Abstract
DNA fragmentation is a hallmark of apoptosis (programmed cell death). However, the biological function of apoptotic DNA fragmentation remains unclear. Here, we show that DNA fragmentation factor plays an important role for maintaining genomic stability. Inhibition or loss of the DNA fragmentation factor (DFF)/caspase-activated DNase (CAD), whose nuclease activity is responsible for digesting genomic DNA during apoptosis, led to significant increases in spontaneous or induced gene mutations, gene amplifications, and chromosomal instability in primary mouse cells and transformed human cell lines. The mechanism underlying genetic instability in DFF/CAD-deficient cells, at least in part, involves a small but significant elevation in the survival of cells exposed to ionizing radiation, suggesting that apoptotic DNA fragmentation factor contributes to genomic stability by ensuring the removal of cells that have suffered DNA damage. In support of this hypothesis are the observations of increased cellular transformation of mouse embryonic cells from the DFF/CAD-null mice and significantly enhanced susceptibility to radiation-induced carcinogenesis in these mice. These data, in combination with published reports on the existence of tumor-specific gene mutations/deletions in the DFF/CAD genes in human cancer samples, suggest that apoptotic DNA fragmentation factor is required for the maintenance of genetic stability and may play a role in tumor suppression.
Keywords: apoptosis, cancer, genetic instability
DNA fragmentation is a hallmark of apoptosis (1, 2). It is now recognized that apoptotic DNA fragmentation is carried out by a heterodimeric protein complex called DNA fragmentation factor (DFF) (3, 4) or caspase-activated DNase (CAD) (5, 6). DFF is a heterodimeric protein complex composed of two subunits, DFF40/CAD and DFF45/inhibitor (I)CAD (3). DFF45/ICAD is an inhibitor as well as a chaperone of DFF40/CAD that ensures its proper folding. Expression of DFF40/CAD in various systems in the absence of coexpressed DFF45 results in generation of DFF40-inactive aggregates. Under normal circumstances, DFF40/CAD is complexed with DFF45/ICAD, so its DNase activity is inhibited (3-5). When apoptosis is initiated, the activated caspase cleaves DFF45/ICAD, and DFF40/CAD is released into the nucleus to carry out DNA fragmentation.
Recently, it was discovered that the genes encoding the nuclease that are responsible for the fragmentation of DNA during apoptosis, DFF40/CAD and DFF45/ICAD, are aberrantly expressed in many tumor types. In addition, the abnormalities in this gene are associated with poor prognosis in cancer patients (7-9). Most significantly, tumor-specific DFF45 gene mutations or deletions were identified in human germ cell tumors and neuroblastoma tumors from patients (10, 11), indicating the involvement of this gene in tumor development.
The present study was initiated to explore the potential relationship between apoptotic DNA fragmentation and tumor development. We reasoned that the process involved in the destruction of genomic DNA might have a direct effect on genomic integrity, especially when cells are under assault from DNA-damaging agents. This effect on genomic instability may be responsible for the observed association between DFF abnormality and cancer. Such a relationship is consistent with the view that genomic instability is an important factor during carcinogenesis/malignant tumor development (12-15). Our results indicate that impairment of DNA fragmentation leads to increased genetic instability, cellular transformation, and susceptibility to radiation carcinogenesis and suggest that DNA fragmentation factor may play important roles in maintaining genetic stability and preventing tumorigenesis.
Results
Efficient Inhibition of Apoptotic DNA Fragmentation by Engineered Mutant ICAD Gene Expression. To examine the potential role of DNA fragmentation in maintaining genetic stability, we created cell lines in which the nuclease activities of DFF/CAD (3, 5) were genetically blocked. This result was achieved by stable transduction of a modified copy of the gene DFF45/ICAD (6), which is an inhibitor as well as a chaperone of DFF40/CAD (4-6), the nuclease that is responsible for fragmentation of genomic DNA during cellular apoptosis. Under normal circumstances, DFF45/ICAD is physically associated with DFF40/CAD, preventing its activation. However, when cellular apoptosis is initiated, DFF45/ICAD is digested by activated caspases (e.g., caspase 3) and dissociates from DFF40/CAD, allowing for the activation of the nuclease activities of the latter and subsequent fragmentation of genomic DNA. We followed a published approach to modify the ICAD protein (6). The two caspase-sensitive sites of DFF45/ICAD were mutagenized (D117E and D224E) to derive the mutated ICAD (mICAD) protein (Fig. 1A). The modification made to the protein rendered it resistant to caspases, thereby inhibiting its dissociation from the CAD protein and preventing the activation of the latter. Overexpression of the gene has been shown to block apoptotic DNA fragmentation in cells of human and murine origin (6).
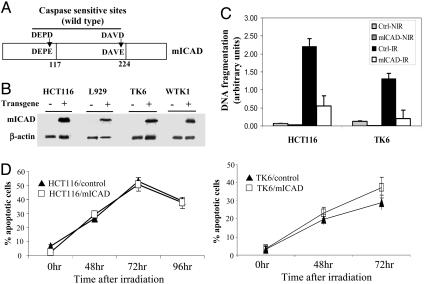
Fig. 1.
Generation of genetically modified cell lines with significantly reduced DNA fragmentation. (A) The structure of the modified ICAD/DFF45 protein. The mutations lead to amino acid changes that render the mutant ICAD protein resistant to caspase cleavage. (B) Western blot analysis of mICAD protein expression in cells that have been stably transduced with a mICAD gene. An antibody (from Roche Molecular Biology) against the hemagglutinin tag that was engineered into the 3′ end of the mICAD gene was used so that only the modified ICAD gene is detected. It is clear that only those cells that were transduced with the mICAD gene express the mutant form of the ICAD protein. (C) Evaluation of the extent of radiation-induced apoptotic DNA fragmentation in cells that were transduced with the mICAD gene. Control and mICAD-transduced cells were irradiated with γ-rays (3 Gy for HCT116 and 1.5 Gy for TK6), and the DNA fragmentation was quantified by an ELISA kit at 72 h after radiation. NIR, nonirradiated; IR, irradiated. (D) Quantification of apoptosis induced by γ-irradiation in mICAD-transduced HCT116 and TK6 cells. Apoptosis was evaluated by an annexin V staining kit. Error bars represent standard deviation.
The mICAD gene was then transduced into three human cell lines HCT116, TK6, and WTK1 and a mouse cell line L929 by a retrovirus-mediated approach. HCT116 is a colon cancer cell line, whereas TK6 and WTK1 are lymphoblastoid cell lines derived from the same individual with differing p53 status (16). L929 is a transformed mouse fibroblast cell line. Stable expression of the mICAD gene was readily detectable in transduced cells (Fig. 1B). The expression of the mICAD gene effectively inhibited the activation of the CAD/DFF40 nuclease, consistent with previous reports (6), evidenced by the fact that ionizing radiation-induced apoptotic DNA fragmentation in mICAD-transduced cells was effectively suppressed by the expression of mICAD (P ≤ 0.001) (Fig. 1C). Inhibition is also confirmed in DNA-ladder-based assays (see Fig. 7, which is published as supporting information on the PNAS web site) and analysis of cells with subG1 DNA content (see Fig. 8, which is published as supporting information on the PNAS web site), an indication of apoptotic DNA fragmentation. Similar to previous studies (6), the inhibition of DNA fragmentation had little effect on the rate of apoptosis measured by annexin V-based apoptosis assays. When externalized cell membrane phosphotidyl serine, an indicator of early apoptosis, was stained (by fluorescence labeled annexin V), no difference was observed between cells transfected with an empty vector and cells transfected with the modified ICAD gene (P > 0.1, Fig. 1D). The lack of effect of CAD deficiency on apoptotic cell death was also confirmed by additional assay (see Figs. 9 and 10B, which are published as supporting information on the PNAS web site).
Increased Frequency of Radiation-Induced Gene Mutations in Mutant ICAD-Expressing Cells. To evaluate the effects of apoptotic DNA fragmentation on nucleotide-level genetic instability, we examined radiation-induced mutation frequencies at the autosomal thymidine kinase (tk) locus and the X-linked hypoxanthine guanine phosphoribosyl transferase (hprt) locus in the TK6 and WTK1 lymphoblastoid cells. These two gene loci were chosen because of the availability of quantitative mutation assays and the abundance of published data in these two cell lines. The hemizygous status of tk and hprt genes in the TK6 and WTK1 cells makes it possible to observe the mutation frequencies at these two gene loci with a reasonable number (<107) of cells. Mutations in the tk gene allow the host cells to be resistant to the cytotoxic drug trifluorothymidine, whereas mutations in the hprt gene allow host cells to be resistant to the cytotoxic drug 6-thioguanine (6TG). The addition of these two drugs will, therefore, allow for the selective outgrowth of cells that have suffered mutations in these two genes, which, in turn, will allow for an evaluation of the mutation frequencies of the two genes in the general cell population.
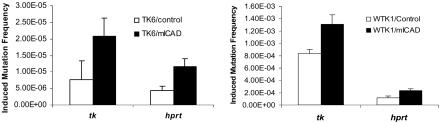
In TK6 cells, mutant ICAD/DFF45 expression significantly enhanced radiation-induced mutation frequencies at both the tk and hprt loci (P ≤ 0.001) (Fig. 2 Left). In WTK1 cells, where the background frequencies of mutation were much higher, the difference was smaller but still significant (P < 0.02) (Fig. 2 Right). One plausible explanation for the smaller difference in WTK1 is the very high background mutation rates in the WTK1 cells, which may overshadow the contribution of DNA fragmentation on induced mutations at these gene loci. These results suggest that apoptotic DNA fragmentation may function to prevent or suppress DNA-damage-induced gene mutations.
Fig. 2.
The frequency of radiation-induced gene mutations at the tk and hprt in TK6 (Left) and WTK1 (Right) cells. Both control and mICAD-transduced cells were used in all experiments. In each experiment, mean and standard deviation were calculated from three independent experiments. Radiation-induced gene mutation frequencies at the tk and hprt gene locus of control and in mICAD-transduced cells were shown. In both TK6 and WTK1 cells, the results were statistically significant (P < 0.05). Background mutation frequencies (from nonirradiated cell populations) were subtracted in each of the experiments shown.
Significantly Elevated Frequency of Gene Amplification in Cells Expressing Mutant ICAD. We next examined the effects of apoptotic DNA fragmentation on spontaneous and DNA-damage-induced gene amplification (17, 18), a commonly used measure of genomic instability in transformed cells (19-22). It has been shown that ionizing radiation can increase the frequency of gene amplification significantly (23, 24). The frequencies of gene amplification were estimated at two gene loci: the cabamyl-P-synthetase, aspartate transcarbarmylase, dihydroorotase (cad) gene and the dihydrofolate reductase (dhfr) gene. Amplifications of these two genes will allow the cells to be resistant to the drugs N-(phosphoacetyl)-l-aspartate (PALA) and methotrexate (MTX), respectively. Therefore, it is possible to estimate the frequency of the cells that have undergone cad or dhfr gene amplifications by quantifying the number of cell colonies that are resistant to PALA or MTX, respectively.
As shown in Table 1, stable mICAD expression in HCT116 caused a significant increase in the level of spontaneously arising PALA-resistant clones compared with parental cells. Furthermore, the frequencies of induced gene amplification in irradiated cells were also much higher (≈5-fold) in mICAD-expressing cells than that in the control cells. An even more impressive increase in PALA-resistant clones was seen in L929 cells. The spontaneously arising frequency of PALA-resistant colonies in mICAD-expressing cells is 670-fold higher than that in control cells (Table 1). In addition, radiation-induced frequency of PALA-resistant clones was ≈16-fold higher in mICAD-expressing cells than in control cells. Amplification of the cad gene in PALA-resistant clones was confirmed by Southern blotting, shown in the supporting data (see Fig. 11, which is published as supporting information on the PNAS web site). In most of the clones examined, cad gene dosage was increased 2- to 3-fold in the cells.
Table 1. Frequency of gene amplification in control or mlCAD-expressing HCT116 and L929 cells.
| PALA selection (for cad gene amplification)
|
MTX selection (for dhfr gene amplification)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug selection/cell types | Irradiation dose | PE, % | LD50, μM | Total no. selected cells (× 106) | PALAr frequency (× 10−5) | PE, % | LD50, μM | Total no. selected cells (× 106) | MTXr frequency (× 10−5) |
| HCT116/control | None | 45.3 | 31.4 | 4.5 | 5.54 | 42.0 | 17.5 | 15.6 | 0.046 |
| HCT116/mlCAD | None | 64.7 | 15.0 | 1.5 | 27.4 | 41.0 | 13.3 | 15.6 | 0.25 |
| HCT116/control | 3 Gy | 59.3 | 31.4 | 2.4 | 8.50 | 13.4 | 17.5 | 14.3 | 0.21 |
| HCT116/mlCAD | 3 Gy | 66.0 | 15.0 | 0.6 | 41.7 | 18.0 | 13.3 | 11.5 | 1.01 |
| L929/control | None | 47.3 | 21.3 | 20 | 0.032 | 47.3 | 31.0 | 10 | 0.15 |
| L929/mlCAD | None | 55.5 | 9.9 | 3 | 21.3 | 55.5 | 16.7 | 12 | 0.81 |
| L929/control | 4.5 Gy | 22.8 | 21.3 | 4.8 | 3.66 | 22.8 | 31.0 | 4.8 | 10.5 |
| L929/mlCAD | 4.5 Gy | 11.8 | 9.9 | 2.4 | 81.6 | 11.8 | 16.7 | 3 | 37.4 |
PE, plating efficiency; LD50, dose at which 50% cell growth was inhibited; PALAr, PALA-resistant clone; MTXr, MTX-resistant clone.
At the dhfr site, a similar pattern was observed. In both HCT116 and L929 cells, mICAD expression caused significant increases in the frequencies of spontaneous and radiation-induced gene amplification (Table 1).
In addition, fluctuation analysis (see Table 2, which is published as supporting information on the PNAS web site) indicated that apoptotic DNA fragmentation is important not only in regulating gene amplification induced by DNA-damaging agents but also under normal growth conditions.
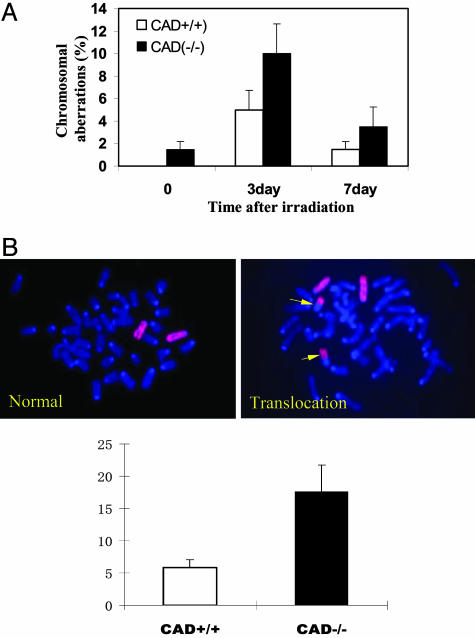
Elevated Chromosome Instability in CAD-Deficient Mice. To examine the effects of the CAD on genetic instability in vivo, we conducted studies in transgenic mice with targeted disruption of the mouse CAD/DFF40 gene (25). Similar to the mICAD-transduced cells, CAD deficiency renders the bone marrow cell derived from CAD-/- mice resistant to apoptotic DNA fragmentation (Fig. 9A) but have minimal effect on overall cellular apoptosis (Fig. 9B). Mutant (CAD-/-) and wild-type (CAD+/+) mice from the same litter were exposed to sublethal doses of whole-body γ-rays. Metaphase chromosomal spreads from bone marrow cells were then prepared and examined for chromosomal aberrations. The aberrations scored included chromatid break, double minute, chromosome terminal deletion, ring chromosome, and dicentric chromosome. Most of these were acute, short-term aberrations that would be lost with cellular proliferation. A significant increase of chromosomal aberrations was seen in the CAD-/- mice when compared with wild-type control cells at both 3 (P < 0.006) and 7 (P < 0.04) days after irradiation of the mice (Fig. 3A; and see Table 3, which is published as supporting information on the PNAS web site). Quantitatively, the fraction of CAD-/- cells with induced aberrations was, on the average, twice as high as that of the wild-type cells. In addition, in nonirradiated CAD-/- animals, ≈1-1.5% metaphase chromosomal spreads prepared from CAD-/- bone marrow cells showed chromosome aberrations in the form of chromatid breaks (Table 3). No chromosomal aberrations were observed in wild-type control cells.
Fig. 3.
Elevation of radiation-induced chromosomal instability in cells with targeted disruption of the CAD gene. (A) Chromosomal aberrations in bone marrow cells derived from irradiated mice. CAD-/- and CAD+/+ mice from the same litter were exposed to sublethal doses of γ-radiation (3 Gy) and were then scored for chromosome aberrations at different time points. (B) Elevated frequency of radiation-induced chromosome 2 translocations in embryonic fibroblast cells derived from CAD-/- mice. (Upper Left and Right) The fluorescent photomicrographs of typical spreads with normal (Left) and translocated (Right) chromosome 2. The yellow arrows show the chromosomes that are involved in translocations. (Lower) The average of chromosome-2-associated translocation frequency from three independent pairs of CAD+/+ and CAD-/- MEFs.
Subsequently, whole-chromosome painting was conducted to evaluate the frequencies of chromosomal translocations, which were indicative of more stable chromosomal abnormalities that will be transmitted to progeny cells. The results revealed abnormally high levels of radiation-induced translocations and rearrangements involving chromosome 2 in mouse embryonic fibroblasts (MEF) isolated from CAD-/- mice when compared with those from wild-type mice (Fig. 3B). In three pairs of CAD-/- and CAD+/+ cells examined, the fractions of cells with chromosome-2-associated translocations were consistently higher in CAD-/- cells after exposure to ionizing radiation (P < 0.03). These data indicate that disruption of the CAD gene leads to elevated frequency of DNA-damage-induced chromosomal instability in vivo and in vitro.
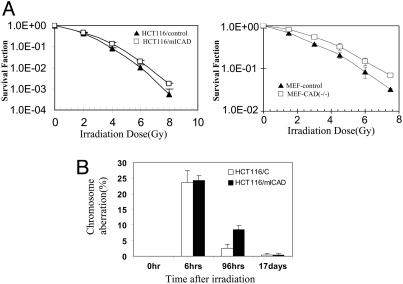
Mechanisms Underlying Genetic Instability in Cells Deficient in DNA Fragmentation. What are the mechanisms responsible for the observed increase in genetic instability in cells with disrupted DFF/CAD genes? A series of experiments was conducted to characterize potential anomalies associated with the disruption of the DFF/CAD genes. Cell cycle analysis indicated that DFF/CAD disruption had no effect on cell cycle progression of the nonirradiated or irradiated cells (data not shown). Subsequently, clonogenic survival assays were carried out to evaluate long-term survival of cells that have suffered DNA damage. In contrast to the results of apoptosis assays (Figs. 1D and 10B), a small but statistically significant (P ≤ 0.02) increase in clonogenic survival in mICAD-expressing HCT116 (Fig. 4A Left), TK6, and WTK1 cells (see Fig. 12, which is published as supporting information on the PNAS web site) were observed. The same difference was also seen in immortalized MEF cells derived from mice with targeted disruption in the CAD gene when they were compared with wild-type control cells (Fig. 4A Right).
Fig. 4.
Increased clonogenic survival and delayed removal of DNA-damaged cells in CAD/DFF impaired cells. (A) Clonogenic survival of mICAD-expressing HCT116 and CAD-/- MEF cells after γ-irradiation. (B) Delayed removal of cells with chromosomal aberrations in mICAD-expressing HCT116 cells. Control and mICAD-transduced HCT116 cells were exposed to ionizing radiation (3 Gy) and examined for chromosomal aberrations at different time points. For each time point, at least 100 metaphases were counted. The error bars represent standard deviation.
The difference in clonogenic survival of the cells after irradiation suggested a potential mechanism for the role of DFF/CAD in maintenance of cellular genomic stability: increased survival of genetically damaged cells that were otherwise destined to die and, therefore, be removed from the cell population. This mechanism was supported by the observations made in HCT116-mICAD cells that were subjected to ionizing radiation exposure. At an earlier time point (6 h) postirradiation, the incidence of radiation-induced chromosomal aberrations were similar between wild-type and CAD/DFF-inhibited cells (P = 0.42, Fig. 4B). However, at a later time point (96 h), the CAD-inhibited cells showed significantly more aberrations than the controls (P < 0.005, Fig. 4B; and see Table 4, which is published as supporting information on the PNAS web site). Similar observations were made in control and mICAD-transduced TK6 and WTK1 cells (data not shown). These data suggest that CAD/DFF plays a role similar to that of the archetypical checkpoint protein p53, namely, the removal of cells that have suffered severe DNA damage. However, we cannot rule out the possibility that other yet unidentified functions of CAD/DFF may also be involved.
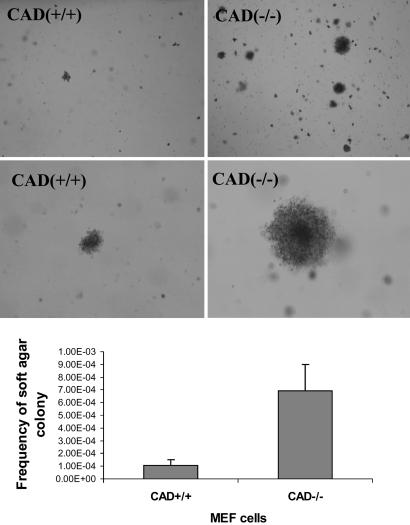
Cellular Transformation in CAD-Null MEF Cells. The increased susceptibility to radiation-induced genetic instability in CAD-deficient cells was reminiscent of similar characteristics observed in mouse cells with disrupted p53 or gadd45 genes (26, 27). To further examine whether the CAD/DFF40 gene possesses functions that are similar to checkpoint genes such as p53 or gadd45, the soft-agar assay, which is a very stringent in vitro assay for malignant cellular transformation, was carried out to examine the ability of MEF cells derived from CAD-/- and wild-type mice (littermates of the CAD-/- mice) to undergo anchorage-independent growth. The observed frequency for the growth of soft-agar colonies in CAD-/- MEF cells was much higher than those observed in CAD+/+ MEF cells (P < 0.01, Fig. 5). When these soft-agar colonies were expanded and inoculated s.c. into athymic nude mice, 100% (20 of 20) of the colonies formed tumors in 3-12 weeks. These results indicated that loss of the CAD gene increased the susceptibility of host MEF cells to oncogenic transformation.
Fig. 5.
Elevated frequency of cellular transformation in embryonic fibroblast cells derived from CAD-/- mice. (Top and Middle) Photomicrographs of typical colony growth in CAD+/+ and CAD-/- plates. The black spots in the photos are the transformed colonies. (Bottom) The average of frequency of soft-agar colonies from three independent pairs of embryonic fibroblasts isolated from CAD+/+ and CAD-/- littermates. The difference between the two groups of mice is statistically significant (P < 0.001).
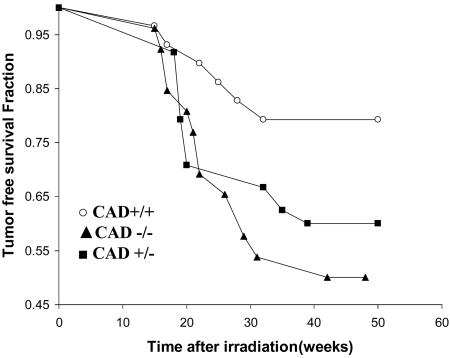
Increased Susceptibility to Radiation Carcinogenesis in CAD-Null Mice. To further examine the potential roles of CAD/DFF in carcinogenesis, radiation-induced carcinogenesis was examined in CAD-/- mice. Wild-type control (CAD+/+), heterozygous (CAD+/-), and CAD-/- mice that were littermates in C57BL/6 background were subjected to the irradiation. One year after the irradiation, significant difference in tumor-free survival was observed between the CAD+/+ and CAD-/- mice (Fig. 6). At 50 weeks posttreatment, the fraction of tumor-free survival in the CAD+/+ group (Fig. 6) was 79% vs. 50% in CAD-/- mice (the difference was statistically significant P < 0.001). These observations indicate a significant role for the DFF/CAD genes in the suppression of radiation carcinogenesis.
Fig. 6.
Increased susceptibility to radiation carcinogenesis in CAD-/- mice. Twenty-nine wild-type, 24 heterozygous, and 26 homozygous littermates were given 6 Gy x-ray whole-body irradiation (given in two fractions at 3 Gy) each). The mice were then observed for 12 months for tumor development. The tumor-free survival of these mice is shown. All tumored mice succumbed to cancers. The difference between the CAD+/+ and CAD-/- groups is significant (P < 0.05).
Discussion
Apoptosis plays an important role in development and homeostasis. Alterations in apoptosis contribute to the pathogenesis of a number of human diseases, including cancer, viral infections, autoimmune diseases, neurodegenerative disorders, and AIDS. However, the biological function for apoptotic DNA fragmentation, a hallmark of the apoptotic process, has not been defined clearly. Our results indicate that apoptotic DNA fragmentation is an important step to maintain genetic stability and suppress cellular transformation/carcinogen-induced tumorigenesis and suggest that DNA fragmentation factor activity is required for the maintenance of genomic stability and prevention of tumorigenesis.
Clinical Relevance of DFF/CAD-Deficiency to Human Cancers. Our results are consistent with previous studies on the potential roles of the DFF/CAD genes in human cancers. In human neuroblastomas, a high percentage (≈28-47%) contain deletions of chromosome 1p (28-31). Analyses of such chromosome deletions suggest that there may be tumor-suppressor gene(s) in the region 1p36.1-1p36.3, which contains both DFF40/CAD and DFF45/ICAD (29, 31-34). In agreement with this finding is a report showing the homozygous deletion of DFF45 in neuroblastoma cell lines (7). Deletions of the DFF45 gene have been shown to disrupt the function of the DFF/CAD complex completely (35). In addition to these studies, tumor-specific mutations from human patients provide additional evidence for the potential role of the loss of DFF/CAD functions in human carcinogenesis (11). An RT-PCR gene-expression study showed that DFF45 is preferably expressed in low-stage neuroblastoma tumors and, to a lesser degree, in high-stage neuroblastomas (10). Decreased expression of DFF45/ICAD is correlated with a poor prognosis in patients with esophageal carcinoma (8). Taken together, the evidence for the involvement of DFF/CAD in human cancer development is strong. The radiation carcinogenesis data provided in this study (Fig. 6) suggested a role of CAD/DFF in tumor suppression, albeit in a murine model.
Discrepancy Between Apoptosis and Clonogenic Survival Assays. How does one reconcile the fact that disruption of apoptotic DNA fragmentation has no measurable effect on apoptosis (Fig. 1D and 10B) but a small and statistically significant influence on clonogenic survival (Fig. 4A; and see Fig. 12)? The discrepancies between the two may reflect different endpoints these assays were measuring. Apoptosis assays measure the fraction of cells undergoing apoptosis at the time of observation, whereas clonogenic assays take into account the cumulative effects of cell death over the entire period of experimentation (2-3 weeks), which is more likely to detect smaller survival differences between two cell populations.
The increase in clonogenic survival after radiation is surprising, given the fact that DNA fragmentation is almost at the end stage of the “execution” phase of the whole apoptotic process. However, each step in the multistep apoptotic process, including those in the execution phase, may influence eventual cell death, as evidenced by two reports that indicated that processes at the end stage of apoptotic death, such as the engulfment (phagocytosis) of apoptotic bodies, could affect the overall death rate (36, 37). Therefore, it is possible that disruption of DNA fragmentation, which occurs before phagocytosis, may also affect the outcome of apoptosis. Our own data (see Fig. 13, which is published as supporting information on the PNAS web site) suggest that, in HCT116 and MEF cells that have been irradiated with ionizing radiation, a small fraction (≈1%) of the cells that have entered into early apoptosis (as evidenced by positive annexin V staining and negative propidium iodide staining, indicating intact membrane structure), can indeed survive. More importantly, deficiencies in the DFF/CAD significantly increased the level of survival cells. These data are consistent with the hypothesis that DFF/CAD functions, even those required only during late stages of apoptosis, can influence eventual cellular survival. It is quite conceivable that this small increase in survival allowed cells with a high amount of DNA damage to recover and influence mutagenesis/carcinogenesis.
Spontaneous and Induced Tumorigenesis. In our studies, spontaneous tumor development has not been observed in CAD-/- mice, in contrast to p53-deficient mice. However, CAD-null mice showed significantly increased susceptibility to tumors when exposed to genotoxic stress. In this respect, the CAD or ICAD genes are similar to quite a few other tumor suppressor genes such as gadd45, p16INK4a, chk2, and ku80. Mice deficient in these genes do not develop tumors spontaneously. However, they do develop tumors at an elevated frequency when the mice are exposed to carcinogens or when the deficiencies are juxtaposed with mutations in other tumor-suppressor genes (27, 38-40).
In summary, the results presented here clearly established the roles of DNA fragmentation factor in regulating mammalian genomic stability and suppressing cancer development.
Materials and Methods
Cell Culture and Plasmid Construction. HCT116 colon cancer cells, TK6, and WTK1 lymphoblastoid cells were obtained from American Type Culture Collection and were cultured as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Plasmid construction and gene transduction were similar to a described method (6) and is detailed in Supporting Materials and Methods.
Transgenic Animal and MEF Cells. The CAD-/- transgenic mice and immortalized CAD-/- MEF cells were obtained from Shigekazu Nagata's group at Osaka University (25). The original CAD-/- mice were in the 129/Sv background (25). All the mice used for experiments were maintained on C57BL/6 background after at least six backcrosses from the original 129Sv/C57BL/6 founder mice. All the experiments involving CAD-/- and wild-type mice were carried out by using littermates.
Early-passage CAD-/- cells were obtained from pregnant female mice at day 14. The cells were cultured in DMEM supplemented with 10% FBS.
Apoptosis Assay and Quantification of Apoptotic DNA Fragmentation. Apoptosis was determined by annexin V staining, as described in Supporting Materials and Methods. To quantify the effect of mICAD expression on the incidence of cells with apoptotic DNA fragmentation, a commercially available sandwich ELISA kit was purchased from Roche Diagnostics (Cat#1 774 425). The kit quantifies cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) in cells undergoing programmed cell death and was chosen because of its high specificity and sensitivity for apoptotic DNA fragments.
Clonogenic Assay of Cellular Survival After Irradiation. To evaluate the effect of apoptotic DNA fragmentation on cellular survival, clonogenic assays were used. For HCT116 cells, different numbers of cells were plated in a 10-cm Petri dish and irradiated with different doses of ionizing radiation. The number of cells plated at different doses was empirically determined so that, in each dish, the surviving cells numbered ≈30-150. Two weeks after irradiation, the colonies that emerged in the Petri dish were stained and counted and used to calculate the surviving fraction of cells under each irradiation dose.
Gene Mutation Assays. To evaluate radiation-induced mutation frequency at the tk and hprt loci, established methods were adopted (41). Please refer to Supporting Materials and Methods for a brief description.
Gene Amplification Assay and Fluctuation Analysis. We followed classic methods of gene amplification assay and fluctuation analysis as described in Supporting Materials and Methods.
Analysis of Chromosomal Aberrations. To analyze in vivo chromosomal aberrations, mice were given 3 Gy whole-body γ-irradiation. Three or 7 days later, the mice were killed. Chromosome metaphase spreads were made from the bone marrow cells for scoring of aberrations (see Supporting Materials and Methods for details).
Soft-Agar Assay for Tumorigenic Transformation. The soft-agar assays were conducted according to established protocols (42). Please refer to Supporting Materials and Methods for a detailed description.
Two-Stage Skin Carcinogenesis Assay. In this assay, 7- to 10-week-old mice were used. The dorsal skin of the mice was treated by dimethylbenz[a]anthracene (DMBA) and TPA(12-O-tetradecanoylphorbol-13-acetate) according to a standard protocol, as described in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Shigekazu Nagata and Kohki Kawane (Osaka University, Osaka, Japan) for carefully reading our manuscript, providing the CAD-/- knockout mice, and insightful input to this work; Drs. Michael Cook and Peter Keng for assistance in flow cytometry analysis; Drs. Takashi Kon and Jiangao Zhu for help with the animal experiments; and Dr. Marco Davia for help in the chromosome painting experiment. This work was supported by U.S. Department of Defense Prostate Cancer Research Program Grant DAMD17-02-1-0052, U.S. Department of Energy Low Dose Research Program Grant DE-FG02-03ER63635, and National Aeronautics and Space Administration Grant NAG2-1629 (to C.-Y.L.).
Author contributions: B.Y. and C.-Y.L. designed research; B.Y., Huili Wang, Y.P., He Wang, X.Z., and Q.C. performed research; J.S.B. and M.W.D. contributed new reagents/analytic tools; B.Y., Huili Wang, and Y.H. analyzed data; and B.Y. and C.-Y.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAD, caspase-activated DNase; DFF, DNA fragmentation factor; ICAD, inhibiting CAD; mICAD, mutant ICAD; MEF, mouse embryonic fibroblast; MTX, methotrexate; PALA, N-(phosphoacetyl)-l-aspartate.
References
- 1.Williams, J. R., Little, J. B. & Shipley, W. U. (1974) Nature 252, 754-755. [DOI] [PubMed] [Google Scholar]
- 2.Wyllie, A. H. (1980) Nature 284, 555-556. [DOI] [PubMed] [Google Scholar]
- 3.Liu, X., Zou, H., Slaughter, C. & Wang, X. (1997) Cell 89, 175-184. [DOI] [PubMed] [Google Scholar]
- 4.Liu, X., Li, P., Widlak, P., Zou, H., Luo, X., Garrard, W. T. & Wang, X. (1998) Proc. Natl. Acad. Sci. USA 95, 8461-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A. & Nagata, S. (1998) Nature 391, 43-50. [DOI] [PubMed] [Google Scholar]
- 6.Sakahira, H., Enari, M. & Nagata, S. (1998) Nature 391, 96-99. [DOI] [PubMed] [Google Scholar]
- 7.Ohira, M., Kageyama, H., Mihara, M., Furuta, S., Machida, T., Shishikura, T., Takayasu, H., Islam, A., Nakamura, Y., Takahashi, M., et al. (2000) Oncogene 19, 4302-4307. [DOI] [PubMed] [Google Scholar]
- 8.Konishi, S., Ishiguro, H., Shibata, Y., Kudo, J., Terashita, Y., Sugiura, H., Koyama, H., Kimura, M., Sato, A., Shinoda, N., et al. (2002) Cancer 95, 2473-2478. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh, S. Y., Liaw, S. F., Lee, S. N., Hsieh, P. S., Lin, K. H., Chu, C. M. & Liaw, Y. F. (2003) Br. J. Cancer 88, 210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel, F., Sjoberg, R. M., Ejeskar, K., Krona, C. & Martinsson, T. (2002) Br. J. Cancer 86, 596-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abel, F., Sjoberg, R. M., Krona, C., Nilsson, S. & Martinsson, T. (2004) Int. J. Oncol. 25, 1297-1302. [PubMed] [Google Scholar]
- 12.Loeb, L. A. (1991) Cancer Res. 51, 3075-3079. [PubMed] [Google Scholar]
- 13.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623-627. [DOI] [PubMed] [Google Scholar]
- 14.Stoler, D. L., Chen, N., Basik, M., Kahlenberg, M. S., Rodriguez-Bigas, M. A., Petrelli, N. J. & Anderson, G. R. (1999) Proc. Natl. Acad. Sci. USA 96, 15121-15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak, M. A., Komarova, N. L., Sengupta, A., Jallepalli, P. V., Shih Ie, M., Vogelstein, B. & Lengauer, C. (2002) Proc. Natl. Acad. Sci. USA 99, 16226-16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little, J. B., Nagasawa, H., Keng, P. C., Yu, Y. & Li, C. Y. (1995) J. Biol. Chem. 270, 11033-11036. [DOI] [PubMed] [Google Scholar]
- 17.Schimke, R. T., Kaufman, R. J., Alt, F. W. & Kellems, R. F. (1978) Science 202, 1051-1055. [DOI] [PubMed] [Google Scholar]
- 18.de Saint Vincent, B. R., Delbruck, S., Eckhart, W., Meinkoth, J., Vitto, L. & Wahl, G. (1981) Cell 27, 267-277. [DOI] [PubMed] [Google Scholar]
- 19.Tlsty, T. D., Margolin, B. H. & Lum, K. (1989) Proc. Natl. Acad. Sci. USA 86, 9441-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livingstone, L. R., White, A., Sprouse, J., Livanos, E., Jacks, T. & Tlsty, T. D. (1992) Cell 70, 923-935. [DOI] [PubMed] [Google Scholar]
- 21.Yin, Y., Tainsky, M. A., Bischoff, F. Z., Strong, L. C. & Wahl, G. M. (1992) Cell 70, 937-948. [DOI] [PubMed] [Google Scholar]
- 22.Smith, K. A., Agarwal, M. L., Chernov, M. V., Chernova, O. B., Deguchi, Y., Ishizaka, Y., Patterson, T. E., Poupon, M. F. & Stark, G. R. (1995) Philos. Trans. R. Soc. London B 347, 49-56. [DOI] [PubMed] [Google Scholar]
- 23.Sharma, R. C. & Schimke, R. T. (1989) Cancer Res. 49, 3861-3866. [PubMed] [Google Scholar]
- 24.Hahn, P., Nevaldine, B. & Morgan, W. F. (1990) Somatic Cell Mol. Genet. 16, 413-423. [DOI] [PubMed] [Google Scholar]
- 25.Kawane, K., Fukuyama, H., Yoshida, H., Nagase, H., Ohsawa, Y., Uchiyama, Y., Okada, K., Iida, T. & Nagata, S. (2003) Nat. Immunol. 4, 138-144. [DOI] [PubMed] [Google Scholar]
- 26.Lowe, S. W., Jacks, T., Housman, D. E. & Ruley, H. E. (1994) Proc. Natl. Acad. Sci. USA 91, 2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollander, M. C., Sheikh, M. S., Bulavin, D. V., Lundgren, K., Augeri-Henmueller, L., Shehee, R., Molinaro, T. A., Kim, K. E., Tolosa, E., Ashwell, J. D., et al. (1999) Nat. Genet. 23, 176-184. [DOI] [PubMed] [Google Scholar]
- 28.Iolascon, A., Lo Cunsolo, C., Giordani, L., Cusano, R., Mazzocco, K., Boumgartner, M., Ghisellini, P., Faienza, M. F., Boni, L., De Bernardi, B., et al. (1998) Cancer Lett. 130, 83-92. [DOI] [PubMed] [Google Scholar]
- 29.Hiyama, E., Hiyama, K., Ohtsu, K., Yamaoka, H., Fukuba, I., Matsuura, Y. & Yokoyama, T. (2001) Med. Pediatr. Oncol. 36, 67-74. [DOI] [PubMed] [Google Scholar]
- 30.Fong, C. T., Dracopoli, N. C., White, P. S., Merrill, P. T., Griffith, R. C., Housman, D. E. & Brodeur, G. M. (1989) Proc. Natl. Acad. Sci. USA 86, 3753-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtsu, K., Hiyama, E., Ichikawa, T., Matsuura, Y. & Yokoyama, T. (1997) Clin. Cancer Res. 3, 1221-1228. [PubMed] [Google Scholar]
- 32.Komuro, H., Valentine, M. B., Rowe, S. T., Kidd, V. J., Makino, S., Brodeur, G. M., Cohn, S. L. & Look, A. T. (1998) J. Pediatr. Surg. 33, 1695-1698. [DOI] [PubMed] [Google Scholar]
- 33.White, P. S., Thompson, P. M., Seifried, B. A., Sulman, E. P., Jensen, S. J., Guo, C., Maris, J. M., Hogarty, M. D., Allen, C., Biegel, J. A., et al. (2001) Med. Pediatr. Oncol. 36, 37-41. [DOI] [PubMed] [Google Scholar]
- 34.Caron, H., Spieker, N., Godfried, M., Veenstra, M., van Sluis, P., de Kraker, J., Voute, P. & Versteeg, R. (2001) Genes Chromosomes Cancer 30, 168-174. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, J., Liu, X., Scherer, D. C., van Kaer, L., Wang, X. & Xu, M. (1998) Proc. Natl. Acad. Sci. USA 95, 12480-12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddien, P. W., Cameron, S. & Horvitz, H. R. (2001) Nature 412, 198-202. [DOI] [PubMed] [Google Scholar]
- 37.Hoeppner, D. J., Hengartner, M. O. & Schnabel, R. (2001) Nature 412, 202-206. [DOI] [PubMed] [Google Scholar]
- 38.Krimpenfort, P., Quon, K. C., Mooi, W. J., Loonstra, A. & Berns, A. (2001) Nature 413, 83-86. [DOI] [PubMed] [Google Scholar]
- 39.Hirao, A., Cheung, A., Duncan, G., Girard, P. M., Elia, A. J., Wakeham, A., Okada, H., Sarkissian, T., Wong, J. A., Sakai, T., et al. (2002) Mol. Cell. Biol. 22, 6521-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Difilippantonio, M. J., Zhu, J., Chen, H. T., Meffre, E., Nussenzweig, M. C., Max, E. E., Ried, T. & Nussenzweig, A. (2000) Nature 404, 510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liber, H. L. & Thilly, W. G. (1982) Mutat. Res. 94, 467-485. [DOI] [PubMed] [Google Scholar]
- 42.Cox, A. D. & Der, C. J. D. (1994) Methods Enzymol. 238, 277-294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.