Abstract
Dendritic spines have been investigated intensively over recent years; however, little is yet known about how they organize on the cell surface to make synaptic contacts with appropriate axons. Here we investigate spine distributions along the distal dendrites of cerebellar Purkinje cells, after biolistic labeling of intact tissue with a lipid-soluble dye. We show that the spines have a preference to form regular linear arrays and to trace short-pitch helical paths. The helical ordering is not determined by external factors that may influence how individual spines develop, because the same periodicities were present in fish and mammalian Purkinje cells, including those of weaver mice, which are depleted of the normal presynaptic partners for the spines. The ordering, therefore, is most likely an inherent property of the dendrite. Image reconstruction of dendrites from the different tissues showed that the helical spine distributions invariably lead to approximately equal sampling of surrounding space by the spineheads. The purpose of this organization may therefore be to maximize the opportunity of different spines to interact with different axons.
Keywords: biolistic labeling, confocal microscopy, dendritic spine, image reconstruction, weaver mouse
Dendritic spines, the small membrane protrusions on the surfaces of nerve cells, are the sites where most rapid synaptic communication takes place in the brain. Spines are typically 0.5-3 μm in length (1) and elongated, creating specialized biochemical microenvironments that receive input from other neurons and compartmentalize the postsynaptic response (2, 3). Recent advanced imaging techniques have highlighted the fact that spines are dynamic structures and can be modified through synaptic activity, a property thought to be central to the development and plasticity of the nervous system (4-6).
Spines have been studied extensively in cerebellar Purkinje neurons by light and electron microscopy, after Cajal's initial discovery and early descriptions (7). One reason for investigating Purkinje spines is that the cerebellum has relatively simple architecture and circuitry. In addition, in the mouse cerebellum, there are several mutations affecting a specific kind of neuron or a single type of synapse, which have illuminated our understanding of the development and maintenance of spines. In the case of the weaver mutant mouse, for example, where most of their normal presynaptic partners (the axons of the granule cells) are absent, the Purkinje cells form essentially normal spines with postsynaptic densities still present (8-10). This result, and corroborating evidence from other mouse mutants, led to the proposal that the spines of Purkinje cells are formed by an “intrinsic mechanism,” independent of interactions with their presynaptic partners (11).
Purkinje dendrites bear a dense network of spines along their distal shafts, beyond the relatively spine-free primary, secondary, and tertiary branches of the dendritic tree. 3D reconstructions have been made from electron micrographs of small regions of this network (12-15), but the long-range organization has not been characterized or described in terms of underlying rules. The present study examines the long-range organization of spines on Purkinje dendrites, with a view to understanding better how a specific dendritic architecture is used to facilitate appropriate synaptic interactions. To simplify our analysis, we directed our attention initially to Purkinje cells of the mormyrid fish, Gnathonemus petersii, which have a lower density of spines along the distal dendrites and a more regular cerebellum than that of mammalian tissue (16). The regularity of the mormyrid cerebellum is caused by a planar “palisade” pattern of straight unbranched Purkinje dendrites passing through the molecular layer and running orthogonal to tightly packed axons (parallel fibers) of the granule cells [Fig. 1A (16)]. We discovered that spine distribution along the palisade dendrites is not random; rather, the spines have a preference to align in regular linear arrays and, in densely populated regions, to trace short-pitch helical paths. We found subsequently that these organizing principles are not unique to Purkinje cells of the mormyrid fish but apply also to the more randomly oriented distal dendrites of the mouse, including the weaver mutant. Image reconstruction showed that the helical ordering of spines gives rise to a set of similar surface lattices, the dimensions of which lead to approximately equal sampling of the surrounding space by the spineheads.
Fig. 1.
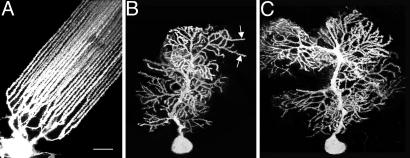
Low-magnification in-plane views of 3,3′-dioctadecyloxacarbocyanine perchlorate-labeled Purkinje cells. Mormyrid fish (A), weaver mouse (B), and wild-type mouse (C) cerebellum. Shown are the typical patterns formed by the spiny dendrites ascending from the cell bodies (bottom) through the molecular layer. The fish dendrites are straighter and less branched than the others, but some weaver dendrites are also straight over lengths of 20-30 μm (arrows). (Scale bar, 50 μm.)
Results
We examined the Purkinje dendrites as they exist in vivo by low-pressure biolistic labeling (17) of the intact cerebellum, using the lipid-soluble fluorescent dye 3,3′-dioctadecyloxacarbocyanine perchlorate and confocal microscopy of wet unfixed slices (see Materials and Methods). The dye highlights the surface of the dendrite, including the protruding spines, by diffusing over the cell membrane of the Purkinje cell that has been labeled (Fig. 1). Thus, the spines and dendritic shafts are shown up clearly against a low-intensity background, while the tissue is maintained under near-physiological conditions with minimal structural damage.
Individual dendritic shafts in the biolistically labeled tissue usually have fairly circular cross sections and, as Fig. 1 shows, fairly uniform diameters beyond the branch points of the dendritic tree. The fish dendrites, as well as being straighter, are thinner and more uniform than those of the mouse. The weaver mouse has a more stunted dendritic tree than the wild-type mouse, but the distal shafts are often straighter and more uniform over longer distances (arrows, Fig. 1B). In both the fish and the mouse, the distal portions of the dendrites are typically (but not always) densely populated with spines, with the fish spines being generally smaller than those of either the weaver or wild-type mouse.
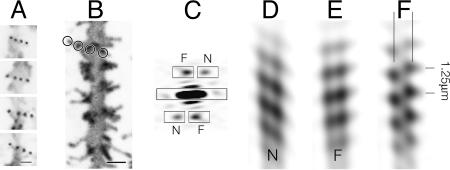
The Mormyrid Fish. The fish spines project by up to ≈2 μm from the surfaces of the palisade dendrites, allowing them to reach as far as axons passing midway between adjacent shafts of the same cell. The spines create an irregular pattern of protrusions, because they vary in size and shape and bend over a range of angles. However, glancing confocal sections, encompassing just the shaft surface and the emerging necks of spines (z thickness, 0.4-0.8 μm), showed that spines were more regularly positioned near their bases. The spine necks thus imaged in cross section appeared as discrete ≈0.2-μm diameter spots (Fig. 2A), which frequently were located at regular intervals along lines traversing the shaft circumference. Measurements made from lines of four spines (the number most frequently observed) yielded a value of 0.54 μm± 0.09 for the average center-to-center separation of the spots (Table 1).
Fig. 2.
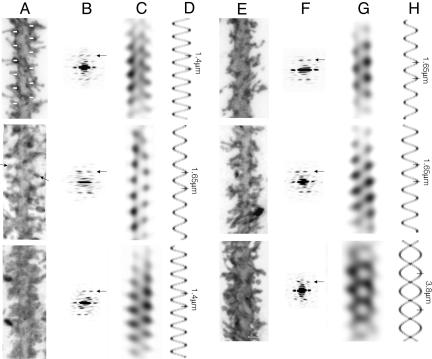
Regular features along the dendritic shaft of the fish. (A) Spine necks forming regular linear arrays over the shaft surface, revealed in glancing confocal sections (shaft axis is vertical). (B) Periodic arrangement incorporating linear arrays of spines (e.g., circles) in a region of high population density. (C) Diffraction pattern of B, showing two pairs of peaks arranged with approximate mirror symmetry about a vertical axis; the distance of these peaks from the equator indicates that the periodicities repeat every 1.25 μm. (D and E) Filtered images revealing the paths traced by lines of spines on the near (N) and far (F) sides of the shaft, made by including only terms associated with the separate pairs of peaks within the masks (see Materials and Methods). (F) Filtered image made by including terms within all of the masked areas in C; the pair of vertical lines indicate the radius at which the modulations have the greatest contrast. Here and in Figs. 3, 4, 5, 6 and 7, the contrast has been inverted so that areas of high fluorescence appear dark. (Scale bars, 1.0 μm.)
Table 1. Measurements of spine separations in linear arrays.
| d, 1-2* | d, 2-3 | d, 3-4 | n‡ | |
|---|---|---|---|---|
| Fish | 0.52 ± 0.09† | 0.57 ± 0.09 | 0.54 ± 0.08 | 30 |
| weaver | 0.55 ± 0.11 | 0.62 ± 0.14 | 0.58 ± 0.13 | 32 |
| WT mouse | 0.58 ± 0.13 | 0.61 ± 0.09 | 0.59 ± 0.10 | 30 |
Center-to-center separation in μm between the first and second spines, measured at the shaft surface.
Mean value ± standard deviation.
Numbers of lines measured.
Linear arrays of spines were most obvious in the regions of low spine density, where they oriented at small angles to the axis of the shaft (typically 10-35°), sometimes continuing around to the opposite surface. They were found mainly on the thicker proximal parts of the palisade dendrites and were also occasionally present on the proximal branches of the dendritic tree. In densely populated regions, linear arrays of spines could not be distinguished readily, because they were integrated into networks. Nevertheless, their presence was suggested by the fact that one could sometimes observe lines of spines aligned roughly side by side and seemingly associated with extended periodic groupings of spines. Fig. 2B is an example of such an image, encompassing the whole thickness of the dendrite. The diffraction pattern computed from this image (see Materials and Methods) displays a pair of strong peaks (Fig. 2C, F and N) superimposed on a relatively low-intensity background, confirming the visual impression that there is a repeating motif. Moreover, the two peaks have similar intensities and are arranged with near-mirror symmetry about a vertical line (parallel to the axis of the shaft), suggesting that the repeating motif arises from equivalent features on the near and far sides of the shaft.
Spatial frequency filtering, by Fourier synthesis of the terms included in masks around selected diffraction peaks, can be used to extract the image of one side of an object from a two-sided image (ref. 18; see Materials and Methods). Filtered images reconstructed from the peaks associated with the near and far sides of the shaft in Fig. 2B are shown in Fig. 2 D and E. They reveal bands of density that can be identified with individual linear arrays of spines (e.g., circles in Fig. 2B), inclined at small angles to the axis of the shaft. When an image is reconstructed by including all of the masked-out diffraction peaks, the bands superimpose to create a zigzag pattern characteristic of a helix (Fig. 2F). The complete filtered image shows, therefore, that the lines of spines not only align on each side of the shaft but also, to a good approximation, trace a continuous helical path (pitch, 1.25 μm in this example) over the shaft surface. The lines of maximum contrast produced by the zigzag modulations (pair of vertical lines in Fig. 2F) would be expected to coincide with the regions of maximal ordering of the spines. These lines are only slightly farther apart than the edges formed by the membrane of the shaft (Fig. 2B), confirming that it is the basal neck portions of the spines where the ordering is most pronounced.
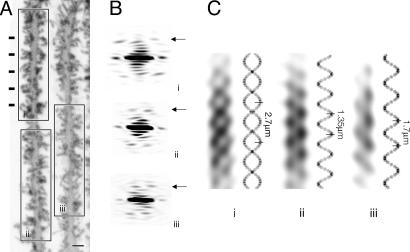
Although the shaft selected in Fig. 2 represents just one example, diffraction analyses of other stretches along the dendrites showed that the periodic features (although not easily detected by eye) are present at all levels throughout the molecular layer. Fig. 3A is an image of two adjacent shafts of the same Purkinje cell where inspection reveals obvious periodicities in only one part (horizontal bars). On the other hand, diffraction patterns of all the straight and relatively uniform stretches along the shafts (boxes) show pairs of peaks like those in Fig. 2C (arrows, Fig. 3B), demonstrating the widespread occurrence of the periodicities. The corresponding filtered images (Fig. 3C) display zigzag patterns, as would be expected from lines of spines winding helically around the shaft.
Fig. 3.
The periodicities on the fish dendrites trace helical paths. (A) Two adjacent shafts from the same Purkinje cell (horizontal bars identify the clearest repeat). (B) Diffraction patterns from the “boxed-off”regions i, ii, and iii in A, showing pairs of peaks (arrows) similar to those in Fig. 2B. The peaks in i are approximately twice as far apart as those in ii and iii, because they are associated with a different (two-start) helical symmmetry. (C) Corresponding filtered images, together with helical projections (see Materials and Methods), simulating the paths followed by the spines. Both one- (ii and iii) and two-start helices (i) are present. The numbers indicate the helical pitches. (Scale bar, 1.0 μm.)
Because the periodicities are associated mainly with the basal regions of the spines, and these regions superimpose in projection depending on their location around the shaft, one would expect the strength of fluorescence (i.e., dark areas in Figs. 2 and 3) to alter (and to be enhanced, for example, at the edges of the shaft). We confirmed that the observed variations in fluorescence are consistent with a helical arrangement by simulating projection images of helices (Fig. 3C). In these images, the fluorescence was assumed to arise from small patches positioned at regular translational and azimuthal increments around the shaft (see Materials and Methods). The simulations predict strongest fluorescence near the edges of the shaft and in regions where helices cross over, matching qualitatively these features in the filtered images.
Fig. 3, which is typical of many examples from the fish, shows two classes of helical ordering of the spines. Most commonly (as in Fig. 3, ii and iii), the spines trace a single path along the shaft surface (one-start helix). But occasionally (as in Fig. 3 i or in thicker regions of the shaft), they arrange so that any given cross section is intercepted by two paths, one rotated by 180° with respect to the other (two-start helix). As indicated by the simulations, the two-start helix has approximately twice the pitch of the one-start helix and gives rise to a symmetrical rather than to a “sine-wave” projection pattern. We found both left- and right-handed examples of these helices, when examining serial confocal sections through the dendrites (data not shown).
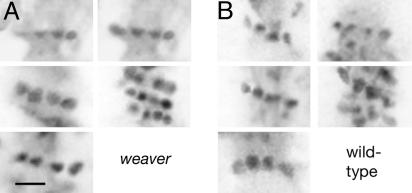
Weaver Mouse. To determine whether the linear and helical periodicities observed in the fish applied also to mammalian tissue, we examined next the distal dendrites of weaver Purkinje cells, where straight shafts occur more frequently than with the wild-type mouse. Only the straight relatively uniform regions of the shafts give rise to discrete diffraction peaks and therefore are amenable to Fourier analysis. The images again revealed examples of lines of regularly spaced spines inclined at small angles to the axis of the shaft (Fig. 4A), as well as periodicities extending over long distances (Fig. 5 A-D). The spines composing the linear arrays had an average center-to-center separation of 0.58 μm ± 0.14 SD, measured in glancing confocal sections encompassing the surface of the shaft (Table 1). This value is essentially the same as that for the fish. Moreover, the diameter of the cross-sectional “spots” was close to 0.2 μm, the diameter of the neck portion of Purkinje spines measured in electron micrographs from the rat (14, 19). The linear arrays appeared with similar frequency as in the fish, each typically containing only four spines but with individual arrays sometimes clustered together in groups (Fig. 4A).
Fig. 4.
Spine necks forming regular linear arrays over the shaft surface. Shown are glancing confocal sections from dendritic shafts of the weaver (A) and the wild-type mouse (B). Direction of the shaft axes is vertical. (Scale bar, 1.0 μm.)
Fig. 5.
Dendrites of both the weaver and wild-type mouse exhibit the same regular features as those of the fish. (A-D) Weaver.(E-H) Wild type. (A and E) Images of straight shafts densely populated with spines. (B and F) Corresponding diffraction patterns. (C and G) Filtered images. (D and H) Helical projections simulating the paths traced by the spines. Arrows point to diffraction peaks associated with periodicities in the images; numbers indicate helical pitches; horizontal bars and pair of arrows in A identify, respectively, repeating features close to the shaft surface and a line of spines. [Scale bar (original and filtered images), 1.0 μm.]
The weaver spines were more variable in shape and size than the fish spines, adding to the difficulty of distinguishing periodic features along the shafts. However, repeating patterns could still be detected along most straight shafts (e.g., bars at the bases of the spines in Fig. 5A) and characterized further by diffraction. The diffraction patterns (Fig. 5B) displayed peaks equivalent to those from the fish but had more contracted equatorial intensities due to the increased thickness of the shafts. The interval between successive helical turns, measured from the filtered images (Fig. 5C), also appeared to be slightly greater. Individual linear arrays could sometimes be distinguished within the densely populated network (pair of arrows in Fig. 5A) and again, both one- and two-start (not shown) helices were present.
Wild-Type Mouse. We examined next the wild-type mouse to determine whether the features observed in the weaver mutant, which is depleted of presynaptic partners for the spines, apply also when the normal complement is present. The dendritic shafts of the wild-type animal were generally more torturous and variable in thickness than those of the weaver mutant. However, we were still able to observe the linear arrays (spine separation, 0.59 μm ± 0.14 SD; Fig. 4B) and in straighter more uniform parts, the extended periodicities (one- and two-start helices, Figs. 5 E-H). Thus, the regular patterning of the spines is not influenced greatly, if at all, by the presence of synaptic contacts.
In both the wild-type and the weaver mouse, the thickness of the spines varied more than in the fish. At one extreme, spines were so thin as to be barely detectable in the light-optical image (e.g., Fig. 5E Top); at the other, spines were so thick that in cross section, they were comparable with the thickness of the shaft (e.g., Fig. 5A Bottom). Despite the size variability, however, we did not observe any obvious correlation between the dimensions of the spines and either the separation of the spines in linear arrays or the interval between successive helical turns.
Discussion
This study investigated the spine distribution along the distal dendrites of Purkinje cells, using a lipid-soluble fluorescent dye to highlight the surface membranes and wet unfixed tissue slices to minimize structural alterations before imaging. We examined three kinds of cerebellar tissue (Fig. 1). The mormyrid fish provided straight and uniform Purkinje dendrites, facilitating analysis of spine distributions over spatially extended regions; the weaver mutant mouse provided Purkinje dendrites in an axon-depleted environment, allowing assessment of the influence, if any, of presynaptic partners for the spines; the wild-type mouse provided an example of the torturous and thicker Purkinje dendrites characteristic of normal mammalian tissue.
First, we showed that the spines, in more sparsely populated regions, have a preference to form regular linear arrays inclined at small angles to the axis of the dendritic shaft (Figs. 2 A and 4). The positioning of spines in these arrays was highly regular, according to measurements of their locations at the dendrite surface (Table 1), with the interval between successive spines (0.5-0.6 μm) being similar, if not identical, in all three animals (despite the fact that mouse spines are larger than those of fish). Second, we showed that the linear arrays of spines, in densely populated regions, trace contiguous lines around the dendritic shafts, giving rise to helical periodicities (Figs. 2, 3, and 5). The intervals measured between successive helical turns were similar in all three animals but varied over a wider range (1.2-1.9 μm) than the interval between successive spines making the helical paths. Third, we found no correlation between the dimensions of the spines and their separation either around or along the shaft.
In other words, spines of the distal dendrites have a preference to arrange regularly in lines and in helical patterns, a property independent of the thickness of the dendritic shaft, the presence or absence of presynaptic partners, the size of the spines, or of the kind of tissue involved. These results imply that the ordering of spines is not controlled significantly by external factors, which may influence how individual spines develop, but is an inherent property of the Purkinje cell itself. It appears the organization of the spines is specified by a common set of molecular processes occurring inside the dendritic shafts.
Helical Ordering of Spines. The helical ordering of dendritic spines has not been described in previous light or electron microscopic studies, which have usually focused on details of the spines themselves. Variations in spine shape and size and localized changes in membrane curvature tend to dominate the overall picture, disguising any periodicities that might be present. The Fourier methods used here are relatively insensitive to local perturbations but highlight spatially extended periodicities by separating out the periodic information into discrete diffraction peaks. In addition, filtered images, synthesized from the terms giving rise to the peaks, revealed the helical nature of the periodicities and their relation to the linear arrays of spines seen in more sparsely populated regions.
The most frequently observed periodicities could be classified in terms of short-pitch one- and two-start helices. Helical arrays having different start numbers are also formed by proteins ordering in tubular membranes (e.g., refs. 20 and 21). The insertion (or elimination) of a line of protein molecules allows the tube to vary in diameter while retaining the quasiequivalent set of intermolecular interactions that generate the continuous surface lattice. Analogous principles might be involved in maintaining a high concentration of spines on the surfaces of varying-diameter shafts. However, the dendritic helices do not have a consistent chirality, indicating that interactions between spines in successive helical turns are not as specific as those between proteins in tubular membranes.
Whereas the helical paths traced by the spines were resolved clearly in the filtered images, the spines themselves were too disordered (and presumably sometimes too small) to be identified individually, as they were in the glancing sections through linear arrays. Nevertheless, it is instructive to estimate spine densities, assuming perfect helical ordering, from the observed values for the helical pitch, the center-to-center spine separation measured at the bases of the spines (Table 1) and the radius of the shaft. The number of spines per μm length of shaft, N, would be:
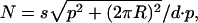 |
where s is the start number of the helix, p is the helical pitch, d is the spacing between successive spines, and R is the radius of the shaft at the base of the spines. From the measurements in Table 1 and the filtered images presented, we estimate densities of spines in Figs. 2, 3, and 5 to be 4.5, 5.6, 4.6, and 3.8 per μm (fish dendrites: Fig. 2E and Fig. 3C i, ii, and iii); 6.9, 4.2, and 7.7 per μm (weaver dendrites: Fig. 5C); and 4.2, 5.0, and 7.2 per μm (mouse dendrites: Fig. 5G). These estimates are in agreement with the range of spine densities on the distal dendrites of Purkinje cells measured from small regions in electron micrographs: 6-14 per μm for the mormyrid fish (16), 7 per μm for the mouse (12), and 5-17 per μm for the rat (13, 14, 19). The lower figures in some of our examples most likely reflect the fact that we have selected preferentially the thinner dendrites, which have smaller numbers of spines per unit length. Thicker dendrites are usually more distorted, making periodicities on them more difficult to analyze.
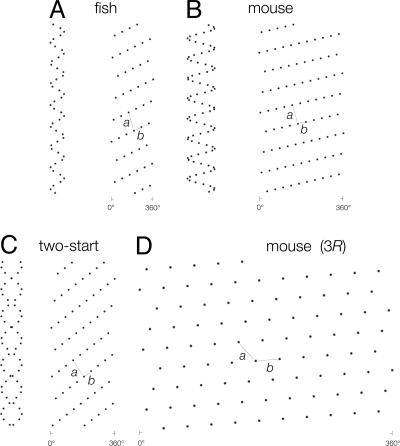
Similar Surface Lattices. The distal dendrites are typically ≈1 and 2 μm in diameter at the bases of the spines in the fish and mouse, respectively, and therefore (with 0.6-μm spine separation) contain respectively ≈6 and 11 spines per helical turn, i.e., per every ≈1.4 μm along their length. Fig. 6 A and B sketch helices constructed according to these parameters and the corresponding helical net plots made by opening up the cylindrical surfaces and laying them out flat. The helical nets show that, although the exact geometrical arrangement of the spines differ, the two surface lattices have similar dimensions. In particular, the axial a dimension of the unit cell is ≈2.3× the circumferential b dimension in either case. The same is true for the two-start helix (Fig. 6C). This helix has a pitch approximately twice that of the one start and so traces paths inclined more steeply to the axis of the shaft; however, the surface lattice is similar to that of the one start, because the distance between successive helical turns (half of the pitch of the two-start helix) is unchanged.
Fig. 6.
Helical representations of (idealized) spine distributions in densely populated regions along the distal dendrites. The dots tracing helices denote the locations of spines on the dendrite surface, scaled in proportion to the experimental measurements; helical nets next to the helices plot the corresponding surface lattices (a and b are the unit cell vectors). (A) Fish dendrite (5.7 spines per turn, 1.0-μm-diameter shaft). (B) Mouse dendrite (10.7 spines per turn, 2.0-μm-diameter shaft). (C) Two-start helix (9.1 spines per turn, 1.5-μm-diameter shaft). (D) Surface lattice in B plotted at a radius of 3 μm (i.e., 3R) from the axis of the shaft; at this radius (corresponding to the spineheads), each lattice point is approximately equidistant from each of its neighbors.
Spines in densely populated regions of the distal dendrites therefore create a set of near-equivalent surface lattices, which are common to all tissues examined. The near equivalence of the lattices means, in turn, that the number of spines per unit area of membrane surface is close to invariant over extended distances, and that the linear density of spines is approximately proportional to the thickness of the shaft.
Equal Sampling by the Spineheads. Although no quantitative measurements have been made, it is clear from visual inspection that the thicker shafts of Purkinje dendrites usually bear larger spines. As the figures suggest, the majority of spineheads extend from the shaft surface by a distance roughly equal to the diameter of the shaft. If we now make this assumption and construct the same surface lattices as in Fig. 6 A, B, or C, but at the radius of the spineheads, we find that the a and b unit cell dimensions become nearly equal to one another (Fig. 6D). As a result, individual lattice points become about the same distance apart. Each spine projecting radially out from the dendrite surface therefore has its head portion positioned approximately equidistant from each of its neighbors.
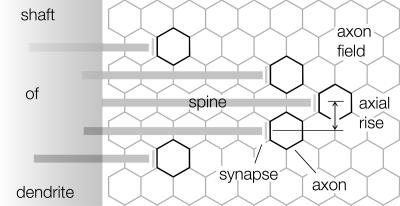
This arrangement, in which the spineheads distribute equally in three dimensions, would ensure the most complete sampling of space around the dendrite under circumstances where the spines are motile. Furthermore, the short-pitch helical organization would ensure that each spine is located at a different level along the shaft. The inclination of the b unit cell vector up the shaft ranges from 10 to 40° at a radius where the spines are 0.6 μm apart (Fig. 6 A, B, and C), so the typical axial rise per spine is 0.1-0.4 μm. These figures for the spine separation at successive levels along the shaft match remarkably well the center-to-center separation of the passing parallel axons, based on measurements of their diameters in close-packed arrays. For the fish, the average separations range from 0.1 to 0.4 μm (16); for the mouse and other mammals, the average separations range from 0.2 to 0.3 μm (22). Hence, each spinehead is not only optimally separated from its neighbors in terms of efficient “scanning” of the surrounding space but also is positioned appropriately to make synaptic contact with a new axon not approached by neighboring spines (Fig. 7). The consequent uniformity in sampling of the parallel fiber input by the dendrites may be fundamental to the correct processing of information by the cerebellum.
Fig. 7.
Schematic drawing of helically arranged spines projecting into a field of parallel-fiber axons (hexagons). The axial rise per spine is similar to the center-to-center separation of the axons, and spineheads are equally separated around the shaft, penetrating by varying amounts into the axon field. This geometry maximizes the opportunity for each spine to contact a new axon not approached by neighboring spines.
Given the cerebellar architecture of densely packed parallel axons running through the septa between each branch of the dendritic tree, one might have expected the preferred spine arrangement to be like that of sleepers on a railroad track, in which the spines project from either side of the dendrite directly into the bordering axon fields. The spine processes and/or axonal boutons would then adjust their lengths by variable amounts to accommodate the range of distances involved. However, spines are designed to function as independent microcompartments that alter their shape and size in response to changes in electrical activity, and these properties would be compromised if large variations in spine length were needed for the appropriate synaptic contacts to be achieved. Also, this alternative arrangement would enforce much greater crowding of the spines to yield a population density equivalent to that attained by helical ordering around the shaft.
Although the present analysis yields a simple semiquantitative explanation for the helical patterning of dendritic spines, it does not provide any insight into how the patterns are formed and maintained. Nevertheless, it seems likely that cytoskeletal proteins in the dendrite would play an important role. The alignment of spines in linear arrays and the highly regular 0.5- to 0.6-μm interval between spines in these arrays suggest that a filamentous protein, such as actin, might be involved. Giant actin-binding proteins, such as nebulin, are more than long enough to span the interval between successive spines and could play a role in creating regularly spaced templates for the growth of filipodia, from which mature spines are thought to develop.
Materials and Methods
Materials. The fish used, Gnathonemus petersii, were adult specimens, 8-15 cm in length, obtained from local aquaria. weaver mutant mice hybrid stock (B6CBACa Aw-j/A-Kcnj6wv) were obtained from The Jackson Laboratory. The heterozygous phenotype was confirmed by the abnormal “weaver” gait patterns. In addition, the weaver cerebellum was severely reduced in size. Wild-type mice (C57 BL/6) were supplied from our own breeding colonies. Only cerebella from adult mice (>8 weeks old) were analyzed.
Specimen Preparation. After decapitation, the fish cerebellum was dissected out and biolistically labeled, using a modified hand-held gene gun and a helium gas pressure of 75 psi (17). The bullets were 1.0-μm-diameter gold particles (Bio-Rad) coated with the lipid-soluble fluorescent dye, 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO; Molecular Probes) (23). The tissue was placed on a membrane support (8.0-μm pore size, Corning) and incubated for 2 h at 25°C (95% O2/5%CO2) in 2 ml of fish culture medium (112 mM NaCl/10 mM KCl/2 mM CaCl2/1 mM MgCl2/2 mM NaHCO3/4 mM Hepes, pH 7.4, supplemented with Medium 199 (Sigma) in a ratio of 1:20). The tissue was transferred to ice-cold PBS, sliced into ≈50-μm-thick sections (Series1000, Vibratome, St. Louis, MO), and mounted on glass slides for direct viewing in the microscope. After overdose with sodium pentobarbital, the mouse cerebellum was exposed and left undisturbed in the cranium. Dye/gold particles (as for the fish) were fired in one shot at the exposed region. The cerebellum was carefully removed and incubated for 30 min at 37°C (95%O2/5%CO2) in sterile PBS. The tissue was transferred to ice-cold PBS, sliced into ≈50-μm-thick sections, and mounted as above. In pilot experiments, whole brains were immersed into fixative (4% paraformaldehyde in PBS, 1 h at room temperature) before sectioning. Although dendrites after this treatment exhibited the same periodic features as found in the unfixed tissue, the shafts were generally more distorted, making them less suitable for analysis. Only results from unfixed tissue are therefore reported.
All experimental manipulations were conducted under license by the Home Office in accordance with the Animals (Scientific Procedures) Act 1986, and the Medical Research Council code of practice for scientific procedures on animals.
Imaging and Analysis. Images were recorded within 24 h after the slices were cut by using a Bio-Rad Radiance 2100 confocal microscope with long-working distance ×60/1.4 numerical aperture (n.a.) or ×40/1.3-n.a. oil-immersion objectives. The laser was set to an excitation wavelength of 484 nm and optimized for collection of emitted light in the 500- to 510-nm band. Images were typically 1,024 × 1,024 pixels. The scanning step size was 0.05 μm, and stacks of z sections were recorded at 0.2-μm intervals to encompass the full width of each dendrite that was analyzed. Glancing-section images for presentation (Figs. 2 and 4) and for measurement of spine separations (Table 1) were computed by summation of between two and four successive sections in the stack. Determination of the handedness of the helical paths traced by spines was made by following the changes in the locations of the spines between successive sections. Full projection images were computed by summation over the whole stacks.
Projection images of the dendrites were analyzed with Medical Research Council image-processing software (24). To examine the extended periodicities, we selected stretches from the distal shafts that had approximately circular uniform cross sections and were relatively straight over distances of at least 10 μm. Slight curvature was sometimes corrected by a spline-fitting procedure applied to a set of equally spaced points identifying the location of the axis of the shaft (25). To obtain filtered images, areas were “boxed-off” as in Fig. 3, “floated” to yield an average density equal to the average value around the box perimeter, and padded to an array size of 512 × 512 pixels (26). Fourier transforms were calculated from the arrays and displayed as diffraction patterns. Filtered images were calculated by Fourier synthesis of terms included in masked-out regions in the transform. The size of mask (which determines the amount of averaging over adjacent regions) was such as to include the entire extent of each diffraction peak. Filtered images from either the near or far sides were obtained by using the appropriately selected peaks together with the peaks on the equator (giving information about the shape of the shaft). Complete filtered images were obtained by using all of the relevant masked out regions. Helical projections, simulating the paths traced by the spines around the dendritic shaft (Figs. 3, 5, and 6), were computed by placing spherical patches of density at regular intervals along a length of helix and summing the densities along the viewing direction.
Author contributions: J.O. and N.U. designed research, performed research, contributed new reagents/analytic tools, and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harris, K. M. & Kater, S. B. (1994) Annu. Rev. Neurosci. 17, 341-371. [DOI] [PubMed] [Google Scholar]
- 2.Nimchinsky, E. A., Sabatini, B. L. & Svoboda, K. (2002) Annu. Rev. Physiol. 64, 313-353. [DOI] [PubMed] [Google Scholar]
- 3.Yuste, R. & Bonhoeffer, T. (2004) Nat. Rev. 5, 24-34. [DOI] [PubMed] [Google Scholar]
- 4.Matus, A. (2000) Science 290, 754-758. [DOI] [PubMed] [Google Scholar]
- 5.Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E. & Svoboda, K. (2002) Nature 420, 788-794. [DOI] [PubMed] [Google Scholar]
- 6.Grutzendler, J., Kasthuri, N. & Gan, W.-B. (2002) Nature 420, 812-816. [DOI] [PubMed] [Google Scholar]
- 7.Ramon, Y. & Cajal, S. (1911) Histologie du Système Nerveux de l'Homme et des Vertèbres (Maloine, Paris).
- 8.Rakic, P. & Sidman, R. L. (1973) J. Comp. Neurol. 152, 133-162. [DOI] [PubMed] [Google Scholar]
- 9.Sotelo, C. (1975) Brain Res. 94, 19-44. [DOI] [PubMed] [Google Scholar]
- 10.Hirano, A., Dembitzer, H. M. & Yoon, C. H. (1977) Acta Neuropathol. 40, 85-90. [DOI] [PubMed] [Google Scholar]
- 11.Sotelo, C. (1978) Prog. Brain Res. 48, 149-170. [DOI] [PubMed] [Google Scholar]
- 12.Spacek, J. & Hartmann, M. (1983) Anat. Embryol. 167, 289-310. [DOI] [PubMed] [Google Scholar]
- 13.Hillman, D. E. & Chen, S. (1984) Brain Res. 295, 325-343. [DOI] [PubMed] [Google Scholar]
- 14.Harris, K. M. & Stevens, J. K. (1988) J. Neurosci. 8, 4455-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martone, M. E., Gupta, A., Wong, M., Qian, X., Sosinsky, G., Ludascher, B. & Ellisman, M. H. (2002) J. Struct. Biol. 138, 145-155. [DOI] [PubMed] [Google Scholar]
- 16.Meek, J. & Nieuwenhuys, R. (1991) J. Comp. Neurol. 306, 156-192. [DOI] [PubMed] [Google Scholar]
- 17.O'Brien, J. A., Holt, M., Whiteside, G., Lummis, S. C. R. & Hastings, M. H. (2001) J. Neurosci. Methods 112, 57-64. [DOI] [PubMed] [Google Scholar]
- 18.DeRosier, D. J. & Klug, A. (1968) Nature 217, 130-134. [DOI] [PubMed] [Google Scholar]
- 19.Napper, R. M. & Harvey, R. J. (1988) J. Comp. Neurol. 274, 158-167. [DOI] [PubMed] [Google Scholar]
- 20.Castellani, L., Hardwicke, P. M. D. & Vibert, P. (1985) J. Mol. Biol. 185, 579-594. [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima, C. & Unwin, N. (1990) J. Cell Biol. 111, 2623-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt, K. D., Tanapat, P. & Wang, S.-H. (2005) Eur. J. Neurosci. 21, 2285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan, W.-B, Grutzendler, J., Wong, W. T., Wong, R. O. L. & Lichtman, J. W. (2000) Neuron 27, 219-225. [DOI] [PubMed] [Google Scholar]
- 24.Crowther, R. A., Henderson, R. & Smith, J. (1996) J. Struct. Biol. 116, 9-16. [DOI] [PubMed] [Google Scholar]
- 25.Smith, J. D. (1999) J. Struct. Biol. 125, 223-228. [DOI] [PubMed] [Google Scholar]
- 26.DeRosier, D. J. & Moore, P. B. (1970) J. Mol. Biol. 52, 355-369. [DOI] [PubMed] [Google Scholar]