for unknown) superimposed on a model of class I α1 and α2 domains with those residues of the major (above) and minor (below) class I sequences that are both polymorphic and potentially peptide contacts indicated as smaller letters; numbering based on HLA-A2 sequence.
Nearly all of the sequences of the individual peptides (Fig. 3B) fit the motifs from the pool sequences exactly, with the exception of a C-terminal overhang in a B12 peptide [as seen for some peptides (18)] and the presence of a residue in the C-terminal anchor position of a B4 peptide, which is present at low levels in the pool sequences.
The peptide motifs are consistent with peptides binding the major but not the minor class I molecules, based on display of the peptide motifs on simple wire models of the peptide-binding domains (Fig. 3C). The model of the major B4 sequence has four positively charged residues in the peptide-binding domain (R9, R81, R114, and R155), consistent with binding the negatively charged residues at peptide positions 2 and 5, and glutamic acid at the C-terminal position. In contrast, the model of the minor B4 sequence has only one of these positively charged residues. Similarly, the model of the major B12 sequence has hydrophobic residues (including L81, W97, Y99, Y114, and M116) that correspond well with the valine or isoleucine at peptide position 5 and valine at the C-terminal position, whereas the minor B12 sequence has polar and charged residues instead. The model of the single B15 sequence has E63 on top of the helix, D24 in the region of pocket B, and a cluster of hydrophobic residues in the region of pocket F (particularly L81), consistent with binding the positively charged residues pointing up at peptide position 1, arginine at peptide position 2, and hydrophobic residues at the C-terminal position.
The Peptide Motifs of the Dominantly Expressed Class I Molecules Can Explain the MHC-Determined Response to RSV. The most important consequence of the expression of a single dominant class I molecule is that each MHC-homozygous chicken strain should be able to bind peptides from, and therefore respond to, some small pathogens and vaccines but not others (3-5). In fact, it has long been known that chicken strains with some MHC haplotypes survive infection by certain pathogens, whereas chicken strains with other MHC haplotypes do not (6-9).
A particularly well studied system is the infection of chickens with RSV, the classic transforming retrovirus, for which the MHC is the major genetic locus responsible for progression or regression of tumors (6-9, 11, 12). The B4 and B15 haplotypes confer susceptibility (progression) to infection by RSV Prague strain C (and many but not all RSV strains), whereas the B12 haplotype confers resistance (regression). We analyzed polypeptide sequences from the four RSV Prague strain C genes for the number of peptides that might bind to the major class I molecules of these haplotypes (Table 2). Some RSV peptides were identified for the susceptible haplotypes, but >10 times more were found for the resistant haplotype.
Table 2. Number of peptides predicted from the sequence of RSV Prague strain C that fit motifs.
| Haplotype | gag | pol | env | src |
|---|---|---|---|---|
| B4 | 3 | 5 | 3 | 2 |
| B12 | 31 | 47 | 43 | 13 |
| B15 | 1 | 3 | 1 | 1 |
For mammals, it is known that only a fraction of peptides that fit the simple kind of motif used here actually bind the corresponding class I molecule with measurable affinities (22). Therefore, we used synthetic peptides from the v-src gene to assess binding to chicken PBL, by an assay analogous in procedure to the RMA-S and T2 stabilization assays using mammalian cells with defective TAP genes (25, 26). None of the few v-src peptides predicted to bind to cells of the susceptible haplotypes B4 and B15 actually did bind above background, whereas three of the v-src peptides predicted to bind B12 cells bound, one of them strongly (Table 3). We have reported that vaccination of B12 chickens with the strongly bound peptide confers protection to RSV (27).
Table 3. Flow cytometry of class I molecules on peripheral white blood cells incubated overnight with peptides.
| Control | Predicted | ||
|---|---|---|---|
| B4 | |||
| None | 1674 | KDAWEIPRE | 1677 |
| ADVEEYEE | 1898 | REVLDQVE | 1698 |
| GTVPVGRV | 1679 | ||
| KRLIGKRY | 1671 | ||
| B12 | |||
| None | 1834 | ANILVGENL | 1711 |
| ADVEEYEE | 1839 | GENLVCKV | 2074 |
| GTVPVGRV | 2424 | GRFTIKSDV | 1857 |
| KRLIGKRY | 1863 | KHYKIYKL | 1874 |
| LAGGVTTFV | 2038 | ||
| LPACVLEV | 2470 | ||
| QEAQVMKKL | 1844 | ||
| RMNYVHRDL | 1784 | ||
| SEEPIYIV | 1727 | ||
| SEEPIYIVI | 1886 | ||
| TRVAIKTL | 1845 | ||
| VNREVLDQV | 1726 | ||
| YPGMVNREV | 1894 | ||
| B15 | |||
| None | 1788 | ARLIEDNEY | 1767 |
| ADVEEYEE | 1739 | ||
| GTVPVGRV | 1742 | ||
| KRLIGKRY | 2005 |
Discussion
We have shown here that there are two class I genes expressed in most chicken MHC haplotypes, that there is a dominantly expressed chicken class I molecule at the level of RNA, protein, and antigenic peptide, and that the peptide motif of this dominantly expressed class I molecule determines resistance and susceptibility to an important poultry pathogen. Taken together, these data are a cornerstone for the concept of a minimal essential MHC, in which the structural organization of the chicken MHC has striking effects on gene function (3-5). Recently, other data that also support this concept have been published (14, 16, 28-30).
Some years ago, we reported the peptide motifs for the class I molecules of the B4, B12, and B15 haplotypes, the only peptide motifs for MHC molecules described outside of mammals (3). In this report we refine and present the evidence for these motifs, which superficially are much like those for mammalian class I molecules, but in fact have the following important differences.
First, the presence of glutamic acid as the dominant residue in the C-terminal position of the B4 motif is unprecedented, because there are no examples in mammals of negatively charged residues at the C-terminal anchor position. It has been suggested that this bias in mammals is due, at least in part, to the specificity of the inducible proteasome components LMP2 and LMP7 (31, 32). Such genes may be different, defective, or absent in chickens, at least in the B4 haplotype. In fact, genes for inducible proteasome components were not found in the chicken MHC sequence (B12 haplotype) or indeed in the draft chicken genome sequence (B21-like haplotype) (5, 33).
Second, even simple models indicate that the way in which chicken class I molecules bind their peptides may differ significantly from what is the norm for mammals. Many if not most human class I molecules have a deep pocket (pocket B) for binding peptide residue 2, which reaches under the α-helix of the α1 domain to interact with class I residue 45, a very polymorphic residue in humans. For instance, the HLA-B27 motif has an arginine at peptide position 2, which interacts with glutamic acid at position 45 at the bottom of pocket B. For the chicken B15 haplotype, the motif also has an arginine at peptide position 2, but there is a small hydrophobic alanine instead of a negatively charged residue at position 45. It is almost certain that the arginine at peptide position 2 interacts with the aspartic acid at position 24, with a side-chain that points up from the β-sheet. In fact, the stringent motifs reported here are not a consequence of specific binding to deep pockets in the class I molecule but are mostly a consequence of the translocation specificity of the polymorphic chicken TAP molecules (ref. 34 and B. Walker, L.G.H., and J.K., unpublished results).
Materials and Methods
Animals. Highly inbred MHC-congenic lines CC and CB originating in Prague, MHC-homozygous lines H.B2, H.B14, H.B15, H.B19, and H.B21 originating in Copenhagen and GB2 originating in Iowa were all bred and maintained at the Gipf (Oberfrick) Farm of the Basel Institute for Immunology.
Determination of Class I Sequences. Blood taken in heparin was centrifuged at slow speed (4 vol of PBS, 60 × g, 15 min, room temperature), and the top 1/3 and the resuspended pellet were separately centrifuged onto Ficoll-Paque cushions (150 × g, 10 min, 4°C), to collect from the interface the gray dispersed leukocytes in the first case, and the yellow clumped thrombocytes in the second case. B cells or T cells were isolated to >95% purity (after reanalysis) by using a FACstar-plus (Becton Dickinson), after staining with rabbit anti-mouse Ig coupled with fluorescein that had been previously absorbed with chicken erythrocytes or with mAb CT3 to chicken CD3 (35), followed by the absorbed anti-mouse reagent. RNA was isolated by guanidinium thiocyanate/acid phenol procedure at 106 cells per ml, and reverse-transcribed by using oligo(dT) or random hexamer primers and Moloney murine leukemia virus reverse-transcriptase (36). Sequences were amplified by using Taq polymerase (Roche Diagnostics, AG), primers 9447 (CGAGCTCCATACCCTGC) and 9451 (CTCCTGCCCAGCTCAG), and AMS Protocol thermocycler (Biometra, Tampa, FL) with 30 cycles of 7 s at 94°C, 15 s at 58°C, and 2 min at 72°C. The resulting single bands were cloned into dT-tailed pCR II plasmid vector as per the manufacturer's instructions (Invitrogen). Minipreps from randomly chosen clones were sequenced by using dideoxy fluorescently labeled terminators with 373A DNA sequencer (Applied Biosystems).
Pulse-Labeling and 2D Gel Electrophoresis. Peripheral blood was treated similar to above (slow speed with equal volume PBS, 60 × g, 15 min, 4°C), with clear upper half Ficolled as above. Dispersed gray PBL were collected at the interface, with care taken to avoid yellow thrombocyte clumps at the side, washed in warm medium (37) lacking Met, resuspended at 3 × 107 cells and 0.5 mCi (1 Ci = 37 GBq) of [35S]Met per ml, and incubated for 20 min at 37°C, 5% CO2. NaN3 was added to 0.1%, and cells were spun down at 4°C. The samples were treated with Nonidet P-40 lysis buffer, precleared, immunoprecipitated with mAb F21-21 directed to chicken β2m followed by protein A-Sepharose, and washed with PBS, 0.1% Nonidet P-40, and 0.1% NaN3 as described (38). Immunoprecipitates were eluted (9.5 M urea/2% Nonidet P-40/5% 2-mercaptoethanol/2.0% ampholines), subjected to isoelectric focusing by using 1.6% pH 5-8 and 0.4% pH 3-10 ampholines (LKB) for 3.5 h followed by SDS gel electrophoresis (12% polyacrylamide gels) by using the miniProtean 2-D gel system as per the manufacturer's instructions (Bio-Rad) (39). Gels were fixed (7.5% acetic acid, 10% methanol), washed with water, and impregnated (1 M sodium salicylate/1% glycerin) before drying and exposing on XR5 film (Kodak).
Isolation of Class I Molecules and Analysis of Peptides. Methods were based on refs. 40 and 41. Peripheral blood (100 ml) was treated similar to above (4 vol of PBS, 60 × g, 15 min, room temperature) with the top 2/3 of the supernatant discarded, and the procedure repeated twice with the resuspended pellet to remove as many PBL as possible. The final pellet (≈1011 cells, >99% erythrocytes) was lysed on ice (PBS, 5% Nonidet P-40), and subcellular material was removed by centrifugation (3,000 × g, 15 min, 4°C), followed by either ultracentrifugation (100,000 × g, 30 min, 4°C) or filtration through 0.45-μm filters. The lysate was treated with unsubstituted Sepharose to remove nonspecific material, and adhered to Sepharose coupled to F21-21 (or to mAb F21-2 directed to class I α chains, and by using either batch or column chromatography, with essentially the same results). Batches were washed (PBS, 1% Nonidet P-40; then PBS, 0.1% Nonidet P-40; then PBS). Columns were washed (PBS, 1% Nonidet P-40; then PBS, 0.1% sodium desoxycholate) before elution of MHC molecules with 50 mM diethylamine, 0.1% sodium desoxycholate; the neutralized eluate was concentrated and washed with PBS by centrifugation on a Centricon 10 filter. Both Sepharose batches and column eluates were treated with 0.1% trifluoroacetic acid (TFA) to dissociate peptides from MHC molecules.
Peptides were recovered from the flow-through of a Centricon 10 filter, dried by using a Speed-Vac, resuspended in 0.1% TFA, and separated by reversed-phase HPLC (Sephasil C18 SC 2.1/10 column on SMART system, Amersham Pharmacia) with a gradient (A, 0.1% TFA in water; B, 0.1% TFA in acetonitrile). Fractions containing peptides were pooled and sequenced by Edman degradation by using an Applied Biosystems 475A protein sequencer. At least three isolations of class I molecules were analyzed by pool sequencing, with similar results. Anchor, strong and weak residues were assigned as described (24, 40, 42).
Fractions containing individual peptide peaks were sequenced by Edman degradation and also by using a Sciex (Thornhill, ON, Canada) API III triple quadrupole mass spectrometer. For mass spectral analysis, dried fractions were resuspended (5% acetic acid/47.5% acetonitrile/47.5% water) and infused at 2 μl/min into the ionspray source. Molecular masses and sequences of selected precursor ions were obtained, aided by programs macbiospec and peptideseq with purity and final mass confirmed. Potential source proteins for each peptide were determined by searching GenBank/EMBL and Uniprot sequence databases by using the fasta program (43).
Peptide Prediction and Stabilization Assay. Predicted amino acid sequences from RSV Prague strain C (44) for the gag (P03322), pol (P03354), env (P03396), and src (P00526) genes were analyzed by using the GCG program findpatterns (43) and relaxed motifs: octamers and nonamers with all residues found in anchor positions by pool sequencing [x(D,E)xx(D,E)x(x,xx)(E,L,I) for B4, xxx(x,xx)(V,I)xx(V,L,I) for B12, and xRxxxx(x,xx)Y for B15]. Peptides were synthesized by f luorenylmethoxycarbonyl (fMOC) chemistry, purified by HPLC, and resuspended at 1 mg/ml in PBS (with up to 1% DMSO). Based on stabilization assays (25, 26), chicken PBL without thrombocytes (isolated as described above) were cultured (107 cells per ml at 40°C, 5% CO2) overnight in DMEM with 0.5 mg/ml BSA and with or without 1 mM synthetic peptides (in triplicate). Then, cells were washed (PBS/0.5% BSA/0.1% NaN3), and flow cytometric analysis using F21-21 followed by FITC-conjugated goat anti-mouse IgG (Silenus, Paris) was performed, gating on small lymphocytes.
Acknowledgments
We thank Mark Dessing and Brit Johansson for excellent technical assistance and Andy van Hateren for critical reading. We thank Hoffmann-LaRoche for many years of financial support and the Biotechnology and Biological Sciences Research Council in the U.K. for support (to J.K.).
Author contributions: H.-J.W. and J.K. designed research; H.-J.W., D.A., L.G.H., T.J.P., P.R., J.S., K.S., O.V., F.V., M.V.W., and J.K. performed research; H.-J.W., D.A., L.G.H., T.J.P., P.R., J.S., K.S., O.V., F.V., and J.K. analyzed data; and J.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RSV, Rous sarcoma virus; β2m, β2-microglobulin; PBL, peripheral blood leukocyte; TFA, trifluoroacetic acid.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. Z54314-Z54326, Z54329, Z54330, and Z54359-Z54363).
References
- 1.Pink, J., Droege, W., Hala, K., Miggiano, V. & Ziegler, A. (1977) Immunogenetics 5, 203-216. [Google Scholar]
- 2.Guillemot, F., Billault, A., Pourquie, O., Behar, G., Chausse, A.-M., Zoorob, R., Kreiblich, G. & Auffray, C. (1988) EMBO J. 7, 2775-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman, J., Völk, H. & Wallny, H.-J. (1995) Immunol. Rev. 143, 63-88. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman, J., Jacob, J., Shaw, I., Walker, B., Milne, S., Beck, S. & Salomonsen, J. (1999) Immunol. Rev. 167, 101-118. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman, J., Milne, S., Goebel, T., Walker, B., Jacob, J., Auffray, C., Zoorob, R. & Beck, S. (1999) Nature 401, 923-925. [DOI] [PubMed] [Google Scholar]
- 6.Dietert, R., Taylor, R. & Dietert, M. (1990) in MHC, Differentiation Antigens and Cytokines in Animals and Birds, Monographs in Animal Immunology, ed. Basta, O. (Bar-Lab, Inc., Backsburg, VA), Vol 1, pp. 7-26. [Google Scholar]
- 7.Schat, K. (1987) in Avian Immunolgy: Basis and Practice, eds. Toivanen, A. & Toivanen, P. (CRC, Boca Raton FL), Vol. 2, pp. 101-128. [Google Scholar]
- 8.Plachy, J., Pink, J. R. L. & Hala, K. (1992) Crit. Rev. Immunol. 12, 47-79. [PubMed] [Google Scholar]
- 9.Bacon, L. & Witter, R. (1993) Avian Dis. 37, 53-59. [PubMed] [Google Scholar]
- 10.Hansen, H., van Zandt, N. & Law, G. (1967) Poultry Sci. 46, 1268. [Google Scholar]
- 11.Plachy, J., Hala, K., Hejnar, J., Geryk, J. & Svoboda, J. (1994) Immunogenetics 40, 257-265. [DOI] [PubMed] [Google Scholar]
- 12.Taylor, R. (1994) Poultry Sci. 83, 638-649. [DOI] [PubMed] [Google Scholar]
- 13.Chausse, A. M., Coudert, F., Dambrine, G., Guillemot, F., Miller, M. M. & Auffray, C. (1989) Immunogenetics 29, 127-130. [DOI] [PubMed] [Google Scholar]
- 14.Jacob, J., Milne, S., Beck, S. & Kaufman, J. (2000) Immunogenetics 51, 138-147. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman, J., Andersen, R., Avila, D., Engberg, J., Lambris, J., Salomonsen, J., Welinder, K. & Skjødt, K. (1992) J. Immunol. 148, 1532-1546. [PubMed] [Google Scholar]
- 16.Hunt, H. & Fulton, J. (1998) Immunogenetics 47, 456-467. [DOI] [PubMed] [Google Scholar]
- 17.Madden, D. (1995) Annu. Rev. Immunol. 13, 587-622. [DOI] [PubMed] [Google Scholar]
- 18.Collins, E., Garboczi, D. & Wiley, D. (1994) Nature 371, 626-629. [DOI] [PubMed] [Google Scholar]
- 19.Madden, D. R., Gorga, J. C., Strominger, J. L. & Wiley, D. C. (1991) Nature 353, 321-325. [DOI] [PubMed] [Google Scholar]
- 20.Madden, D. R., Gorga, J. C., Strominger, J. L. & Wiley, D. C. (1992) Cell 70, 1035-1048. [DOI] [PubMed] [Google Scholar]
- 21.Garboczi, D., Ghosh, P., Utz, U., Fen, Q., Biddison, W. & Wiley, D. (1996) Nature 384, 134-141. [DOI] [PubMed] [Google Scholar]
- 22.Ruppert, J., Sidney, J., Celis, E., Kubo, R., Grey, H. & Sette, A. (1993) Cell 74, 929-937. [DOI] [PubMed] [Google Scholar]
- 23.Møller, L., Kaufman, J., Verland, S., Salomonsen, J., Avila, D., Lambris, J. & Skjødt, K. (1991) Immunogenetics 34, 110-120. [DOI] [PubMed] [Google Scholar]
- 24.Rammensee, H.-G., Friede, T. & Stevanovic, S. (1995) Immunogenetics 41, 178-228. [DOI] [PubMed] [Google Scholar]
- 25.Baas, E., van Santen, H., Kleijmeer, M., Geuze, H., Peters, P. & Ploegh, H. (1992) J. Exp. Med. 176, 147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elvin, J., Potter, C., Elliot, T., Cerundolo, V. & Townsend, A. (1993) J. Immunol. Methods 158, 161-171. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, A., Plachy, J., Hunt, L., Kaufman, J. & Hala, K. (2003) Vaccine 21, 4694-4699. [DOI] [PubMed] [Google Scholar]
- 28.Juul-Madsen, H., Dalgaard, T. & Afanassieff, M. (2000) Anim. Genet. 31, 252-261. [DOI] [PubMed] [Google Scholar]
- 29.Livant, E., Brigati, J. & Ewald, S. (2004) Anim. Genet. 35, 18-27. [DOI] [PubMed] [Google Scholar]
- 30.Mesa, C, Thulien, K., Moon, D., Veniamin, S. & Magor, K. (2004) Immunogenetics 56, 192-203. [DOI] [PubMed] [Google Scholar]
- 31.Driscoll, J., Brown, M., Finley, D. & Monaco, J. (1993) Nature 365, 262-264. [DOI] [PubMed] [Google Scholar]
- 32.Gaczynska, M., Rock, K. & Goldberg, A. (1993) Nature 365, 264-267. [DOI] [PubMed] [Google Scholar]
- 33.International Chicken Genome Sequencing Consortium (2004) Nature 432, 695-716. [DOI] [PubMed] [Google Scholar]
- 34.Walker, B., van Hateren, A., Milne, S., Beck, S. & Kaufman, J. (2005) Immunogenetics 57, 232-247. [DOI] [PubMed] [Google Scholar]
- 35.Chen, C., Ager, L., Gartland, G. & Cooper, M. (1986) J. Exp. Med. 164, 375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller, G., Kennedy, M., Papayannopolous, T. & Wiles, M. (1993) Mol. Cell. Biol. 13, 473-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vainio, O., Mansikka, A. & Lassila, O. (1991) in Immunological Methods (Academic, San Diego), Vol. 4, 265-280. [Google Scholar]
- 38.Kaufman, J., Ferrone, S., Flajnik, M., Kilb, M., Völk, H. & Parisot, R. (1990) J. Immunol. 144, 2258-2272. [PubMed] [Google Scholar]
- 39.O'Farrell, P. (1975) J. Biol. Chem. 250, 4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 40.Falk, K., Rötzschke, O., Stevanovic, S., Jung, G. & Rammensee, H.-G. (1991) Nature 351, 290-296. [DOI] [PubMed] [Google Scholar]
- 41.Hunt, D., Henderson, R., Shabanowitz, J., Sakaguchi, K., Michel, H., Sevilir, N., Cox, A., Appella, E. & Engelhard, V. (1992) Science 255, 1261-1263. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura, M., Fremont, D., Peterson, P. & Wilson, I. (1992) Science 25, 927-934. [DOI] [PubMed] [Google Scholar]
- 43.Anonymous (1991) GCG Users Guide (Genetics Computer Group, Madison, WI).
- 44.Schwartz, D., Tizard, R. & Gilbert, W. (1983) Cell 32, 853-869. [DOI] [PubMed] [Google Scholar]