Abstract
The use of retroviral vectors in gene therapy has raised safety concerns for the genotoxic risk associated with their uncontrolled insertion into the human genome. We have analyzed the consequences of retroviral transduction in T cells from leukemic patients treated with allogeneic stem cell transplantation and donor lymphocytes genetically modified with a suicide gene (HSV-TK). Retroviral vectors integrate preferentially within or near transcribed regions of the genome, with a preference for sequences around promoters and for genes active in T cells at the time of transduction. Quantitative transcript analysis shows that one fifth of these integrations affect the expression of nearby genes. However, transduced T cell populations maintain remarkably stable gene expression profiles, phenotype, biological functions, and immune repertoire in vivo, with no evidence of clonal selection up to 9 yr after administration. Analysis of integrated proviruses in transduced cells before and after transplantation indicates that integrations interfering with normal T cell function are more likely to lead to clonal ablation than expansion in vivo. Despite the potentially dangerous interactions with the T cell genome, retroviral integration has therefore little consequence on the safety and efficacy of T cell transplantation.
Keywords: donor lymphocyte infusion, gene therapy, graft-versus-host disease, insertional mutagenesis, retroviral integration
The clinical use of retroviral vectors has raised safety concerns due to the potential genotoxic consequences of uncontrolled insertion into the human genome. Gamma-retroviral vectors have a propensity to integrate into active chromatin regions and around promoters (1–4), where insertion of LTR transcriptional enhancers may interfere with normal gene regulation (5) and eventually contribute to malignant transformation (6–8). The occurrence of leukemia in patients treated by gene therapy for X-linked severe combined immunodeficiency (X-SCID) has been correlated with insertional activation of a T cell proto-oncogene (9). No malignancy has been reported so far in other preclinical (10) or clinical (11, 12) trials, suggesting the existence of specific risk factors in the X-SCID case (13). In-depth analysis on the consequences of retroviral vector integration is therefore necessary, to provide risk-benefit assessments in different biological and clinical contexts.
We have analyzed retroviral integrations in genetically modified donor lymphocytes administered to patients undergoing allogeneic hematopoietic stem cell (HSC) transplantation for the treatment of leukemia/lymphoma. Donor lymphocyte infusion (DLI) effectively promotes immune reconstitution and antitumor activity in these patients, although its efficacy is limited by the risk of graft-versus-host disease (GvHD). The infusion of lymphocytes transduced with a retroviral vector expressing a suicide gene [the herpes simplex virus-derived timidine kinase (TK)] and a surface marker (ΔLNGFR) allows an efficient control of GvHD by administration of ganciclovir (14–17), with no apparent side effect (18). TK+ DLI is currently profiled to provide a graft-versus-leukemia effect to patients in relapse after HLA-identical HSC transplantation (14), or to promote immune reconstitution and prevent relapse in patients undergoing HLA-haploidentical HSC transplantation. In 46 patients treated since 1994 in both contexts, we were able to control GvHD in 100% of the cases, while preserving antiviral and antitumor activity (14–16).
Proviral integrations were analyzed by sequencing the vector-genome junctions in T cells before and after infusion in four different patients. A vast majority of the integrations occurred within or around genes, with a preference for those active in T cells at the time of transduction. Quantitative transcript analysis in individual T cell clones showed that almost 1/5 of the promoter-proximal integrations leads to gene activation. However, transduced T cell populations maintained remarkably stable gene expression profiles, phenotype, biological functions, and immune repertoire in vivo, with no evidence of clonal selection up to 9 yr after administration. Analysis of vector integrations before and after transplantation indicates that integrations in growth-controlling genes or interfering with normal T cell function are counterselected in vivo. This study indicates that retroviral transduction of mature T cells is clinically safe, and associated with undetectable risk of insertional oncogenesis.
Results
Clinical Trials. In a first clinical trial, patients experiencing disease relapse after HLA-identical HSC transplantation received up to 108 donor TK+ cells per kg to provide a graft-versus-leukemia effect (14). In a second trial, patients with high-risk malignancies and lacking an HLA-identical donor received a T cell-depleted HSC transplantation from a haploidentical relative and one or more infusions of TK+ cells early after transplantation, to promote immune reconstitution and prevent infections and relapse. Cumulatively, the 46 patients treated since 1994 received a total of >1011 genetically modified T cells generated by >90 independent transductions. Expansion (up to 40% of circulating cells) and long-term persistence (>10 yr) of transduced T cells were observed in these patients in the absence of any adverse or toxic effects related to the gene transfer procedure (14–16, and C. Bonini, F.C., and C. Bordignon, unpublished data).
Analysis of Retroviral Integrations in T Cells. A genome-wide analysis of the retroviral integration sites was carried out on lymphocytes obtained from four patients (HTK7, MMTK1, MMTK8, and TK8) 1 month to 9 yr after DLI. In the first 3 patients, transduced T cells ranged from 10% (HTK7) to 52% (MMTK8) of circulating T lymphocytes and were purified by sorting for ΔLNGFR expression. Both transduced and untransduced T cells were of donor origin, as indicated by HLA genotyping (data not shown). Vector–genome junctions were cloned and sequenced by a linker-mediated nested PCR approach (1). The same analysis was carried out on transduced, uninfused T cells from MMTK1, MMTK8, and an unrelated donor, and from individual T cell clones obtained ex vivo from MMTK1, MMTK8, and HTK7. Cumulatively, we sequenced 312 independent insertions, 300 of which were unambiguously mapped on human chromosomes. Thirty-nine integrations (13%) occurred at an arbitrarily chosen distance of >30 kb from any known gene and were classified as intergenic. One hundred fifty-five integrations (52%) were within the transcribed portion of 160 RefSeq genes, whereas 106 integrations (35%) were at a distance of ≤30 kb upstream or downstream of 248 genes (a complete list is in Table 3, which is published as supporting information on the PNAS web site). Compared with a collection of computer-generated random insertions (4), the overrepresentation of intragenic integrations is highly significant (52.0% vs. 31.6%, P < 10-5). Clusters of integrations were observed in correspondence of known gene-dense regions of the human genome (19). Recurrent integration sites, showing at least two independent hits <50 kb apart, were observed within or upstream of genes in 11 instances (3.7%), 6 in the gene-dense regions on chr1q32.1, 6p21.1, and 14q32.2 (Fig. 5, which is published as supporting information on the PNAS web site).
A functional classification of all hit genes by the Gene Ontology criteria (20) showed no statistically significant difference between the expected and the observed distribution of genes among the different classes in T cells before and after infusion, except for a moderate overrepresentation of genes involved in signal transduction and cell proliferation in uninfused T cells (32.7 vs. 22.6% and 8.9 vs. 5.0%, respectively, P < 0.01), which disappeared in T cells obtained ex vivo from patients (Table 4, which is published as supporting information on the PNAS web site).
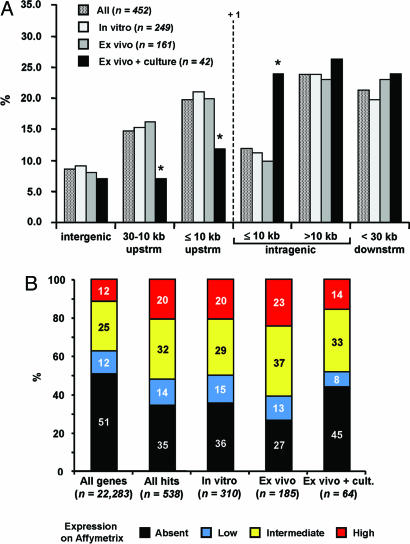
The position of the integrated proviruses with respect to RefSeq genes is shown in Fig. 1A, which considers the total number of vector–genome interactions (i.e., 452, many integrations hit more than one gene). Over 31% of the integrations occurred ≤10 kb upstream or downstream of transcription start sites, with no significant difference between uninfused T cells and T cells obtained ex vivo 1–11 months after DLI (Fig. 1A). Compared with a collection of random insertions (4), clustering around transcription start sites is highly significant (31.6% vs. 5.5%, P < 10-10). Interestingly, T cells expanded and/or cloned ex vivo showed significantly fewer integrations <30 kb upstream of (19.0% vs. 36.0% in ex vivo cells, P < 10-3) and more <10 kb downstream of (23.8% vs. 9.9%, P < 10-6) transcription start sites (Fig. 1A).
Fig. 1.
Retroviral integration sites in human T cells. (A) Distribution of integration sites within an interval of ±30 kb upstream (upstrm) or downstream (dwnstrm) RefSeq genes. A +1 indicates the transcription start site. Bars of different shadings represent integration events in all cells (All), uninfused cells (In vitro), cells obtained ex vivo (Ex vivo), or cells obtained ex vivo and expanded/cloned in vitro (Ex vivo + culture). *, P < 10-3. (B) Correlation between retroviral integration and gene activity in T cells. The bars show the percent distribution of expression values from an Affymetrix HG-U133A microarray of mock-transduced T cells, subdivided into absent (black), low (blue), intermediate (yellow), and high (red). The first bar shows the distribution of the >16,000 genes on the microarray (all genes), the other bars the expression values of genes hit by a retroviral vector in all T cells (All hits), uninfused T cells (In vitro), T cells obtained ex vivo (Ex vivo), and T cells obtained ex vivo and expanded/cloned in vitro (Ex vivo + cult.). The n values indicate the number of analyzed probesets for each group of genes.
Retroviral Vectors Integrate Preferentially in Genes Expressed in T Cells at the Time of Transduction. To correlate vector integration with gene activity, we determined the expression profile of >16,000 genes in mock-transduced lymphocytes by microarray analysis. As shown in Fig. 1B, 65% of 538 probesets representing 409 vector–gene interactions had a “present” call, and, among them, 14% were classified as lowly abundant, 32% as intermediately abundant, and 20% as highly abundant, compared with a 49% present call on the whole microarray and a 12%, 25%, and 12% breakdown in the three abundance classes. All differences were statistically significant (P < 10-9), indicating that retroviral vectors integrate preferentially into genes active in T cells at the time of transduction, and particularly in the fraction of genes expressed at higher levels. The same distribution was observed with probeset lists representing 226 and 148 genes hit in T cells transduced in vitro or ex vivo from MMTK1, MMTK8, and HTK7 (Fig. 1B). On the contrary, the fraction of hit genes active in T cells expanded ex vivo was significantly lower (55% vs. 64 and 73%, P < 10-4), with a distribution more similar to that of the entire microarray (P = 0.2) (Fig. 1B).
Integrated Proviruses Influence the Expression of Hit Genes. To provide a direct estimate of the capacity of integrated proviruses to influence the expression of nearby genes, we analyzed 40 randomly chosen, individual T cell clones harboring an integrated vector within or around a RefSeq gene (Table 5, which is published as supporting information on the PNAS web site). The expression of the 44 hit genes was analyzed by real-time RT-PCR on two microfluidic cards designed to measure gene expression in quadruplicate in the clones interested by each hit and in all other clones as controls. Expression levels were measured as relative mRNA quantity (RQ) after normalization for the level of GAPDH, and plotted in Fig. 2A as log2 variation from the median levels. By this analysis, 8 of 44 genes (18%) showed significantly higher expression when hit by a provirus, ranging from 4-fold the average value in the control clones for BRCA2 (Z score: 2.12, P < 0.05) to >2,000-fold for LY64 (Z score: 3.63, P < 10-3). Four up-regulated genes (KCNN1, LY64, MIA2, and THRAP2) are not expressed in T cells, three (BRCA2, SNARK, and ZNF217) are expressed at low level, and one (CD5) is expressed at intermediate level. One of the activating insertions (BRCA2) was in T cells cloned from MMTK1 5 months after DLI. We were unable to retrieve this insertion in a library of 69 independent integrations at 5 and 11 months (Table 2), suggesting that T cells overexpressing BRCA2 were present at very low level in the patient's circulation.
Fig. 2.
Retroviral vector integration interferes with normal gene regulation. (A) Quantitative PCR analysis of the expression of genes hit by a retroviral integration event (x axis) in randomly selected, individual T cell clones in vitro and ex vivo, as analyzed on two Applera microfluidic cards (one gene was represented in both cards). Expression levels (y axis) were measured as relative mRNA quantity (RQ) after normalization for the level of GAPDH, and plotted as log2 variations from the median level (0) in all analyzed clones. The expression value in the clone interested by the hit is indicated by a red dot. Values departing from the mean by 2 or more standard deviations (Z score >1.96) are indicated by arrows. (B) Schematic representation of the eight proviral insertions leading to activation of gene expression. Integrated proviruses are shown above each gene locus, where black boxes represent exons and a vertical bar indicates the insertion site. SD, splice donor site. Arrows indicate transcription start sites.
Table 2. Immune repertoire of T cells from patients MMTK1 and MMTK8.
| MMTK1
|
MMTK8
|
|||
|---|---|---|---|---|
| Immune repertoire | CD3+ | ΔLNGFR+ | CD3+ | ΔLNGFR+ |
| TCR Vβ families detected | 22 | 21 | 22 | 22 |
| TCR Vβ families, polyclonal | 4 | 6 | 5 | 3 |
| TCR Vβ families, oligoclonal | 15 | 15 | 15 | 15 |
| TCR Vβ families, monoclonal | 3 | 0 | 2 | 4 |
The functional immune repertoire of total (CD3+) and transduced (ΔLNGFR+) peripheral T cell populations was evaluated by spectratype analysis of TCR Vβ-chain rearrangement. Analysis was carried out on cells harvested 2 months after DLI.
The eight activating integrations are shown in Fig. 2B. All but one of the vectors were inserted in opposite transcriptional orientation, at a distance from promoters ranging from 25 kb upstream (LY64) to 105 kb downstream (THRAP2). Overall, 61% of the proviruses integrated upstream or within genes in T cell clones were in opposite transcriptional orientation, compared with 51% in ex vivo T cells and 52% in uninfused T cells (Table 2). One provirus up-regulated the expression of KCNN1 4.5 kb downstream but had no effect on the activity of the IFI30 promoter 38 kb upstream.
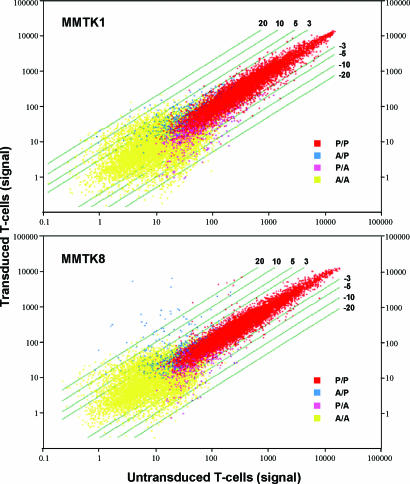
Retroviral Integration Has No Effect on the Biology and Function of T Cells in Vivo. To analyze the consequences of vector integration on the biology of transduced T cells, we studied the gene expression profile, phenotype, and functional immune repertoire of transduced and untransduced CD8+ T cells obtained from MMTK1 and MMTK8 6 and 4 months, respectively, after DLI. Transduced (ΔLNGFR+) cells represented >40% of circulating T lymphocytes at these time points and were mostly CD8+. As shown in Fig. 3, there was little difference between the expression profiles of transduced vs. untransduced cells: 220 (0.99%) and 243 (1.09%) probesets showed a significant, 2-fold or greater increase [signal log2 ratio (s.l.r.) ≥ 1] in expression in MMTK1 and MMTK8, respectively, whereas 493 (2.21%) and 256 (1.15%) showed a similar decrease (s.l.r. ≤ 1). Only 7 genes showed a concordant increase and 20 a concordant decrease in expression in both patients (Fig. 3). None of the genes hit by a proviral insertion in MMTK1 and MMTK8 showed differences in expression between transduced and nontransduced T cell populations by this analysis (a complete data set is in Table 6, which is published as supporting information on the PNAS web site).
Fig. 3.
Analysis of T cell gene expression profiles. Comparative analysis of the expression levels of >16,000 genes in untransduced (y axis) and TK-transduced (x axis) CD8+ T cells obtained from patients MMTK1 and MMTK8 as analyzed by Affymetrix microarrays. Each gene is represented by a dot, where the color code indicates genes present in both samples (P/P, red), absent in TK+ and present in TK- cells (A/P, blue), present in TK+ and absent in TK- cells (P/A, cyan), or absent in both samples (A/A, yellow). Bars represent transcript level fold increase (3, 5, 10, and 20) or fold decrease (-3, -5, -10, and -20) intervals. Seven genes (LTF, PRNPIP, CAPG, KIR3DL2, RFP, KATNB1, and FSTL1) showed a 2-fold or higher increase, and 20 genes (PHC3, ZAP3, NKTR, DDX17, SLC4A7, FLJ20856, PCBP2, SFRS11, CDC14A, RAD21, RBL2, MLL, KIAA1172, KIAA0675, PA200, SMARCC1, KIF5B, SMA5, AP3D1, and BTEB1) showed a 2-fold or lower decrease in transcript levels in TK+ T cells in both patients (a complete set of data is available in Table 6).
In both patients, the majority of circulating T cells were effector memory (CD54RA-/CCR7-) and terminally differentiated effector (CD54RA+/CCR7-) cells, with no evidence of central memory or naive cells (Table 1). The activation status of circulating T cells was determined by analysis of HLA-DR and IL-2R (CD25) expression. Although differences were observed in the relative T cell subset distribution between the two patients, no significant difference was detected between untransduced and transduced cells from the same patient (Table 1). Upon polyclonal stimulation, the majority of circulating lymphocytes produced high levels of IFN-γ in the absence of IL-4 (Th1/Tc1 polarization), with no difference between ΔLNGFR+ and ΔLNGFR- cells (Table 1). Spectratype analysis carried out on total (CD3+) and transduced T cell fractions showed rearrangement of >20 of 27 T cell receptor (TCR) Vβ families tested, documenting a wide immune repertoire in all T cell populations (Table 2).
Table 1. Immune phenotype of T cells from patients MMTK1 and MMTK8.
| T cells, %
|
||||
|---|---|---|---|---|
| MMTK1
|
MMTK8
|
|||
| Phenotype | ΔLNGFR+ | ΔLNGFR– | ΔLNGFR+ | ΔLNGFR– |
| T cell differentiation | ||||
| CD45RA+/CCR7+ | 1.7 | 4.5 | 0.2 | 1.0 |
| CD45RA–/CCR7+ | 1.4 | 4.2 | 0.1 | 1.5 |
| CD45RA–/CCR7– | 58.1 | 43.3 | 63.2 | 35.8 |
| CD45RA+/CCR7– | 38.8 | 48.0 | 36.5 | 61.7 |
| T cell activation | ||||
| CD25+/HLA-DR+ | 0.3 | 0.2 | 0.6 | 0.1 |
| CD25+/HLA-DR– | 0.7 | 0.6 | 0.1 | 1.4 |
| CD25–/HLA-DR+ | 84.3 | 86.2 | 65.9 | 65.5 |
| CD25–/HLA-DR+ | 14.7 | 13.0 | 33.4 | 33.0 |
| T cell polarization | ||||
| IFN-γ+/IL4– | 9.6 | 10.2 | 43.2 | 43.3 |
| IFN-γ–/IL4+ | 1.1 | 1.1 | 1.0 | 1.8 |
| IFN-γ+/IL4+ | 0.4 | 0.4 | 7.2 | 3.6 |
Immune phenotype was determined on cells stained with anti-CD3 and LNGFR antibodies, and events were gated on the transduced (CD3+/ΔLNGFR+) and untransduced (CD3+/ΔLNGFR–) T cell populations. Expression of differentiation and activation markers was detected by FACS analysis. T cell polarization was evaluated by intracytoplasmic staining upon in vitro activation with PMA and ionomycin.
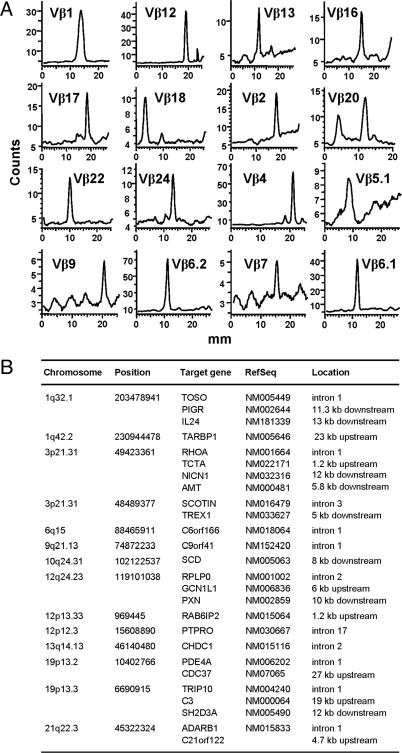
Analysis of Long-Term Surviving T Cells. TK8 was the longest survivor among patients receiving TK+ DLI (14). Nine years after DLI, transduced T cells were below detection limits in peripheral blood and were obtained as a G418-resistant culture. They were predominantly CD4+, expressed ΔLNGFR, and were sensitive to ganciclovir (data not shown). Spectratype analysis showed rearrangement of 22 of 27 tested TCR Vβ chains (Fig. 4A), indicating a wide T cell repertoire. Fourteen different vector integrations were identified at a frequency of 1.5–21% by sequencing to saturation a library of vector–genome junctions. All integrations occurred within or around RefSeq genes, half of which are expressed in cultured T cells. Although a statistically significant analysis is prevented by the small size of the sample, there seems to be no obvious bias for any functional class in the list of hit genes.
Fig. 4.
Analysis of long-term surviving T cells, obtained from patient TK8 9 yr after DLI. (A) Spectratype analysis of TCR Vβ-chain rearrangements, after PCR amplification with primers specific for 16/27 different Vβ chains analyzed. (B) List of the 14 different integration sites cloned from the TK8 T cells. The position of the junctions between the vector 5′ LTR on the human genome is indicated (position), together with its chromosomal mapping (chromosome) and location with respect to a hit gene (location). Genes are listed by name (target gene) and RefSeq identifier (RefSeq).
Discussion
We report an integration and gene expression analysis on T cells transduced by Moloney leukemia virus (MLV)-derived retroviral vectors in the context of two clinical trials, in which TK-modified lymphocytes were administered to patients undergoing allogeneic HSC transplantation. Most proviruses integrated within or in close proximity to genes, an expected result based on previous studies on retroviral integration in human and nonhuman primates (1–4). Clusters of integrations varying in size from a few hundreds of kb to several Mb were observed in correspondence of known gene-rich regions of the human genome (19), previously reported as favored sites for γ-retroviral vectors (1p36, 1q32, 3p21, 19p13, and 19q13) (21), WT HIV (1p36, 6p21, 6q23, 19p13, and 19q13) (22), and lentiviral vectors (2, 23). Almost 4% of the integrations occurred at recurrent sites containing at least two hits at a distance of <50 kb from each other, within or around active genes. Similar “hot spots” were previously found in hematopoietic cells but at different sites (4, 21), suggesting that they are induced by cell-specific transcriptional complexes and/or chromatin configurations. In fact, 2/3 of the integrated proviruses were found within or around genes active in T cells at the time of transduction.
A crucial parameter in the calculation of the potential risks of gene transfer is the frequency by which an integrated provirus leads to activation, or deregulation, of gene expression. We directly addressed this issue by evaluating the expression of targeted genes in randomly selected T cell clones, and found that almost 1/5 of them are up-regulated independently from the distance of the integrated vector, which varied from 25 kb upstream to >100 kb downstream of transcription start sites, and its transcriptional orientation. Most up-regulated genes are expressed at very low levels or not expressed in T cells, suggesting that proviral insertion is more likely to affect genes that are not already engaged in high-level transcription. Interestingly, insertions as close as 0.5 kb upstream or 0.7 kb downstream of transcription start sites caused no perturbation of gene expression. In one case, a single provirus caused up-regulation 4.5 kb downstream from the insertion site, but had no effect 38 kb upstream. These findings suggest that insertional gene activation is not simply a function of relative distance or orientation of integrated proviruses but may depend on the permissivness of gene regulatory elements to the activity of transcription factors recruited by the LTRs.
Despite the observed frequency of insertional gene activation, there was no evidence of clonal selection or preferential survival of transduced T cells in a relatively long follow-up of four patients. In all cases, including long-term surviving T cells analyzed 9 years after infusion, we found polyclonal T cell populations with a normal phenotype and gene expression profile, containing many different proviral integrations and a wide functional immune repertoire. In particular, <2% of >16,000 analyzed genes were differentially expressed in transduced T cells in two different patients, and only a handful of genes were consistently over- or underexpressed in both. These data point to a substantial biological identity of T cell populations developed in the patients after HSC transplantation and those administered as DLI and show that genetic modification and expression of TK and ΔLNGFR have little or no effect on the average T cell-expressed genome.
The representation of different Gene Ontology categories in the hit genes in T cells before and after infusion was similar and not significantly different from the expected values for a random distribution. Interestingly, a slight overrepresentation of proliferation-related genes in uninfused T cells disappeared in T cells obtained from patients, suggesting that such hits have, if anything, a negative effect on in vivo survival of transduced T cells. In cultured T cells, integrations into active genes, in direct transcriptional orientation, or upstream from transcription start sites, are significantly underrepresented, suggesting that, in cells subjected to strong selective pressure, interference with transcription, splicing, and polyadenylation is more likely to lead to clonal ablation than expansion.
Overall, our analysis indicates that genetic modification of T cells with retroviral vectors is safe and is not associated to a measurable risk of insertional oncogenesis. This finding is in contrast with that observed in the X-linked severe combined immunodeficiency trial and reinforces the concept that the risk of gene therapy in general, and of the use of retroviral vectors in particular, must be assessed in each specific experimental and clinical context. This analysis should include the biology of the target cell, the cell activation and transduction procedure, and the nature and function of the transgene, as well as the vector design. Given the therapeutic potential of gene transfer technology, there is no alternative to its testing in carefully designed clinical trials, where the risk-benefit balance must remain the central consideration.
Materials and Methods
Patients. The clinical trials were authorized by Italian regulatory authorities and approved by the Institutional Review Board of the San Raffaele Hospital under informed consent. Patient TK8 (F, 18 yr) was treated with HLA-identical HSC transplantation for chronic myelomonocytic leukemia. She received escalating infusions (2 × 107 total) of T cells transduced with the SFCMM-2 retroviral vector, encoding a TK/Neo fusion suicide gene and ΔLNGFR. She underwent complete remission and developed a chronic GvHD that was resolved by ganciclovir administration (14). Patients MMTK1 (M, 54 yr), MMTK8 (F, 53 yr), and HTK7 (M, 30 yr) were transplanted with HSCs from HLA-haploidentical donors and received respectively 1.1, 1.0, and 2.5 × 107 donor lymphocytes transduced with the SFCMM-3 vector, encoding only TK and ΔLNGFR (15). TK+ lymphocytes were purified after transduction by immunomagnetic sorting for ΔLNGFR and immediately infused (14–16).
T Cell Isolation and Cloning. peripheral blood mononuclear cells (PBMCs) from MMTK1, MMTK8, and HTK7 were isolated by Ficoll-Hypaque gradient separation. TK+/ΔLNGFR+ cells were quantified by flow cytometry and immunoselected as described (14, 15). For transcriptional profiling, CD8+ lymphocytes were selected with a CD8+ T cell isolation kit (Miltenyi Biotec, Auburn, CA) and further fractionated in a CD8+/ΔLNGFR+ and a CD8+/ΔLNGFR- population by immunomagnetic sorting. PBMCs from TK8 were cultured in the presence of 0.7 mg/ml G418 to select TK+ cells (16). Individual TK+ cell clones were obtained by plating TK+ cells in 96-well plates at a concentration of 0.3–3 cells per well as described (16).
T Cell Phenotype Analysis. Phenotype and cytokine production was analyzed by flow cytometry by using FITC-conjugated mAbs to CD25, CD45RA, and IFN-γ, and phycoerythrin (PE)-conjugated mAbs to LNGFR and IL-4 (Pharmingen). CCR7 and, in some experiments, ΔLNGFR expression were revealed by PerCP-conjugated streptavidin after staining with an anti-CCR7 mAb and biotinylated anti-mouse IgM or with a biotinylated anti-LNGFR mAb, respectively. Expression of IL-4 and IFN-γ was evaluated by intracytoplasmic staining upon in vitro activation with 50 ng/ml phorbol myristate acetate (PMA) and 1 μg/ml ionomycin (Sigma). After 4 h, brefeldin A (10 μg/ml, Sigma) was added for an additional 2 h to inhibit cytokine secretion. Spectratype analysis of TCR Vβ families was performed on cDNA obtained from 2 × 106 T cells as described (24).
Analysis of Gene Expression Profiles. RNA was isolated from 1–2 × 106 T cells, transcribed into biotinylated cRNA, purified on Affymetrix spin columns, and hybridized to Affymetrix HG-U133A Gene Chip arrays. Scanned images were processed by the Affymetrix mas 5.0 suite, and transcript levels were determined with the mas 5.0 absolute analysis algorithm. Gene expression levels were compared by the mas 5.0 comparison analysis algorithm and global scaling option, and differences were expressed as signal log2 ratio (s.l.r.). To correlate retroviral integration and gene activity, expression values from a mock-transduced T cell microarray were divided in four classes, i.e., absent, low (below the 25th percentile in a normalized distribution), intermediate (between the 25th and the 75th percentile), and high (above the 75th percentile). Differences in the expression classes were evaluated by a parametric likelihood test, distributed as χ2. To account for the different distributions between each sample, an asymptotic approximation was considered by a Monte-Carlo method based on 10,000 replications.
Analysis of Retroviral Vector Integration Sites. Integration sites were cloned by linker-mediated (LM)-PCR, as described (1). Briefly, genomic DNA was extracted from 1–5 × 106 cells, digested with MseI and PstI to prevent amplification of internal 5′ LTR fragments, and ligated to an MseI double-strand linker. LM-PCR was performed with nested primers specific for the LTR and the linker (1). PCR products were shotgun-cloned by the TOPO TA cloning kit (Invitrogen) into libraries of integration junctions, which were sequenced to saturation. Sequences were mapped onto the human genome by the blat genome browser (University of California Santa Cruz Human Genome Project Working Draft, October 2005). A genuine integration contained both LTR- and linker-specific sequences and a genomic sequence featuring a unique best hit with ≥95% identity to the human genome. A two-sample test for proportions was used for pairwise comparison of the integration site distribution groups within the different classes, corrected by the Holm (wise error rate) and Benjamini and Yekutieli (false discovery rate) methods. Functional classification of the hit genes was carried out following the Gene Ontology criteria (20) by the ease bioinformatic software (25).
Real-Time PCR Analysis of Gene Expression. cDNAs were reverse transcribed from total RNA samples (100 ng) by the Archive kit (Applied Biosystems). TaqMan PCRs were carried out onto custom 7900 TaqMan low-density arrays on an ABI PRISM 7900 HT (Applied Biosystems). TaqMan strategies for each gene were developed as “Assay on Demand” by Applied Biosystems. Gene expression profiling was achieved by using the comparative cycle of threshold (CT) method of relative quantification. To normalize data, ΔCT was calculated for each detector by using the median of ΔCTs in all samples as calibrator. The relative quantity (RQ) of each mRNA was calculated as 2-ΔCT and plotted as log2 values.
Supplementary Material
Acknowledgments
We thank Giulia Facchini for RT-PCR data analysis and Alessandro Ambrosi and Clelia Di Serio for statistical analysis. This work was supported by grants from the European Commission (V and VI Framework Programs), the Italian Association for Cancer Research (AIRC), the Italian Ministry of Health (MIUR-FIRB), and Fondazione Cariplo.
Author contributions: C. Bonini, C. Bordignon, and F.M. designed research; A.R., C. Bonini, Z.M., F.U., D.S., S.M., A.B., M.T.L.S., M.B., A.P., and F.C. performed research; E.T. contributed new reagents/analytic tools; A.R., C. Bonini, F.U., E.T., and F.M. analyzed data; and C. Bordignon and F.M. wrote the paper.
Conflict of interest statement: C. Bordignon and F.M. are paid consultants of MolMed S.p.A., an Italian biotechnology company based in Milan, which sponsors one of the two clinical trials described in this paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSC, hematopoietic stem cell; DLI, donor lymphocyte infusion; GvHD, graft-versus-host disease; TK, timidine kinase; TCR, T cell receptor; CT, cycle of threshold.
References
- 1.Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell, R. S., Beitzel, B. F., Schroder, A. R., Shinn, P., Chen, H., Berry, C. C., Ecker, J. R. & Bushman, F. D. (2004) PloS Biol. 2, e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laufs, S., Nagy, K. Z., Giordano, F. A., Hotz-Wagenblatt, A., Zeller, W. J. & Fruehauf, S. (2004) Mol. Ther. 10, 874-881. [DOI] [PubMed] [Google Scholar]
- 4.Hematti, P., Hong, B. K., Ferguson, C., Adler, R., Hanawa, H., Sellers, S., Holt, I. E., Eckfeldt, C. E., Sharma, Y., Schmidt, M., et al. (2004) PloS Biol 2, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelman, A. (2005) Proc. Natl. Acad. Sci. USA 102, 1275-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum, C., Dullmann, J., Li, Z., Fehse, B., Meyer, J., Williams, D. A. & von Kalle, C. (2003) Blood 101, 2099-2114. [DOI] [PubMed] [Google Scholar]
- 7.Williams, D. A. & Baum, C. (2003) Science 302, 400-401. [DOI] [PubMed] [Google Scholar]
- 8.McCormack, M. P. & Rabbitts, T. H. (2004) N. Engl. J. Med. 350, 913-922. [DOI] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]
- 10.Kiem, H. P., Sellers, S., Thomasson, B., Morris, J. C., Tisdale, J. F., Horn, P. A., Hematti, P., Adler, R., Kuramoto, K., Calmels, B., et al. (2004) Mol. Ther. 9, 389-395. [DOI] [PubMed] [Google Scholar]
- 11.Aiuti, A., Slavin, S., Aker, M., Ficara, F., Deola, S., Mortellaro, A., Morecki, S., Andolfi, G., Tabucchi, A., Carlucci, F., et al. (2002) Science 296, 2410-2413. [DOI] [PubMed] [Google Scholar]
- 12.Gaspar, H. B., Parsley, K. L., Howe, S., King, D., Gilmour, K. C., Sinclair, J., Brouns, G., Schmidt, M., Von Kalle, C., Barington, T., et al. (2004) Lancet 364, 2181-2187. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, A. & Cavazzana-Calvo, M. (2005) PloS Med. 2, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonini, C., Ferrari, G., Verzeletti, S., Servida, P., Zappone, E., Ruggieri, L., Ponzoni, M., Rossini, S., Mavilio, F., Traversari, C. & Bordignon, C. (1997) Science 276, 1719-1724. [DOI] [PubMed] [Google Scholar]
- 15.Verzeletti, S., Bonini, C., Marktel, S., Nobili, N., Ciceri, F., Traversari, C. & Bordignon, C. (1998) Hum. Gene Ther. 9, 2243-2251. [DOI] [PubMed] [Google Scholar]
- 16.Marktel, S., Magnani, Z., Ciceri, F., Cazzaniga, S., Riddell, S. R., Traversari, C., Bordignon, C. & Bonini, C. (2003) Blood 101, 1290-1298. [DOI] [PubMed] [Google Scholar]
- 17.Tiberghien, P., Ferrand, C., Lioure, B., Milpied, N., Angonin, R., Deconinck, E., Certoux, J. M., Robinet, E., Saas, P., Petracca, B., et al. (2001) Blood 97, 63-72. [DOI] [PubMed] [Google Scholar]
- 18.Bonini, C., Grez, M., Traversari, C., Ciceri, F., Marktel, S., Ferrari, G., Dinauer, M., Sadat, M., Aiuti, A., Deola, S., et al. (2003) Nat. Med. 9, 367-369. [DOI] [PubMed] [Google Scholar]
- 19.Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M. C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laufs, S., Gentner, B., Nagy, K. Z., Jauch, A., Benner, A., Naundorf, S., Kuehlcke, K., Schiedlmeier, B., Ho, A. D., Zeller, W. J. & Fruehauf, S. (2003) Blood 101, 2191-2198. [DOI] [PubMed] [Google Scholar]
- 22.Han, Y., Lassen, K., Monie, D., Sedaghat, A. R., Shimoji, S., Liu, X., Pierson, T. C., Margolick, J. B., Siliciano, R. F. & Siliciano, J. D. (2004) J. Virol. 78, 6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder, A. R., Shinn, P., Chen, H., Berry, C., Ecker, J. R. & Bushman, F. (2002) Cell 110, 521-529. [DOI] [PubMed] [Google Scholar]
- 24.Verfuerth, S., Peggs, K., Vyas, P., Barnett, L., O'Reilly, R. J. & Mackinnon, S. (2000) Blood 95, 3990-3995. [PubMed] [Google Scholar]
- 25.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.