Abstract
Armadillo/β-catenin and related proteins have important functions during animal and Dictyostelium development, regulating cell differentiation, proliferation, and adhesion. Armadillo-repeatcontaining proteins also exist in plants, but the majority have unknown roles. The Arabidopsis genes that show greatest sequence homology to Armadillo/β-catenin are called ARABIDILLO-1 and -2. Here, we demonstrate that ARABIDILLO-1 and -2 promote lateral root development. arabidillo-1/-2 mutants form fewer lateral roots, and ARABIDILLO-1-overexpressing lines produce more lateral roots than wild-type seedlings. ARABIDILLO-yellow fluorescent protein fusions are nuclear. ARABIDILLO proteins contain an F-box motif, and thus may target other proteins for proteasomal degradation. Overexpression of ARABIDILLO-1 protein fragments, including F-box fragments, in wild-type seedlings reduces lateral root formation to the level of the arabidillo-1/-2 mutant. We have shown that plant β-catenin-related proteins regulate root development. We suggest that ARABIDILLO proteins may target an inhibitor of lateral root development for degradation and propose that Arabidopsis β-catenin-related proteins define a previously uncharacterized pathway that promotes root branching.
Keywords: plant, β-catenin, arabidillo
Mammalian β-catenin and its Drosophila homologue Armadillo (Arm) are multifunctional proteins required for embryonic development and with roles throughout adult life (1, 2). These proteins specify cell fates by regulating gene expression and are regulated posttranscriptionally. Cytosolic β-catenin/Arm protein is targeted for destruction by the proteasome unless stabilized by extracellular Wnt signals. Wnt signals allow β-catenin to translocate to the nucleus and interact with LEF/TCF transcription factors. This protein complex can activate (or relieve the repression of) target genes (3, 4). β-catenin can also lead indirectly to transcriptional activation. For instance, the Caenorhabditis elegans β-catenin WRM-1 relocates the TCF-related repressor POP-1 from the nucleus to the cytosol (5). β-catenin/Arm is also required for the formation of adherens junctions, sites of cell-cell adhesion where transmembrane cadherin molecules are linked to the actin cytoskeleton by α- and β-catenin (1). This adhesive function of β-catenin is required for development (6-8).
In the multicellular protist Dictyostelium, a β-catenin-related protein, Aardvark, promotes prespore gene expression and is also found in actin-containing cell junctions (9). Thus, β-catenin functions are conserved outside the animal kingdom. However, the detailed mechanisms of Wnt/β-catenin signaling have changed during evolution, with “noncanonical” signaling pathways defined in C. elegans and Dictyostelium (10-12).
β-catenin/Arm is part of a superfamily of metazoan Arm-repeatcontaining proteins that regulate cell signaling, the cytoskeleton, and protein-protein interactions (13). A superfamily of Arm-repeat-containing proteins is also present in the plant kingdom (13, 14). Very little is known about the functions of the majority of these proteins. Plant Arm-repeat proteins have known roles in light/gibberellin signaling (15), self-incompatibility (15), trichome development (16, 17), abscisic acid signaling, (18) and receptor-kinase signaling (19). However, apart from Brassica ARC1 (20), the mechanisms of protein function are largely unknown.
We wanted to discover the functions of ARABIDILLO-1 and -2, the two Arabidopsis proteins with greatest sequence homology to metazoan and Dictyostelium β-catenin (13). ARABIDILLO-1 and -2 are unique among Arabidopsis Arm-repeat proteins in having an F-box motif (13, 14) and fall into a phylogenetically distinct subgroup from other plant Arm-repeat proteins (14). Here, we show that ARABIDILLO-1 and -2 regulate root architecture by promoting root branching. Root architecture determines a plant's ability to acquire water and nutrients and is regulated by many signals, including plant hormones (21) and nutrient status (22, 23), allowing the plant to adapt dynamically to a changing environment.
arabidillo-1/-2 mutants and ARABIDILLO-overexpressing seedlings can respond normally to exogenous lateral root-regulating signals such as auxin, suggesting that they regulate root branching by a previously undefined mechanism. We show that full-length ARABIDILLO-1 and -2 localize to the nucleus but that some parts of ARABIDILLO-1 can localize to the cytosol, suggesting that ARABIDILLO-1's localization may depend on the availability of interaction partners, similar to β-catenin. Overexpression of ARABIDILLO protein fragments reduces lateral root formation, possibly by competing for endogenous ARABIDILLO-interacting proteins. We propose that ARABIDILLO-1 and -2 may target an inhibitor of lateral root development for proteasomal degradation.
Thus, we have shown that Arabidopsis proteins related to β-catenin/Arm define a previously uncharacterized pathway that promotes root branching, an important developmental process.
Results
Arabidopsis Genes Related to β-Catenin/Arm Are Expressed Throughout the Plant. As described previously, ARABIDILLO-1 and -2 show the greatest sequence homology of any Arabidopsis genes to Dictyostelium and metazoan β-catenin/Armadillo (13). There is a single ARABIDILLO homologue in the model monocot rice (accession no. AAP55033).
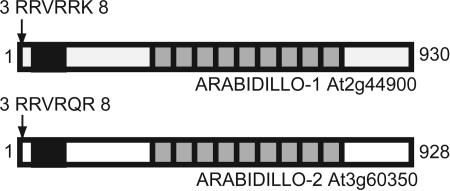
ARABIDILLO-1 and -2 contain nine Arm repeats (Fig. 1) and share 80% amino acid identity (92% within the Arm-repeat region). Dictyostelium β-catenin (Aardvark), ARABIDILLO-1 and -2, and Oryza ARABIDILLO have an F-box motif (residues 45-93 in ARABIDILLO-1, 38-85 in ARABIDILLO-2) (ref. 13 and Fig. 1). ARABIDILLO-1 is also annotated as AtFBX5 (24). The F-box/Arm-repeat-domain structure is unique to these four proteins (www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi). ARABIDILLO-1 and -2 contain a basic nuclear-localization signal (NLS) at their N termini (Fig. 1; and see Fig. 6).
Fig. 1.
ARABIDILLO-1 and -2 are Arabidopsis Arm-related proteins. Shown are ARABIDILLO-1 (At2g44900) and ARABIDILLO-2 (At3g60350) proteins. Fboxes (residues 45-93 in ARABIDILLO-1, 38-85 in ARABIDILLO-2), black; Arm repeats, light gray. Nuclear-localization sequences (residues 3-8) are marked with an arrow. Numbers indicate amino acids.
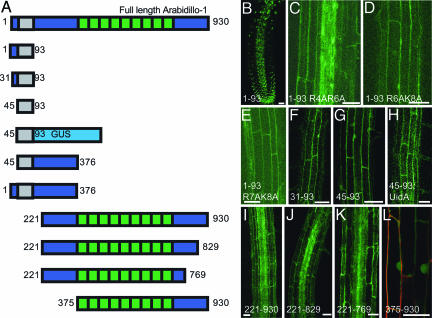
Fig. 6.
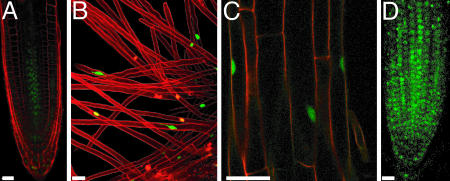
ARABIDILLO-1 protein targeting. (A) Truncated YFP fusion proteins. Green boxes, Arm repeats; gray box, F-box (residues 45-93); dark blue, remainder of ARABIDILLO-1 protein; light blue, GUS. (B) Nuclear localization of residues 1-93. (C-E) Loss of nuclear localization in mutants (residues 1-93) R4AR6A, R6AK8A, and R7AK8A. (F and G) Cytosolic localization of residues 31-93 and 45-93. (H) Cytosolic localization of 45-93-GUS. (I-K) Cytosolic localization of residues 221-930 (I), 221-829 (J), and 221-769 (K). (L) Cytosolic and nuclear localization of residues 375-930. Representative root confocal sections are shown. Green, YFP; red, propidium iodide. (Scale bars, 25 μm.)
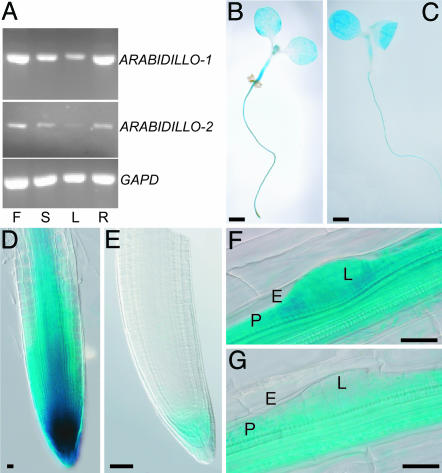
ARABIDILLO-1 and -2 mRNA is detected in all plant organs tested by RT-RCR (Fig. 2A). The spatial expression of ARABIDILLO-1 and -2 was analyzed by fusing their putative promoters to the β-glucuronidase (GUS) gene. pARABIDILLO-1::GUS is active in both roots and shoots of seedlings (Fig. 2B). GUS activity is strongest in the root tip, pericycle, and vasculature (Fig. 2D). pARABIDILLO-2::GUS is also detected in both root and shoot (Fig. 2C), although strong expression is absent from mature primary and lateral root tips, where GUS activity is detected only in the columella (Fig. 2E). Both ARABIDILLO promoters are active in all cells of lateral root primordia (Fig. 2 F and G), with mature lateral root expression patterns refining to resemble primary root patterns as the roots elongate and develop.
Fig. 2.
ARABIDILLO genes are expressed throughout the plant. (A) RT-PCR of ARABIDILLO-1 and -2. F, S, L, and R, cDNA amplified from flowers, stems, leaves, and roots, respectively. GAPD, loading control. (B) pARABIDILLO-1::GUS. (C) pARABIDILLO-2::GUS 5-day seedling expression. (D) pARABIDILLO-1::GUS. (E) pARABIDILLO-2::GUS 5-day primary root-tip expression. (F) pARABIDILLO-1::GUS. (G) pARABIDILLO-2::GUS 8-day expression in lateral root primordia. E, endodermis; P, pericycle; LR, lateral root. [Scale bars, 1 mm (B and C), 25 μm (D), 50 μm (E), and 20 μm (F and G).]
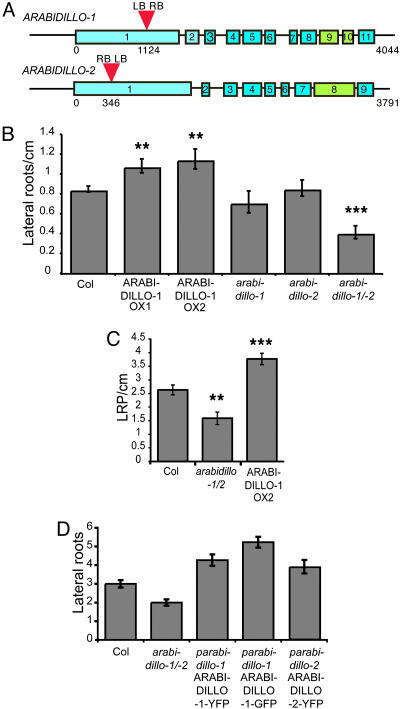
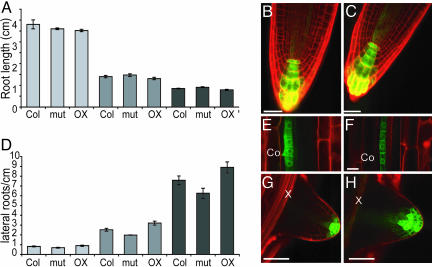
ARABIDILLO-1 and -2 Act Redundantly to Promote Lateral Root Development. To examine the in planta functions of ARABIDILLO-1 and -2, Arabidopsis T-DNA insertion lines were obtained (ref. 25 and Fig. 3A). Neither the arabidillo-1 nor -2 single mutant showed any obvious phenotype. ARABIDILLO-1 and -2 are very similar to each other and show overlapping expression patterns and, thus, could be functionally redundant. Accordingly, when grown on vertical plates, arabidillo-1/-2 double mutants develop fewer lateral roots than wild-type plants or either single mutant, despite having similar primary root lengths to wild type (Fig. 3 B and C). The same result was obtained by using medium with 1% sucrose or basal medium (data not shown). This phenotype could be rescued by stably expressing ARABIDILLO fluorescent proteins from their own promoters (Fig. 3D), suggesting that ARABIDILLO-1 and -2 have a positive function during lateral root development. To test this hypothesis further, Arabidopsis seedlings expressing 35S::ARABIDILLO-1-yellow fluorescent protein (YFP) (Fig. 5) were assayed on vertical plates. ARABIDILLO-1-YFP overexpression increases lateral root formation (Fig. 3 B and C).
Fig. 3.
arabidillo-1 and -2 T-DNA insertion mutants. (A) T-DNA insertions (red) in ARABIDILLO-1 and -2. Blue/green boxes show corresponding exons (numbered) in each gene. Numbers below represent base pairs from the start codon. LB/RB, left/right T-DNA border. (B) Mean lateral root density in 11-day-old seedlings. Left to right, wild type; two independent 35S::ARABIDILLO-1-YFP lines (ARABIDILLO-1 OX1 and ARABIDILLO-1 OX2); arabidillo-1 mutant; arabidillo-2 mutant; arabidillo-1/-2 mutant. Error bars ± SEM. Mean root lengths: Col, 4.00 cm ± 0.05 cm; ARABIDILLO-1 OX1 3.96 cm ±0.11 cm; ARABIDILLO-1 OX2 4.07 cm ± 0.09 cm; arabidillo-1 4.21 cm ± 0.11 cm; arabidillo-2 4.33 cm ± 0.10 cm; and arabidillo-1/2 3.58 cm ± 0.06 cm. t test, ***, P < 0.001 for arabidillo-1/-2 and **, P < 0.01 for ARABIDILLO-1 OX1 and -2. (C) Mean lateral root primordia density at high magnification in 11-day-old seedlings. t test, **, P < 0.01 for arabidillo-1/-2 and ***, P < 0.001 for ARABIDILLO-1 OX2. (D) Lateral roots are restored to arabidillo-1/-2 by ARABIDILLO-fluorescent proteins. Left to right, mean lateral root number in wild type, arabidillo-1/-2, and independent arabidillo-1/-2 lines expressing pARABIDILLO-1::ARABIDILLO-1-YFP, pARABIDILLO-1::ARABIDILLO-1-GFP, and pARABIDILLO-2::ARABIDILLO-2-YFP, respectively.
Fig. 5.
ARABIDILLO-YFP fusion proteins are nuclear. (A) 35S::ARABIDILLO-1-YFP, root tip, confocal section. (B) 35S::ARABIDILLO-1-YFP, root hairs, confocal projection. (C) 35S::ARABIDILLO-2-YFP, root epidermal cells, confocal section. (D) 35S::ARABIDILLO-2-YFP, root tip confocal projection. Green, YFP; red, propidium iodide cell wall counterstain. (Scale bars, 25 μm.)
To assay the stage of lateral root development affected by ARABIDILLO-1 and -2, the number of lateral root primordia (LRP) in wild type, arabidillo-1/-2 and ARABIDILLO-1 overexpressing seedlings was observed at high magnification (Fig. 3C). arabidillo-1/-2 and ARABIDILLO-1 overexpressors show a significant decrease and increase, respectively, in LRP density compared with wild type. No significant changes in later stages of lateral root development were observed, indicating that ARABIDILLO-1 and -2 function redundantly to promote lateral root development in Arabidopsis by controlling lateral root initiation rather than subsequent stages of lateral root development.
ARABIDILLO-1 and -2 Do Not Act Directly in Known Lateral Root-Regulatory Pathways. To test whether ARABIDILLO-1 and -2 act within known signaling networks to promote lateral root development, responses to extracellular signals were assayed. The plant hormone auxin is required for many developmental processes, including lateral root formation. Auxin-signaling mutants have reduced lateral root density (26-29). However, mutation of auxinsignaling proteins also affects additional processes, such as shoot size, primary root length, gravitropism, and floral development. Auxin-signaling mutants are less sensitive to the effects of exogenous auxins and auxin-transport inhibitors (26-29).
arabidillo-1/-2 mutants and ARABIDILLO-overexpressing lines have primary root lengths similar to wild type (Figs. 3 and 4) and do not have auxin-related shoot phenotypes. The arabidillo-1/-2 mutant and ARABIDILLO-1-YFP-overexpressing lines were grown on the auxins indole-3 acetic acid (IAA) and 2,4 dichlorophenoxyacetic acid (2,4D) for 7 days (Fig. 4). Both the arabidillo-1/-2 mutant and the ARABIDILLO-overexpressing lines displayed wild-type root-growth-inhibition responses, indicating that ARABIDILLO-1 and -2 do not change auxin perception or response (Fig. 4A). In addition, arabidillo-1/-2 mutants and ARABIDILLO-1 overexpressors show an increased lateral root density when treated with IAA and 2,4D, similar to wild type (Fig. 4D). Expression of the auxin-responsive reporter DR5rev::GFP (30) is identical in wild type and arabidillo-1/-2 mutants in primary roots (Fig. 4 B and C), dividing pericycle cells that form very early lateral root primordia (Fig. 4 E and F) and emerged lateral roots (Fig. 4 G and H). These data suggest that ARABIDILLO-1 and -2 are not directly involved in auxin signaling during lateral root development.
Fig. 4.
Effects of auxin on arabidillo mutants and overexpressing lines. (A) Root lengths of wild type (Col), arabidillo-1/-2 (mut), and ARABIDILLO-1-overexpressing (OX) seedlings grown on 0.5× MS (light gray), 1 μM IAA (medium gray), and 20 nM 2,4D (dark gray). (B and C) Expression of DR5rev::GFP in the primary root tip of 2-day-old wild-type (B) and arabidillo-1/-2 (C) seedlings. (D) Lateral root density in wild-type (Col), arabidillo-1/-2 (mut), and ARABIDILLO-1-overexpressing (OX) seedlings grown on 0.5× MS (light gray), 1 μM IAA (medium gray), and 20 nM 2,4D (dark gray). (E and F) Expression of DR5rev::GFP in dividing pericycle cells (sites of lateral root initiation) of wild-type (E) and arabidillo-1/-2 (F) seedlings. Co, cortex. (G and H) Expression of DR5rev::GFP in an emerged lateral root of wild-type (G) and arabidillo-1/-2 (H) seedlings. X, xylem. [Scale bars, 25 μm (B, C, G, and H) and 10 μM (E and F).]
arabidillo-1/-2 mutant and ARABIDILLO-1-overexpressing seedlings show a similar response to wild type when treated with 0.5 μM abscisic acid (ABA) (data not shown). ABA is postulated to affect lateral root elongation (31) rather than initiation and so is likely to be working at a later developmental stage than ARABIDILLO-1 and -2. This also suggests that nitrate- or osmoticasensing pathways do not depend on ARABIDILLO-1 and -2 (32, 33). arabidillo-1/-2 mutants and ARABIDILLO-1-overexpressing seedlings responded similarly to wild type when depleted of nutrients (nitrate, phosphate, or sulfate; data not shown). Thus, ARABIDILLO function defines a previously uncharacterized mechanism for promoting lateral root development.
ARABIDILLO-1 and -2 Proteins Localize to the Nucleus. Animal Arm/β-catenin has both nuclear and cytoskeletal functions. To determine where ARABIDILLO-1 and -2 might function in cells, the full-length proteins were expressed as YFP fusions driven from the cauliflower mosaic virus 35S promoter. Both ARABIDILLO-1-YFP and ARABIDILLO-2-YFP are detected exclusively in nuclei (Fig. 5). Despite being expressed from the 35S promoter, the fusion proteins are detected only in root cells. This result could be due to spatial regulation of ARABIDILLO translation, cell-specific ARABIDILLO protein degradation, or low levels of protein expression coupled with the relative ease of fluorescent-protein detection in root cells compared with shoot.
ARABIDILLO-1 Protein Targeting. To understand how the various ARABIDILLO sequence motifs contribute to protein function, a series of truncated ARABIDILLO-1-YFP fusions was generated (Fig. 6A) and expressed stably in Arabidopsis. For each protein, the subcellular localization of YFP was identified.
The first 93 aa (1-93) of ARABIDILLO-1 fused to YFP localize to the nucleus (Fig. 6B). Mutation of single residues in the NLS has no effect on the localization of residues 1-93, but mutation of pairs of residues (R4AR6A, R6AR7A, R6AK8A, or R7AK8A) moves localization away from the nucleus and into the cytosol (Fig. 6 C-E). Deletion of the first 31 or 45 aa of this sequence [proteins 31-93 and 45-93 (the F-box), respectively] also leads to a partial relocation of the fusion protein to the cytosol (Fig. 6 F and G). Addition of a GUS protein module to the F-box upstream of YFP (45-93:GUS, ≈77 kDa) relocates the F-box to the cytosol (Fig. 6H), suggesting that the partial nuclear localization seen with proteins 1-93 (mutated), 31-93, and 45-93 is because their small size allows diffusion into the nucleus.
Expression of the entire N terminus of ARABIDILLO-1, up to the start of the Arm-repeat domain, (residues 1-376) or the N terminus without the NLS (residues 45-376) does not lead to YFP expression. This result might be due to a degradation signal in ARABIDILLO-1 between residues 93 and 221.
A protein consisting of partial N terminus, the Arm-repeat domain and C-terminal region (residues 221-930) is cytosolic, as are similar constructs with truncated C termini (residues 221-769 and 221-829; Fig. 6 I-K). A protein containing only the Arm domain and C terminus (residues 375-930, >80 kDa) localizes to the nucleus and cytosol (Fig. 6L). The partial nuclear localization of residues 375-930 shows that ARABIDILLO-1 protein can localize to the nucleus in the absence of a classical NLS, presumably through its Arm-repeat domain, as is seen with β-catenin (34). There are two putative nuclear export signals (consensus L/VXXL/VXL/V) between residues 221 and 375, but the localization of residues 221-930 is not sensitive to Leptomycin B (J.C.C., unpublished work).
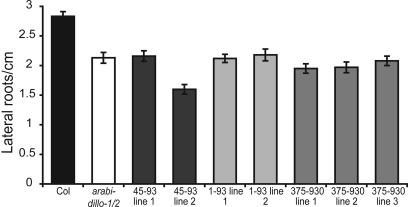
Overexpression of ARABIDILLO-1-YFP Protein Fragments Reduces Lateral Root Development. To understand how ARABIDILLO proteins promote lateral root development, we observed the effect of truncated ARABIDILLO-1-YFP protein overexpression on lateral root number. Wild-type seedlings expressing F-box fragments (1-93 and 45-93) form a similar number of lateral roots to arabidillo-1/-2 mutants (Fig. 7), suggesting that F-box fragments block endogenous ARABIDILLO activity by disrupting the interaction between the full-length proteins and a binding partner(s), for example, component(s) of the proteasome.
Fig. 7.
ARABIDILLO-1-YFP protein fragments reduce lateral root density. Lateral root density of 11-day-old seedlings. Left to right, Wild type (Col, black); arabidillo-1/-2 (white); wild-type seedlings expressing ARABIDILLO-1 (45-93)-YFP, 2 independent lines (dark gray); wild-type seedlings expressing ARABIDILLO-1 (1-93)-YFP, 2 lines (light gray); wild-type seedlings expressing ARABIDILLO-1 (375-930)-YFP, 3 lines (medium-gray). t test, P < 0.001 for all lines compared with wild type.
Seedlings overexpressing ARABIDILLO-1 (residues 375-930)-YFP form a reduced number of lateral roots (Fig. 7), suggesting that this protein also inhibits endogenous ARABIDILLO function. None of the seedlings expressing ARABIDILLO protein fragments show additional phenotypes, suggesting that the fragments act as ARABIDILLO-specific “dominant negatives.”
Thus, we conclude that both the F-box and the C-terminal (Arm-repeat) domain are required for in vivo ARABIDILLO protein function and that both regions of the protein may interact with specific protein partners.
Discussion
β-Catenin-Related Proteins Promote Lateral Root Development in Arabidopsis.` Arabidopsis possesses >100 Arm-repeat-containing proteins (14), of which ARABIDILLO-1 and -2 are most similar to metazoan and Dictyostelium β-catenins (13). Here, we show that ARABIDILLO-1 and -2 promote lateral root development. arabidillo-1/-2 mutant seedlings form fewer lateral roots than wild type and arabidillo single mutants, whereas plants overexpressing ARABIDILLO-1 have an increased lateral root number.
ARABIDILLO-1 and -2 Interact with Other Proteins to Promote Lateral Root Development. Overexpression of ARABIDILLO-1 protein fragments reduces lateral root formation, suggesting that the protein fragments act as dominant negatives, perturbing the function of endogenous ARABIDILLO proteins. Given their nuclear localization, ARABIDILLO-1 and -2 are likely to function by regulating gene expression either directly (as seen with nuclear β-catenin during “canonical” Wnt signaling) or indirectly. Overexpression of the C-terminal fragment (residues 375-930) of ARABIDILLO-1 could block lateral root formation by blocking a signaling function of the Arm domain, for example by forming a nonfunctional transcriptional complex on a target promoter, if N-terminal sequences are also required for transcriptional activation. However, N-terminally truncated animal β-catenin is able to activate transcription in certain contexts.
ARABIDILLO-1 and -2 contain an F-box motif. F-box proteins are specificity factors for multisubunit E3 ubiquitin ligases that degrade target proteins (35). Thus, ARABIDILLO-1 and -2 could target a protein(s), for example, an inhibitor of lateral root development, for proteasomal degradation. The Arm domain may, therefore, inhibit ARABIDILLO function by blocking the interaction of a lateral root inhibitor with the Arm domain of endogenous ARABIDILLO proteins, whereas F-box fragments would inhibit the degradation of target inhibitor(s) by blocking the interaction of endogenous ARABIDILLO with the proteasome.
ARABIDILLO-1 and -2 Do Not Function Within Known Lateral Root Signaling Networks. arabidillo-1/-2 mutants formed fewer lateral roots than wild type, whereas ARABIDILLO-1-overexpressing plants formed more, under all conditions tested. The fact that neither mutant nor overexpressing plants were hypersensitive or resistant to hormonal or nutritional signals suggests that ARABIDILLO-1 and -2 can be added to the small number of known “intrinsic” lateral root regulatory genes (23).
ARABIDILLO-1 and -2 may regulate a competence or likelihood of pericycle cells to divide to form lateral roots. It is known that β-catenin stimulates cell proliferation through transcription of G1 cyclins (36-38). Overexpression of Kip-Related Protein-2, a G1-S-specific cyclin-dependent kinase inhibitor, reduces, but does not abolish, lateral root formation in Arabidopsis (39), similar to the
arabidillo-1/-2 mutant and, thus, could be a candidate for a lateral root inhibitor that ARABIDILLO-1 and -2 degrade independently of auxin signaling.
The arabidillo mutant and overexpressing phenotypes also resemble the root phenotypes of Arabidopsis heterotrimeric G protein-subunit mutants (40). Mutants in the β-subunit, agb-1, have widespread developmental effects, including a doubling of lateral root number. Conversely, gpa-1 α-subunit mutants have half the lateral roots of wild type. Neither GPA-1 nor AGB-1 interact directly with auxin signaling. GPA-1 and AGB-1 may integrate multiple signals controlling cell proliferation (40, 41).
ARABIDILLO-1 Protein Targeting. We have shown that different regions of ARABIDILLO-1 can target YFP to different parts of the cell. The ARABIDILLO-1 N terminus contains a NLS. β-catenin does not contain a classical NLS, but the Arm-repeat domain allows nuclear targeting in an NLS-independent manner (34). The same is true of the Arm domain of ARABIDILLO-1. Whether the ARABIDILLO-1 NLS is regulated in vivo is not known.
Are ARABIDILLO-1 and -2 True Homologues of Animal and Dictyostelium β-Catenin? The phenotype of the arabidillo-1/-2 mutant is subtle compared with the lethal effects of Arm/β-catenin loss of function in animals, which leads to cell patterning defects and developmental arrest (42-46). C. elegans has three β-catenin homologues, wrm-1, hmp-2, and bar-1. Loss of wrm-1 function is embryo-lethal because of a failure to specify endoderm (47). Loss of hmp-2 arrests embryo development during gastrulation because of failed cell-cell adhesion (6). However, loss of bar-1 is not lethal but causes postembryonic cell-fate defects in the vulva (48) and certain neurons (49).
Dictyostelium and plants reprogram their development in response to a changing environment. The Dictyostelium β-catenin (aardvark) mutant is not lethal. Although Aardvark functions analogously to β-catenin (9, 50), the aardvark mutant is able to complete multicellular development, as is arabidillo-1/-2.
Our data suggest that the mechanism by which ARABIDILLO-1 and -2 promote lateral root development may be distinct from the direct transcriptional function of animal β-catenin during canonical Wnt signaling. However, indirect transcriptional functions for β-catenin during Wnt signaling also exist; for example, C. elegans WRM-1 promotes the nuclear export of the LEF/TCF transcription factor POP-1 (5). Dictyostelium Aardvark, like ARABIDILLO-1 and -2, contains an F-box. The mechanism by which Aardvark regulates transcription and cell fate is unknown; Aardvark protein localizes to the cytosol in growing cells (9), suggesting that it may affect transcription indirectly.
Whether ARABIDILLO-1 and -2 resemble metazoan β-catenin by directly regulating transcription and/or cytoskeletal processes or whether they were coopted in plants for a distinct developmental mechanism remains to be established.
Methods
Cloning and Construct Generation. ARABIDILLO promoter sequences were amplified from Col-0 genomic DNA and inserted into pBI101 to make pAR ABIDILLO::GUS reporters. 35S::ARABIDILLO-YFP fusions were made in pGreen0029 (51). Full-length and truncated ARABIDILLO-1 and -2 coding regions were amplified from Col-0 genomic DNA. The ARABIDILLO-1 NLS was mutagenized by using QuikChange XL (Stratagene). Constructs in Agrobacterium tumefaciens GV3101 were transformed into Arabidopsis by floral dip (52).
Plant Growth Conditions. Arabidopsis seedlings were grown in sterile long-day conditions on 0.5× Murashige and Skoog (MS) medium (Sigma M0404), 1% agar, pH 5.7. Mature plants were grown in Levington M3 compost/vermiculite in the greenhouse.
RT-PCR. Total RNA was prepared from whole seedlings, mature aerial tissues, and seedling root tissue (Qiagen RNeasy plant miniprep). Gene-specific cDNA products were amplified from 50 ng of RNA by One-Step RT-PCR (Qiagen) with 32 cycles of PCR. The GAPD gene was amplified as a control.
GUS Staining and Microscopy. Seedlings were assayed for GUS activity according to standard protocols (53). Tissue was cleared through an ethanol series and mounted in Hoyer's medium. For live tissue imaging, Leica TCS and BioRad Radiance 2400 confocal microscopes were used. Seedlings were incubated in 5 mg/liter propidium iodide for 1.5 min and rinsed in distilled water before imaging.
Mutant Analysis. Homozygous, single-insert arabidillo-1 and -2 lines were identified from the Syngenta SAIL collection (25) and screened by segregation analysis, PCR with the recommended primers (25), and Southern blotting (DIG system; Roche). The Nucleon Phytopure kit (Amersham Pharmacia Biotech) was used for genomic DNA extraction. arabidillo-1 and -2 lines were crossed to generate double mutants.
Root Assays. Seedlings were grown vertically as a single row of seeds planted 1.5 cm from the top of 0.5× MS plates. Visible lateral roots were counted 7-13 days after germination. Root length was measured from digital photographs by using the program imagej (http://rsb.info.nih.gov/ij). Seedling lateral root density was defined as lateral roots per cm of primary root.
For growth regulator experiments, 0.5× MS plates were supplemented with 1 μM IAA, 20 nM 2,4D, 0.5-1 μM abscisic acid, or the relevant solvent control.
Nutrient experiments were based on several protocols (54-56). Control plates (pH 5.7) were 20 mM NH4NO3, 3 mM CaCl2, 1.5 mM MgSO4, 19 mM KNO3, 1.25 mM KH2PO4, 1× micronutrients (Bio-101 systems 5100-414), 0.5% sucrose, and 1% agar. Low-nitrogen plates contained 19 mM KCl instead of KNO3 and 10 μM NH4NO3; MgSO4 was substituted with MgCl2 (low sulfur). Low phosphorous macronutrients were 2 mM NH4NO3, 0.3 mM CaCl2, 1.9 mM KNO3, and 1.25 mM KCl.
Acknowledgments
J.C.C. thanks Chris Franklin, Adrian Harwood, Sarah Hodge, Smita Kurup, John Runions, Mark Tester, Elisabeth Truernit, and Mike Wheeler for support and discussion. J.C.C. is funded by the Gatsby Charitable Foundation, including an Interdisciplinary Postdoctoral Fellowship. L.L. was supported by European Molecular Biology Organization Fellowship ALTF110-1999.
Author contributions: J.C.C. designed research; J.C.C. and L.L. performed research; J.C.C. contributed new reagents/analytic tools; J.C.C., L.L., and J.H. analyzed data; and J.C.C. and L.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Arm, Armadillo; GUS, β-glucuronidase; IAA, indole-3 acetic acid; MS, Murashige and Skoog medium; NLS, nuclear-localization signal; YFP yellow fluorescent protein; 2,4D, 2,4 dichlorophenoxyacetic acid.
References
- 1.Conacci-Sorrell, M., Zhurinsky, J. & Ben-Ze'ev, A. (2002) J. Clin. Invest. 109, 987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logan, C. Y. & Nusse, R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781-810. [DOI] [PubMed] [Google Scholar]
- 3.Brannon, M., Gomperts, M., Sumoy, L., Moon, R. T. & Kimelman, D. (1997) Genes Dev. 11, 2359-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riese, J., Yu, X., Munnerlyn, A., Eresh, S., Hsu, S. C., Grosschedl, R. & Bienz, M. (1997) Cell 88, 777-787. [DOI] [PubMed] [Google Scholar]
- 5.Lo, M. C., Gay, F., Odom, R., Shi, Y. & Lin, R. (2004) Cell 117, 95-106. [DOI] [PubMed] [Google Scholar]
- 6.Costa, M., Raich, W., Agbunag, C., Leung, B., Hardin, J. & Priess, J. R. (1998) J. Cell Biol. 141, 297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, R. T., Kirkpatrick, C. & Peifer, M. (1996) J. Cell Biol. 134, 133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haegel, H., Larue, L., Ohsugi, M., Fedorov, L., Herrenknecht, K. & Kemler, R. (1995) Development (Cambridge, U.K.) 121, 3529-3537. [DOI] [PubMed] [Google Scholar]
- 9.Grimson, M. J., Coates, J. C., Reynolds, J. P., Shipman, M., Blanton, R. L. & Harwood, A. J. (2000) Nature 408, 727-731. [DOI] [PubMed] [Google Scholar]
- 10.Coates, J. C. & Harwood, A. J. (2001) J. Cell Sci. 114, 4349-4358. [DOI] [PubMed] [Google Scholar]
- 11.Korswagen, H. C. (2002) BioEssays 24, 801-810. [DOI] [PubMed] [Google Scholar]
- 12.Thorpe, C. J., Schlesinger, A. & Bowerman, B. (2000) Trends Cell Biol. 10, 10-17. [DOI] [PubMed] [Google Scholar]
- 13.Coates, J. C. (2003) Trends Cell Biol. 13, 463-471. [DOI] [PubMed] [Google Scholar]
- 14.Mudgil, Y., Shiu, S. H., Stone, S. L., Salt, J. N. & Goring, D. R. (2004) Plant Physiol. 134, 59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amador, V., Monte, E., Garcia-Martinez, J. L. & Prat, S. (2001) Cell 106, 343-354. [DOI] [PubMed] [Google Scholar]
- 16.Downes, B. P., Stupar, R. M., Gingerich, D. J. & Vierstra, R. D. (2003) Plant J. 35, 729-742. [DOI] [PubMed] [Google Scholar]
- 17.El Refy, A., Perazza, D., Zekraoui, L., Valay, J. G., Bechtold, N., Brown, S., Hulskamp, M., Herzog, M. & Bonneville, J. M. (2003) Mol. Genet. Genom. 270, 403-414. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S., Choi, H. I., Ryu, H. J., Park, J. H., Kim, M. D. & Kim, S. Y. (2004) Plant Physiol. 136, 3639-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, M., Cho, H. S., Kim do, M., Lee, J. H. & Pai, H. S. (2003) Biochim. Biophys. Acta 1651, 50-59. [DOI] [PubMed] [Google Scholar]
- 20.Stone, S. L., Anderson, E. M., Mullen, R. T. & Goring, D. R. (2003) Plant Cell 15, 885-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G. & Bennett, M. J. (2003) Trends Plant Sci. 8, 165-171. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Bucio, J., Cruz-Ramirez, A. & Herrera-Estrella, L. (2003) Curr. Opin. Plant Biol. 6, 280-287. [DOI] [PubMed] [Google Scholar]
- 23.Malamy, J. (2005) Plant Cell Environ. 28, 67-77. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, W. & Jang, J. (2000) Trends Plant Sci. 5, 454-457. [DOI] [PubMed] [Google Scholar]
- 25.Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., et al. (2002) Plant Cell 14, 2985-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie, Q., Frugis, G., Colgan, D. & Chua, N. H. (2000) Genes Dev. 14, 3024-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J. & Estelle, M. (1998) Genes Dev. 12, 198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfluger, J. & Zambryski, P. (2004) Development (Cambridge, U.K.) 131, 4697-4707. [DOI] [PubMed] [Google Scholar]
- 29.Fukaki, H., Tameda, S., Masuda, H. & Tasaka, M. (2002) Plant J. 29, 153-168. [DOI] [PubMed] [Google Scholar]
- 30.Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R. & Jurgens, G. (2003) Nature 426, 147-153. [DOI] [PubMed] [Google Scholar]
- 31.De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C. H. & Zhang, H. (2003) Plant J. 33, 543-555. [DOI] [PubMed] [Google Scholar]
- 32.Deak, K. I. & Malamy, J. E. (2005) Plant J. 43, 17-28. [DOI] [PubMed] [Google Scholar]
- 33.Signora, L., De Smet, I., Foyer, C. H. & Zhang, H. (2001) Plant J. 28, 655-662. [DOI] [PubMed] [Google Scholar]
- 34.Fagotto, F., Gluck, U. & Gumbiner, B. M. (1998) Curr. Biol. 8, 181-190. [DOI] [PubMed] [Google Scholar]
- 35.Moon, J., Parry, G. & Estelle, M. (2004) Plant Cell 16, 3181-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botrugno, O. A., Fayard, E., Annicotte, J. S., Haby, C., Brennan, T., Wendling, O., Tanaka, T., Kodama, T., Thomas, W., Auwerx, J. & Schoonjans, K. (2004) Mol. Cell 15, 499-509. [DOI] [PubMed] [Google Scholar]
- 37.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96, 5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tetsu, O. & McCormick, F. (1999) Nature 398, 422-426. [DOI] [PubMed] [Google Scholar]
- 39.Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inze, D. & Beeckman, T. (2002) Plant Cell 14, 2339-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullah, H., Chen, J. G., Temple, B., Boyes, D. C., Alonso, J. M., Davis, K. R., Ecker, J. R. & Jones, A. M. (2003) Plant Cell 15, 393-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullah, H., Chen, J. G., Young, J. C., Im, K. H., Sussman, M. R. & Jones, A. M. (2001) Science 292, 2066-2069. [DOI] [PubMed] [Google Scholar]
- 42.Wieschaus, E. & Riggleman, R. (1987) Cell 49, 177-184. [DOI] [PubMed] [Google Scholar]
- 43.Wikramanayake, A. H., Huang, L. & Klein, W. H. (1998) Proc. Natl. Acad. Sci. USA 95, 9343-9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imai, K., Takada, N., Satoh, N. & Satou, Y. (2000) Development (Cambridge, U.K.) 127, 3009-3020. [DOI] [PubMed] [Google Scholar]
- 45.Huelsken, J., Vogel, R., Brinkmann, V., Erdmann, B., Birchmeier, C. & Birchmeier, W. (2000) J. Cell Biol. 148, 567-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heasman, J., Crawford, A., Goldstone, K., Garner-Hamrick, P., Gumbiner, B., McCrea, P., Kintner, C., Noro, C. Y. & Wylie, C. (1994) Cell 79, 791-803. [DOI] [PubMed] [Google Scholar]
- 47.Rocheleau, C. E., Downs, W. D., Lin, R., Wittmann, C., Bei, Y., Cha, Y. H., Ali, M., Priess, J. R. & Mello, C. C. (1997) Cell 90, 707-716. [DOI] [PubMed] [Google Scholar]
- 48.Eisenmann, D. M., Maloof, J. N., Simske, J. S., Kenyon, C. & Kim, S. K. (1998) Development (Cambridge, U.K.) 125, 3667-3680. [DOI] [PubMed] [Google Scholar]
- 49.Maloof, J. N., Whangbo, J., Harris, J. M., Jongeward, G. D. & Kenyon, C. (1999) Development (Cambridge, U.K.) 126, 37-49. [DOI] [PubMed] [Google Scholar]
- 50.Coates, J. C., Grimson, M. J., Williams, R. S., Bergman, W., Blanton, R. L. & Harwood, A. J. (2002) Mech. Dev. 116, 117-127. [DOI] [PubMed] [Google Scholar]
- 51.Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. (2000) Plant Mol. Biol. 42, 819-832. [DOI] [PubMed] [Google Scholar]
- 52.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 53.Weigel, D. & Glazebrook, J. (2002) Arabidopsis: A Laboratory Manual (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 54.Zhang, H. & Forde, B. G. (1998) Science 279, 407-409. [DOI] [PubMed] [Google Scholar]
- 55.Lopez-Bucio, J., Hernandez-Abreu, E., Sanchez-Calderon, L., Nieto-Jacobo, M. F., Simpson, J. & Herrera-Estrella, L. (2002) Plant Physiol. 129, 244-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kutz, A., Muller, A., Hennig, P., Kaiser, W. M., Piotrowski, M. & Weiler, E. W. (2002) Plant J. 30, 95-106. [DOI] [PubMed] [Google Scholar]