Abstract
Sexually transmitted human papillomaviruses (HPVs) are the primary cause of cervical cancer. Recent advances in techniques for production of papillomaviral vectors [known as pseudoviruses (PsVs)] have made it possible to perform high-throughput screens for compounds that might block the initial stages of papillomavirus infection. We have used PsVs to screen a variety of compounds that might function as inhibitors of HPV infection, with emphasis on human peptides previously implicated in innate antimicrobial immunity. Little is known about the possible activity of these peptides against nonenveloped viruses, such as HPVs. Our screen revealed that human α-defensins 1-3 [known as human neutrophil peptides (HNPs) 1-3] and human α-defensin 5 (HD-5) are potent antagonists of infection by both cutaneous and mucosal papillomavirus types. In contrast, human β-defensins 1 and 2 displayed little or no anti-HPV activity. HD-5 was particularly active against sexually transmitted HPV types, with 50% inhibitory doses in the high ng/ml range. Microscopic studies of PsV inhibition by the α-defensins revealed that they block virion escape from endocytic vesicles but not virion binding or internalization. Consistent with this finding, PsVs remained susceptible to inhibition by α-defensins for many hours after initial binding to cells. HNPs 1-3 and HD-5 have been reported to be present in the female genital tract at levels that overlap those that inhibit HPVs in vitro, suggesting that they could present a natural barrier to the sexual transmission of HPV and could serve as the basis of a broad-spectrum topical microbicide.
Keywords: cathelicidin, hCAP18, lactoferrin, LL-37, microbicide
Vertebrates have evolved a variety of innate defenses against viral infections. These defenses include sophisticated detection systems such as those involving the toll-like receptors, which detect common microbial structures, and effector signaling molecules such as the interferons, which stimulate cellular defenses directly, targeting the common molecular functions of viruses (1, 2). In humans, a number of secreted antiviral defensive proteins have been described, including lactoferrin, secretory leukocyte protease inhibitor, lysozyme, complement factors, LL-37, and various defensins (reviewed in refs. 3 and 4). These secreted antiviral effectors have been proposed to inactivate enveloped viruses by disrupting virion lipids, by interfering with virion/receptor binding, or by affecting cellular signaling events. The activity of innate antimicrobial peptides against nonenveloped viruses is not well documented, and their mode of action is not understood.
The goal of this study was to determine whether innate human antimicrobial peptides can interfere with the establishment of papillomavirus infection in cultured cells. Papillomaviruses are a diverse group of nonenveloped DNA tumor viruses that infect the epithelial tissues of a range of different vertebrate species. Whereas many of the >100 known human papillomavirus (HPV) species are adapted to infection of nongenital skin surfaces, where they may cause benign warts (papillomas), a subset of HPV species tend to infect genital and mucosal sites. Although some mucosotropic HPV types cause entirely asymptomatic or benign infections, about a dozen sexually transmitted HPV genotypes have collectively been shown to be essential etiologic agents responsible for nearly all cases of cancer of the uterine cervix and a fraction of anogenital and head and neck cancers (reviewed in refs. 5 and 6). Sexually transmitted HPV infections are very common, but most infections do not progress to cancer. Adaptive cell-mediated immunity is thought to be involved in control and elimination of HPV infection. However, a possible role for antimicrobial peptide-based innate immunity in inhibiting initial HPV transmission or subsequent viral spread has not been investigated.
Recent metaanalyses have suggested that, in contrast to other types of sexually transmitted infections, condoms probably afford relatively little protection against the initial sexual transmission of HPVs, perhaps because of the fact that mucosotropic HPVs also infect cornified anogenital skin that is not covered by condoms (7, 8). The ability of HPVs to circumvent condom prophylaxis may be one factor in the high prevalence of sexually transmitted HPVs worldwide. Although prophylactic HPV vaccines currently in clinical development will likely be effective in protecting women against the most common cancer-associated HPV types, the current generation of vaccines probably will not protect against all cancer-associated HPV types (reviewed in ref. 9). Thus, there is a need for development of broad-spectrum antiviral compounds that might be used as topical microbicides to block the sexual transmission of HPV.
Because the papillomavirus life cycle is closely tied to epithelial differentiation, HPVs are difficult to propagate in culture. The limited availability of a tractable cell-culture model for HPV infection has constrained past efforts to identify compounds that might block HPV transmission (10-15). Recently, our group has developed a system for rapidly producing high-titer papillomavirus-based gene-transfer vectors, known as pseudoviruses (PsVs) (16, 17). Recombinant PsVs are a powerful tool for investigation of the initial infection stage of the papillomavirus life cycle. In this article, we have used a high-throughput PsV-based screen to show that several types of human innate antimicrobial peptides have the ability to block HPV infection in vitro.
Results
HPV-Inhibition Assay. Because HPV16 is the type most commonly associated with malignant cervical lesions worldwide (reviewed in ref. 29), it was chosen as a model HPV to screen various compounds for their capacity to inhibit infection. We generated an HPV16 PsV stock carrying a GFP reporter plasmid using methods reported in refs. 16 and 17. Selected compounds were subjected to serial dilution and applied to HeLa cell cultures in 96-well plates. After a brief incubation, the cells were inoculated with HPV16 PsV stock at a multiplicity of infection of 0.1. After 48 h, the cells were subjected to flow-cytometric analysis to determine the percentage of cells transduced with GFP under each condition. For compounds that inhibited the HPV16 PsV, a curve was fitted to determine the 50% inhibitory dose (IC50) (Table 1).
Table 1. Inhibition of HPV16 PsV transduction of HeLa cells.
| α-defensins | Supplier* | IC50† | 95% CI | Toxicity |
|---|---|---|---|---|
| HNP-1 | — | 5 | 3.2-6.6 | >100 |
| HNP-1 | American Peptide | 6 | 5.2-7.7 | >32 |
| HNP-1 | Bachem | 6 | 3.6-8.5 | >100 |
| HNP-1 | Pepnet | 5 | 3.8-7.3 | >100 |
| HNP-1 (HaCaT cells) | Pepnet | 9 | 3.7-18.6 | >100 |
| HNP-1 (293TT cells) | Pepnet | 8 | 6.0-10.1 | >100 |
| HNP-1 (serum-free medium) | — | 3 | 1.1-6.1 | 50 |
| HNP-2 | American Peptide | 4 | 2.5-7.6 | >100 |
| HNP-3 | Pepnet | 10 | 6.9-13.1 | >100 |
| HNP-4 | — | 21 | 6.9-60.6 | 50 |
| HD-5 | — | 0.6 | 0.55-0.75 | >100 |
| HD-5 | Pepnet | 0.6 | 0.59-0.63 | >100 |
| HD-5 (serum-free medium) | — | 0.5 | 0.30-0.72 | 50 |
| HD-6 | — | — | — | 50 |
| β-defensins | ||||
| HBD-1 | Pepnet | — | — | >390 |
| HBD-2 | Pepnet | 190 | 178-203 | >430 |
| Other human antimicrobials | ||||
| Heparin (H4784) | Sigma Aldrich | 2 | 1.7-1.9 | >160 |
| Histatin-5 | Bachem | — | — | >30 |
| Lactoferrin (L4894) | Sigma Aldrich | 62 | 50.7-76.8 | 780 |
| Lactoferrin N-lobe | Bachem | — | — | >100 |
| Liver-expressed antimicrobial peptide-1 | American Peptide | — | — | >100 |
| Liver-expressed antimicrobial peptide-2 | American Peptide | — | — | >100 |
| LL-37 | Phoenix Peptides | 9 | 6.3-13.7 | 15 |
| Lysozyme (neutrophil) | Sigma Aldrich | — | — | >100 |
| Secretory leukocyte protease inhibitor | R&D Systems | — | — | >100 |
All values are given in μg/ml; CI, confidence interval.
Dashes imply that compound was synthesized in-house.
Dashes imply that no inhibitory effect was observed at highest noncytotoxic dose.
To validate the assay, we initially tested high-molecular-weight heparin (30-32) and human lactoferrin protein (15), both of which have been proposed to block HPV entry. Both compounds displayed inhibitory effects at noncytotoxic doses (Table 1).
Several previously untested compounds were also found to be effective in blocking the HPV16 PsV (Table 1). Although the α helical antimicrobial peptide LL-37 (also known as human cathelicidin antimicrobial peptide-18) was relatively cytotoxic, it inhibited PsV transduction at doses for which no gross cytotoxicity was observed (Table 1). LL-37 has been shown to be involved in cellular signaling (33) at cutaneous wound sites (34) and has been found to be expressed in HPV-induced common skin warts (35) and in genital tissues (36). Although the cytotoxic effects of LL-37 would limit its utility as a topical microbicide, its potential involvement in the natural history of papillomavirus infection might merit further investigation.
Five of the six known human α-defensins stood out as potent inhibitors of PsV transduction. The closely related α-defensins 1-3 [also known as human neutrophil peptides (HNPs) 1-3] had IC50s against the HPV16 PsV in the low μg/ml range. HNP-4 was nearly an order of magnitude less active, and human α-defensin 5 (HD-5) was about one order of magnitude more active relative to HNPs 1-3. Only α-defensin 6 failed to inhibit the HPV16 PsV at noncytotoxic concentrations.
HNP-1 and HD-5 were selected for follow-up study. They were both found to inhibit HPV16 PsV transduction of other cell types, such as the human embryonic kidney line 293TT, the spontaneously immortalized keratinocyte line HaCaT, and murine C127 fibroblast cells (Table 1 and data not shown). A separate set of experiments showed that HNP-1 was equally effective against mature and immature (17) PsVs (data not shown). The presence of serum has previously been reported to interfere with the ability of HNP-1 to inhibit viral infection of some cell types in vitro (25, 37). Therefore, we performed the PsV-inhibition assay using serum-free medium. HNP-1 and HD-5 were only slightly more effective against the HPV16 PsV in serum-free conditions (Table 1).
As a further validation of the PsV-based screen, HNP-1 was tested in a standard bovine papillomavirus type (BPV1) focal transformation assay using murine C127 cells (38, 39). Treatment of the cultures with 25 μg/ml HNP-1 for 48 h after virus inoculation resulted in a >95% reduction in the formation of transformed foci, confirming that HNP-1 can prevent infection of cells with an authentic papillomavirus.
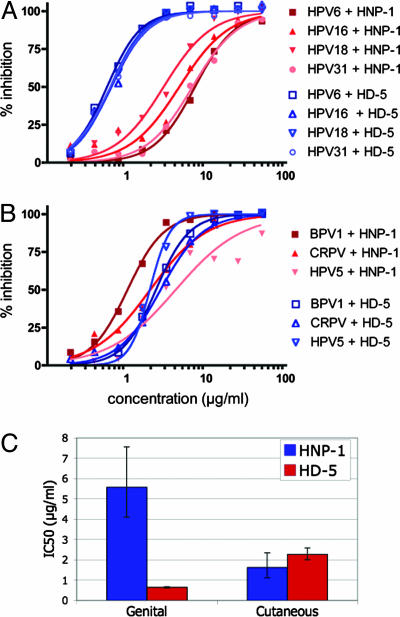
Differential Effects of HNP-1 and HD-5 Against Genital and Cutaneous Papillomaviruses. In addition to HPV16 PsV, we have recently developed PsVs representing papillomaviruses from various phylogenic branches (40). Like HPV16, HPVs 18 and 31 are mucosotropic types associated with a high risk of cervical cancer. HPV6 is a low-risk sexually transmitted type that belongs to the same genus as the high-risk types. HPV6 typically infects genital skin, where it can cause genital warts. Several cutaneous PsV types have also been developed. HPV5 commonly infects nongenital skin and does not cause discernable lesions in most individuals. Two animal papillomaviruses, BPV1 and cottontail rabbit papillomavirus (CRPV), cause nongenital skin lesions in their natural hosts.
The particle-to-infectivity ratios of stocks of the different PsV types varied. We therefore performed control inhibition assays in which “cold” HPV16 PsV particles (produced in the absence of a GFP reporter plasmid) were added at a 50-fold excess over GFP-expressing HPV16 PsV. The presence of cold HPV16 particles did not significantly alter the inhibition curves for HNP-1 or HD-5 (data not shown), suggesting that the particle-to-infectivity ratio of PsV stocks does not have a major influence on inhibition by these compounds.
Each PsV type was tested for inhibition by serial dilutions of HNP-1 or HD-5. The PsVs differed in their susceptibility to inhibition by the two α-defensins in a manner correlating with the epithelial region they typically infect (Fig. 1). The genital HPV types were highly susceptible to inhibition by HD-5, with IC50 values of ≈0.6 μg/ml but were much less susceptible to inhibition by HNP-1, with IC50 values ranging from 2.9 to 7.2 μg/ml. In contrast, the nongenital papillomavirus types showed an intermediate degree of inhibition by both HNP-1 and HD-5 (Fig. 1C).
Fig. 1.
Inhibition of various papillomavirus types by HNP-1 and HD-5. Inhibition curves are shown for genital (A) and cutaneous (B) papillomavirus types. (C) The IC50 for HNP-1 or HD-5 inhibition curves fitted to combined data points for genital or cutaneous types. Error bars represent the 95% confidence intervals for the combined curves. BPV1, bovine papillomavirus 1; CRPV, cottontail rabbit papillomavirus.
We investigated several different methods for modeling defensin inhibition of the different PsV types and found that a sigmoidal dose-response curve with variable slope gave the best fit. The overall steepness of such curves is reflected in a coefficient known as the Hill slope. For each of the PsV types, the Hill slope of the calculated curve was significantly steeper for HD-5 than for HNP-1, with average values of 2.1 and 1.5, respectively. For conventional interactions obeying the law of mass action, a steeper Hill slope implies a greater degree of cooperativity. Because inhibition of viral infection is a complex multifactorial process, the significance of an altered Hill slope is unclear, but potentially consistent with the concept that HD-5 and HNP-1 block papillomavirus infection through differing inhibitory mechanisms.
Time-Course Experiments. HNP-1 is thought to prevent some families of enveloped viruses from infecting cultured cells, at least in part, by interfering with initial virion binding to cells. We therefore determined, by flow-cytometric analysis, whether HNP-1 could block the binding of GFP-tagged HPV16 virions to HeLa cells. No impairment of binding of the GFP-tagged virions was observed in the presence of HNP-1 (data not shown).
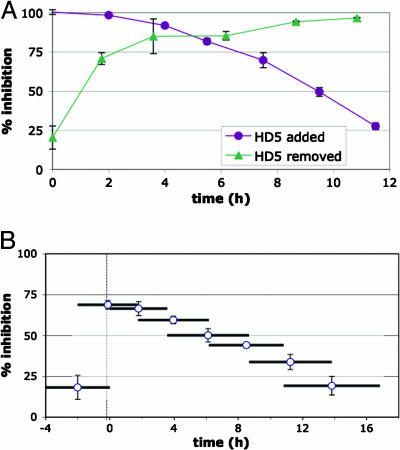
To begin to address which postbinding step in the papillomavirus entry process might be blocked by α-defensins, we performed time-course experiments in which HD-5 was added to or removed from cultured HeLa cells at various intervals after HPV16 PsV binding (Fig. 2A). Similar results were observed for cells treated with HNP-1 (data not shown). These experiments demonstrate that HD-5 and HNP-1 can block the majority of PsV transduction, even when applied many hours after initial virion binding to cells. Fig. 2B shows an experiment in which PsV-transduced cells were incubated with HD-5 for partially overlapping 4-h intervals, represented by horizontal bars. This experiment confirms that the majority of the PsV titer is susceptible to irreversible inhibition by HD-5 during the first 6 h after binding to cells.
Fig. 2.
HD-5 time-course experiments. The percent inhibition of HPV16 PsV transduction of HeLa cells treated with 5 μg/ml HD-5 is shown. (A) HD-5 was added or removed at different time points. (B) Cells were treated with HD-5 for overlapping 4-h intervals (represented by horizontal bars).
Interestingly, pretreatment of cells with defensin before PsV inoculation had minimal impact on transduction (Fig. 2). Likewise, pretreatment of concentrated PsV with various doses of the defensins had essentially no effect on transduction (data not shown).
To begin to address the possibility that the known effects of HNP-1 on receptor-mediated signaling might be responsible for its inhibition of papillomavirus infection, we performed inhibition assays using HeLa cells pretreated with a variety of signaling agonists and antagonists (see Materials and Methods). None of the drugs targeting G protein-coupled receptors, protein kinase C, or MAP kinase significantly altered the anti-HPV16 inhibition curves of HNP-1 or HD-5 (data not shown), suggesting that their HPV-inhibitory effects do not depend on known HNP-1 signaling pathways.
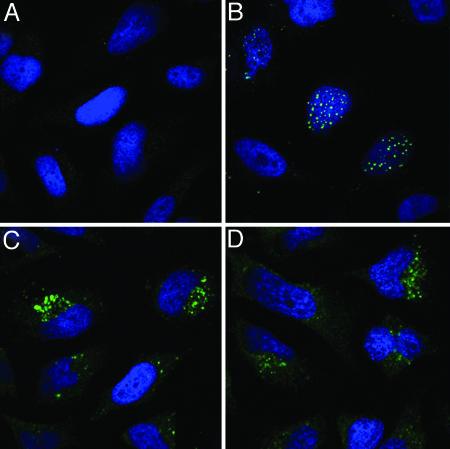
Microscopy. After cell binding, most papillomavirus types, including HPV16, are thought to infect permissive cells by an exceptionally slow clathrin-dependent endocytic pathway (41-43). During the entry process, the viral capsid undergoes a series of structural rearrangements that ultimately result in exposure of the viral genome and certain epitopes of the minor capsid protein L2. After this partial uncoating within endosomes, L2 and the viral genome (or reporter plasmid) escape from a vesicular compartment, allowing them to traffic together to a subnuclear site known as an ND-10 domain (28).
To examine which step in the viral entry process HNP-1 and HD-5 might block, we performed immunofluorescent confocal microscopy on HeLa cells treated with HNP-1 or HD-5, followed by inoculation with PsVs that had been tagged by incorporation of bromo-deoxy uridine (BrdUrd) into the PsV DNA or by HA-tagging of the carboxyl terminus of L2 (28). Staining of PsV-treated cells with an antibody to the major capsid protein, L1, at early time points confirmed that the defensins do not block initial binding of the virion to cells (data not shown).
The trafficking of the BrdUrd-labeled genome or HA-tagged L2 into the nucleus is first detectable at about 16 h after initial binding of capsids to cells. Therefore, to allow measurable accumulation of viral components in the nucleus, we stained cells treated with labeled PsVs in the presence or absence of defensins 24 h after inoculation. In the presence of defensins, PsVs underwent endocytosis and uncoating but failed to escape from cytoplasmic vesicles (Fig. 3). This result suggests that defensins act to block papillomavirus infection at a stage after virion-cell binding but before escape from endosomes.
Fig. 3.
Microscopic analysis of HNP-1 and HD-5 inhibition. HeLa cells were mock treated (A) or treated with BrdUrd-labeled HPV16 PsV (B-D) in the presence of HNP-1 (C) or HD-5 (D). At 24 h after virus inoculation, the cells were stained for BrdUrd to reveal uncoated viral DNA (green). Cells were counter-stained with the DNA stain DAPI (blue).
HNP Mutants. The ability of defensins to disrupt membranes is thought to depend, at least in part, on charge-charge interactions between the cationic defensin and anionic lipid headgroups. A recent study of HNP-2 mutants with altered overall charges showed that various mutants displayed dramatic changes in antibacterial activity and altered ability to interact with synthetic membranes (22). We examined the ability of these mutants to antagonize HPV16 PsV transduction and found that, in contrast to their altered antibacterial effects, these mutants all retained similar anti-HPV effects (Table 2).
Table 2. Analysis of HNP mutants.
| Peptide | IC50 | 95% CI |
|---|---|---|
| HNP-2 WT | 4 | 2.5-7.6 |
| HNP-2 D-Ala | 5 | 2.3-4.6 |
| HNP-2 D-Arg | 7 | 4.8-8.9 |
| HNP-2 D-Glu | 10 | 7.0-15.2 |
| HNP-2 D-Phe | 10 | 6.9-15.2 |
| HNP-2 D-Thr | 7 | 4.7-9.1 |
| HNP-2 D-Tyr | 11 | 8.1-14.6 |
| HNP-2 D-Val | 8 | 6.3-10.5 |
All values are given in μg/ml.
Discussion
The most effective compounds identified in our PsV-based screen for innate peptide inhibitors of papillomavirus infection were α-defensins, a group of cysteine- and arginine-rich antimicrobial peptides. Defensins have been implicated in the innate antibacterial and antiviral defenses of multicellular organisms ranging from plants to humans (reviewed in refs. 4, 44, and 45). Consistent with their proposed role in guarding against invading pathogens, human defensins are typically expressed in leukocytes and epithelial tissues. The highly homologous HNPs 1-3 are abundant in neutrophil granules, along with the more distantly related and less-abundant HNP-4. In addition to participating in the destruction of endocytosed microbes, HNPs can be released into extracellular fluids, such as cervicovaginal secretions, where they can reach effective microbicidal concentrations (36, 46, 47).
HD-5 and human α-defensin 6 are secreted by specialized Paneth cells resident in the crypts of the small intestine. HD-5 is also known to be expressed in other epithelial tissues. One report observed HD-5 peptide at various sites in the human female genital tract and found high ng/ml levels of HD-5 in vaginal wash material (48). Other studies have reported HD-5 expression in the testes and male urethra, as well as in breast milk and keratinocytes of the oral and airway mucosa (49-53). In some of these studies, HD-5 expression appeared to correlate with local inflammation. In contrast, HD-5 mRNA was reported to be absent in skin keratinocytes, even after treatment with inflammatory stimuli (54).
Like other α-defensins, HD-5 is expressed as a propeptide that is proteolytically processed to yield a variety of mature HD-5 isoforms. In Paneth cells, this processing is accomplished by tryptic digestion during or just after secretion into the intestinal lumen (55). A recent report has shown that, in the male urethra, HD-5 processing may depend on proteases secreted by neutrophils (50). Whether alternative HD-5 isoforms, other than the dominant tryptic form we have studied here, are active against papillomaviruses remains to be determined.
Our results raise the possibility that α-defensins may contribute to host defenses against papillomavirus infection, particularly in the female genital tract. From a teleological perspective, it seems puzzling that genital HPV types, which might be predicted to encounter HD-5 in vivo, are acutely sensitive to inhibition by HD-5 compared to cutaneous papillomavirus types (Fig. 1). However, Quayle and colleagues (48) found HD-5 peptide to be absent in the undifferentiated basal-layer keratinocytes that are thought to be the initial target of productive HPV infection. Thus, HD-5 may function to dampen nonproductive infection of more superficial keratinocytes, which could trigger a protective adaptive immune response to the virus. Alternatively, women expressing high levels of α-defensins (56, 57) (or defensin-processing proteases) may be preferentially resistant to initial HPV infection and persistence, particularly if persistence requires multiple rounds of autoinoculation (58, 59). Interestingly, high-level HNP-1 expression was recently reported to be associated with resistance to HIV-1 infection in multiply exposed HIV-1 seronegative women (60). Thus, women with a high risk of HPV exposure but no virologic or serologic evidence of infection might be of particular interest in natural history studies.
In principle, innate barriers to genital HPV infection could be augmented by exogenous application of synthetic defensin peptides. The prospect that α-defensins might function as safe, nonimmunogenic, and noninflammatory topical microbicides is bolstered by the fact that the defensins can normally be found at virucidal or nearly virucidal concentrations in the female genital tract. In contrast, heparin would not be considered an attractive candidate for use as a topical microbicide because of its anticoagulant activity. The ability of α-defensins to substantially inhibit in vitro HPV infection, even 6 h after cell-surface binding, implies that they might be effective as a postcoital microbicide.
Because α-defensins have been found to inhibit HIV-1, herpes simplex viruses, Neisseria gonorrhoeae, and Chlamydia trachomatis, they could potentially function as broad-spectrum topical microbicides to simultaneously block multiple sexually transmitted pathogens (37, 50, 61, 62). Prevention of the sexual transmission of HPV might be a particularly useful endpoint for preliminary efficacy trials of broad-spectrum topical microbicides. The high incidence of initial HPV infection in young adults and its relative resistance to condom prophylaxis (which must be encouraged in clinical trials) might make it possible to determine microbicide efficacy by using relatively small populations of study subjects.
It is thought that defensins exert antibacterial effects primarily by interacting with and destabilizing bacterial membranes. Soon after their initial identification as bactericidal effectors, it was found that α-defensins could also inactivate certain types of enveloped viruses (37, 63). In contrast, these reports found the defensins to be inactive against two nonenveloped viruses, echovirus type 11 and reovirus type 3. Therefore, it was proposed that defensins exert antimicrobial effects primarily by direct interaction with viral or bacterial membranes. However, more recent reports have shown that, in addition to these effects, HNP-1 can block HIV-1 infection by interfering with cellular signaling in some cell types (25). Furthermore, synthetic HNP-1 (64) and stably expressed pro-HD-5 (65) were recently found to block the infection process for a nonenveloped virus, adenovirus type 5, at an unknown step in the viral entry process. These results, together with our finding that α-defensins inhibit papillomavirus infection, clearly demonstrate that additional antiviral mechanisms beyond defensin interaction with microbial envelopes are at play.
It remains to be determined whether the effects of defensins against papillomaviruses are mediated by binding to a virion or cellular component. It is unlikely that the critical inhibitory interaction is a durable one, because the effect requires maintenance of an extracellular defensin pool during the initial period of infection. Inhibitory concentrations of defensins did not impair virion binding, internalization from the cell surface, or uncoating, but PsV in defensin-treated cells failed to escape from vesicles and traffic to the nucleus. It is, therefore, clear that defensins interfere with a postbinding step before endosome escape.
Our group has recently shown that cleavage of the papillomavirus minor capsid protein, L2, by the cellular protease furin is critically required for papillomavirus infection of cultured cells (68). Virions applied to furin-deficient cells or to cells treated with furin protease inhibitors fail to escape from endosomes in a manner reminiscent of our findings with defensin-treated cells. However, a recent study identifying HNP-1 as an inhibitor of anthrax lethal toxin (LeTx) found that HNP-1 does not inhibit the proteolytic activity of furin, which is required for LeTx function (66). Intriguingly, HNP-1 was found to noncompetitively inhibit the metalloproteinase activity of LeTx, showing that HNP-1 can function as a type of protease inhibitor. Although a panel of metalloproteinase-inhibitor drugs failed to inhibit HPV16 PsV transduction of HeLa cells (data not shown), the concept that HNP-1 might block papillomavirus infection by reversibly inhibiting a required cellular protease other than furin remains a possibility.
One common feature of the infectious pathways of adenoviruses and papillomaviruses is their ability to destabilize cellular membranes after endocytosis. Both viruses have recently been shown to carry membrane-destabilizing domains within their capsid structural proteins (67, 69). Because defensins are known to interact with membranes, it is possible to imagine that they act against papillomaviruses and adenoviruses by interacting with cellular membranes in a manner that blocks the effects of membrane-destabilizing viral peptides. To begin to address this question, we performed a series of experiments involving a previously described panel of HNP-2 mutant peptides (22). Although the various mutant peptides have altered ability to bind synthetic membranes and to disrupt bacteria, none of the mutations had a major effect on inhibition of HPV16 PsV (Table 2). The fact that mutant HNPs with reduced antibacterial activity retain the ability to inhibit papillomaviruses could be useful in the setting of a topical microbicide, where preservation of a healthy vaginal bacterial flora would be a desirable goal.
Materials and Methods
Cell Culture and Pseudovirus Production. Cell cultures of 293TT, HeLa, HaCaT and C127 were maintained in DMEM (Invitrogen) supplemented with 10% heat-inactivated FCS (HyClone), 1% nonessential amino acids (Invitrogen), and 1% Glutamax-I (Invitrogen) (DMEM-10). OptiPro SFM (Invitrogen) was used for experiments involving serum-free medium.
Optiprep-purified PsV stocks were produced and titered as described in refs. 16-19. Except for HPV5 (see below), all PsVs carried a 7.9-kb GFP reporter plasmid, p8fwB. Nucleotide maps and detailed protocols for PsV production and inhibition assays are available at http://ccr.cancer.gov/staff/links.asp?profileid=5637.
To construct an HPV5-based PsV, the nucleotide sequences of the L1 and L2 genes of a genomic clone of HPV5 were verified by direct sequencing (GenBank accession no. DQ080001). The verified L1 and L2 ORFs were then reconstructed synthetically (Blue Heron Biotechnology, Bothell, WA) according to a previously reported “as different as possible” codon-modification strategy (constructs p5L1h and p5L2w) (16, 18). Because the titer yield for the HPV5 PsV was lower than for other PsV types, a somewhat more efficient 5.9-kb target plasmid, pfwB, was used for production of HPV5 PsV stocks.
GFP-tagged HPV16 capsids were produced by using a plasmid, puL2f, expressing an HPV16 L2-GFP fusion protein. The L2-GFP fusion protein was incorporated into capsids at an L1-to-L2 ratio similar to that seen when capsids were produced by using wild-type L2. Binding-inhibition studies were performed by applying ≈5,000 capsid equivalents of L1 per cell at 37°C for 2 h in the presence or absence of varying doses of HNP-1, followed by flow-cytometric analysis.
Inhibition Assay. Compounds were purchased from commercial suppliers, as listed in Table 1, or synthesized and folded according to procedures reported in refs. 20-22. Compounds were dissolved in water, typically at 1 mg/ml.
HeLa cells were preplated overnight in 96-well plates at 6,000 cells per well in 50 μl of DMEM-10. The various candidate inhibitors were serially diluted in DMEM-10 in a separate 96-well plate at three times their final dose. Fifty microliters of each dilution were then added to the preplated HeLa cells, followed by 50 μl of virus stock diluted in DMEM-10 supplemented with 3× antibiotic-antimycotic mixture (Invitrogen). Control experiments showed that the antibiotic mixture had no effect on PsV transduction (data not shown). PsV doses were calibrated such that between 5% and 25% of cells registered as GFP+ in control conditions. In some instances, the cells were fed by adding of 100 μl of DMEM-10 24 h after PsV inoculation. The cells were incubated for a total 48-56 h after inoculation and then trypsinized and subjected to flow-cytometric analysis. For HNP-1 and HD-5, similar inhibition curves were observed when cells were analyzed 75 h after inoculation (data not shown). Percent inhibition was calculated by using the formula 100 × (1 - (net percentage of cells GFP+ in test/net percentage of cells GFP+ in mock)). The IC50 and 95% confidence interval for inhibition curves were calculated by using the program prism 4 (GraphPad) to fit a variable slope sigmoidal dose-response curve with top and bottom values constrained at 100 and 0, respectively.
Cytotoxicity was defined as a >50% reduction in turnover of WST-1 metabolic substrate (Roche) at the time of cell harvest. Cytotoxicity was confirmed by microscopic inspection of cell morphology and density or by the appearance of alterations in forward scatter, side scatter, and the autofluorescence profile of the cells on the flow cytometer.
Time-course studies (Fig. 2) were performed by using fixed doses of HD-5 or HNP-1 (5 μg/ml and 30 μg/ml, respectively). For the defensin addition experiments, the PsV inoculum was removed after 2 h of incubation at 37°C and replaced with fresh medium supplemented with defensin, as appropriate. For the zero time point in the defensin-removal time-course experiments, the cells were pretreated with defensin for 2 h.
To test the effects of various signaling agonists and antagonists on the HPV-inhibitory effects of HNP-1 and HD-5 (see Discussion), the inhibition assay was performed by using HeLa cells preincubated for 2 h with standard doses of various drugs. Drugs were removed 24 h after PsV inoculation. Tested compounds included the G protein-coupled receptor antagonist pertussis toxin (23, 24), the protein kinase C agonists bryostatin-I (25) and phorbol 12-myristate 13-acetate (26), the PKC antagonists bisindolylmaleimide-I (26) and Gö6976 (25), and the MAPK-kinase/MEK-1/2 antagonist U0126 (27). Except for bryostatin-I (Sigma), drugs were purchased from Calbiochem.
Microscopy. Immunofluorescent microscopy was performed as described in ref. 28. Where noted, HNP-1 (30 μg/ml) or HD-5 (5 μg/ml) were added together with PsV inoculum for 24 h. All images were acquired by using a Zeiss LSM 510 confocal system.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Author contributions: C.B.B., D.R.L., and J.T.S. designed research; C.B.B., P.M.D., and C.D.T. performed research; P.M.D., J.L., and W.L. contributed new reagents/analytic tools; C.B.B., P.M.D., C.D.T., D.R.L., and J.T.S. analyzed data; and C.B.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HD-5, human α-defensin 5; HNP, human neutrophil peptide; HPV, human papillomavirus; PsV, pseudovirus.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ080001).
References
- 1.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197-216. [DOI] [PubMed] [Google Scholar]
- 2.Platanias, L. C. (2005) Nat. Rev. Immunol. 5, 375-386. [DOI] [PubMed] [Google Scholar]
- 3.Braff, M. H., Bardan, A., Nizet, V. & Gallo, R. L. (2005) J. Invest. Dermatol. 125, 9-13. [DOI] [PubMed] [Google Scholar]
- 4.Selsted, M. E. & Ouellette, A. J. (2005) Nat. Immunol. 6, 551-557. [DOI] [PubMed] [Google Scholar]
- 5.Schiffman, M. & Kjaer, S. K. (2003) J. Natl. Cancer Inst. Monogr. 14-9. [DOI] [PubMed]
- 6.Gillison, M. L. & Shah, K. V. (2003) J. Natl. Cancer Inst. Monogr. 57-65. [DOI] [PubMed]
- 7.Manhart, L. E. & Koutsky, L. A. (2002) Sex. Transm. Dis. 29, 725-735. [DOI] [PubMed] [Google Scholar]
- 8.Holmes, K. K., Levine, R. & Weaver, M. (2004) Bull. W.H.O. 82, 454-461. [PMC free article] [PubMed] [Google Scholar]
- 9.Schiller, J. T. & Davies, P. (2004) Nat. Rev. Microbiol. 2, 343-347. [DOI] [PubMed] [Google Scholar]
- 10.Sokal, D. C. & Hermonat, P. L. (1995) Sex. Transm. Dis. 22, 22-24. [DOI] [PubMed] [Google Scholar]
- 11.Christensen, N. D., Reed, C. A., Culp, T. D., Hermonat, P. L., Howett, M. K., Anderson, R. A. & Zaneveld, L. J. (2001) Antimicrob. Agents Chemother. 45, 3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howett, M. K., Neely, E. B., Christensen, N. D., Wigdahl, B., Krebs, F. C., Malamud, D., Patrick, S. D., Pickel, M. D., Welsh, P. A., Reed, C. A., et al. (1999) Antimicrob. Agents Chemother. 43, 314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermonat, P. L., Daniel, R. W. & Shah, K. V. (1992) Sex. Transm. Dis. 19, 203-205. [DOI] [PubMed] [Google Scholar]
- 14.Bousarghin, L., Touze, A., Yvonnet, B. & Coursaget, P. (2004) J. Med. Virol. 73, 474-480. [DOI] [PubMed] [Google Scholar]
- 15.Drobni, P., Naslund, J. & Evander, M. (2004) Antiviral Res. 64, 63-68. [DOI] [PubMed] [Google Scholar]
- 16.Buck, C. B., Pastrana, D. V., Lowy, D. R. & Schiller, J. T. (2004) J. Virol. 78, 751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck, C. B., Thompson, C. D., Pang, Y. Y., Lowy, D. R. & Schiller, J. T. (2005) J. Virol. 79, 2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastrana, D. V., Buck, C. B., Pang, Y. Y., Thompson, C. D., Castle, P. E., FitzGerald, P. C., Kruger Kjaer, S., Lowy, D. R. & Schiller, J. T. (2004) Virology 321, 205-216. [DOI] [PubMed] [Google Scholar]
- 19.Pastrana, D. V., Gambhira, R., Buck, C. B., Pang, Y. Y., Thompson, C. D., Culp, T. D., Christensen, N. D., Lowy, D. R., Schiller, J. T. & Roden, R. B. (2005) Virology 337, 365-372. [DOI] [PubMed] [Google Scholar]
- 20.Wu, Z., Powell, R. & Lu, W. (2003) J. Am. Chem. Soc. 125, 2402-2403. [DOI] [PubMed] [Google Scholar]
- 21.Wu, Z., Ericksen, B., Tucker, K., Lubkowski, J. & Lu, W. (2004) J. Pept. Res. 64, 118-125. [DOI] [PubMed] [Google Scholar]
- 22.Xie, C., Prahl, A., Ericksen, B., Wu, Z., Zeng, P., Li, X., Lu, W. Y., Lubkowski, J. & Lu, W. (2005) J. Biol. Chem. [DOI] [PubMed]
- 23.Yang, D., Chen, Q., Chertov, O. & Oppenheim, J. J. (2000) J. Leukoc. Biol. 68, 9-14. [PubMed] [Google Scholar]
- 24.Befus, A. D., Mowat, C., Gilchrist, M., Hu, J., Solomon, S. & Bateman, A. (1999) J. Immunol. 163, 947-953. [PubMed] [Google Scholar]
- 25.Chang, T. L., Vargas, J., Jr., DelPortillo, A. & Klotman, M. E. (2005) J. Clin. Invest. 115, 765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charnaux, N., Brule, S., Chaigneau, T., Saffar, L., Sutton, A., Hamon, M., Prost, C., Lievre, N., Vita, C. & Gattegno, L. (2005) Glycobiology 15, 119-130. [DOI] [PubMed] [Google Scholar]
- 27.Aarbiou, J., Verhoosel, R. M., Van Wetering, S., De Boer, W. I., Van Krieken, J. H., Litvinov, S. V., Rabe, K. F. & Hiemstra, P. S. (2004) Am. J. Respir. Cell Mol. Biol. 30, 193-201. [DOI] [PubMed] [Google Scholar]
- 28.Day, P. M., Baker, C. C., Lowy, D. R. & Schiller, J. T. (2004) Proc. Natl. Acad. Sci. USA 101, 14252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch, F. X. & de Sanjose, S. (2003) J. Natl. Cancer Inst. Monogr. 3-13. [DOI] [PubMed]
- 30.Joyce, J. G., Tung, J. S., Przysiecki, C. T., Cook, J. C., Lehman, E. D., Sands, J. A., Jansen, K. U. & Keller, P. M. (1999) J. Biol. Chem. 274, 5810-5822. [DOI] [PubMed] [Google Scholar]
- 31.Giroglou, T., Florin, L., Schafer, F., Streeck, R. E. & Sapp, M. (2001) J. Virol. 75, 1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selinka, H. C., Giroglou, T., Nowak, T., Christensen, N. D. & Sapp, M. (2003) J. Virol. 77, 12961-12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chertov, O., Michiel, D. F., Xu, L., Wang, J. M., Tani, K., Murphy, W. J., Longo, D. L., Taub, D. D. & Oppenheim, J. J. (1996) J. Biol. Chem. 271, 2935-2940. [DOI] [PubMed] [Google Scholar]
- 34.Dorschner, R. A., Pestonjamasp, V. K., Tamakuwala, S., Ohtake, T., Rudisill, J., Nizet, V., Agerberth, B., Gudmundsson, G. H. & Gallo, R. L. (2001) J. Invest. Dermatol. 117, 91-97. [DOI] [PubMed] [Google Scholar]
- 35.Conner, K., Nern, K., Rudisill, J., O'Grady, T. & Gallo, R. L. (2002) J. Am. Acad. Dermatol. 47, 347-350. [DOI] [PubMed] [Google Scholar]
- 36.Valore, E. V., Park, C. H., Igreti, S. L. & Ganz, T. (2002) Am. J. Obstet. Gynecol. 187, 561-568. [DOI] [PubMed] [Google Scholar]
- 37.Daher, K. A., Selsted, M. E. & Lehrer, R. I. (1986) J. Virol. 60, 1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dvoretzky, I., Shober, R., Chattopadhyay, S. K. & Lowy, D. R. (1980) Virology 103, 369-375. [DOI] [PubMed] [Google Scholar]
- 39.Pastrana, D. V., Vass, W. C., Lowy, D. R. & Schiller, J. T. (2001) Virology 279, 361-369. [DOI] [PubMed] [Google Scholar]
- 40.De Villiers, E. M., Fauquet, C., Broker, T. R., Bernard, H. U. & Zur Hausen, H. (2004) Virology 324, 17-27. [DOI] [PubMed] [Google Scholar]
- 41.Day, P. M., Lowy, D. R. & Schiller, J. T. (2003) Virology 307, 1-11. [DOI] [PubMed] [Google Scholar]
- 42.Selinka, H. C., Giroglou, T. & Sapp, M. (2002) Virology 299, 279-287. [DOI] [PubMed] [Google Scholar]
- 43.Bousarghin, L., Touze, A., Sizaret, P. Y. & Coursaget, P. (2003) J. Virol. 77, 3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganz, T. (2003) Nat. Rev. Immunol. 3, 710-720. [DOI] [PubMed] [Google Scholar]
- 45.Lehrer, R. I. (2004) Nat. Rev. Microbiol. 2, 727-738. [DOI] [PubMed] [Google Scholar]
- 46.Panyutich, A. V., Panyutich, E. A., Krapivin, V. A., Baturevich, E. A. & Ganz, T. (1993) J. Lab. Clin. Med. 122, 202-207. [PubMed] [Google Scholar]
- 47.Hein, M., Valore, E. V., Helmig, R. B., Uldbjerg, N. & Ganz, T. (2002) Am. J. Obstet. Gynecol. 187, 137-144. [DOI] [PubMed] [Google Scholar]
- 48.Quayle, A. J., Porter, E. M., Nussbaum, A. A., Wang, Y. M., Brabec, C., Yip, K. P. & Mok, S. C. (1998) Am. J. Pathol. 152, 1247-1258. [PMC free article] [PubMed] [Google Scholar]
- 49.Com, E., Bourgeon, F., Evrard, B., Ganz, T., Colleu, D., Jegou, B. & Pineau, C. (2003) Biol. Reprod. 68, 95-104. [DOI] [PubMed] [Google Scholar]
- 50.Porter, E., Yang, H., Yavagal, S., Preza, G. C., Murillo, O., Lima, H., Greene, S., Mahoozi, L., Klein-Patel, M., Diamond, G., et al. (2005) Infect. Immun. 73, 4823-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armogida, S. A., Yannaras, N. M., Melton, A. L. & Srivastava, M. D. (2004) Allergy Asthma Proc. 25, 297-304. [PubMed] [Google Scholar]
- 52.Dunsche, A., Acil, Y., Siebert, R., Harder, J., Schroder, J. M. & Jepsen, S. (2001) J. Oral Pathol. Med. 30, 154-158. [DOI] [PubMed] [Google Scholar]
- 53.Frye, M., Bargon, J., Dauletbaev, N., Weber, A., Wagner, T. O. & Gropp, R. (2000) J. Clin. Pathol. 53, 770-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, L., Roberts, A. A. & Ganz, T. (2003) J. Immunol. 170, 575-580. [DOI] [PubMed] [Google Scholar]
- 55.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T. & Bevins, C. L. (2002) Nat. Immunol. 3, 583-590. [DOI] [PubMed] [Google Scholar]
- 56.Linzmeier, R. M. & Ganz, T. (2005) Genomics 86, 423-430. [DOI] [PubMed] [Google Scholar]
- 57.Aldred, P. M., Hollox, E. J. & Armour, J. A. (2005) Hum. Mol. Genet. 14, 2045-2052. [DOI] [PubMed] [Google Scholar]
- 58.Winer, R. L., Lee, S. K., Hughes, J. P., Adam, D. E., Kiviat, N. B. & Koutsky, L. A. (2003) Am. J. Epidemiol. 157, 218-226. [DOI] [PubMed] [Google Scholar]
- 59.Winer, R. L., Kiviat, N. B., Hughes, J. P., Adam, D. E., Lee, S. K., Kuypers, J. M. & Koutsky, L. A. (2005) J. Infect. Dis. 191, 731-738. [DOI] [PubMed] [Google Scholar]
- 60.Trabattoni, D., Caputo, S. L., Maffeis, G., Vichi, F., Biasin, M., Pierotti, P., Fasano, F., Saresella, M., Franchini, M., Ferrante, P., et al. (2004) J. Acquired Immune Defic. Syndr. 35, 455-463. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., Yu, W., He, T., Yu, J., Caffrey, R. E., Dalmasso, E. A., Fu, S., Pham, T., Mei, J., Ho, J. J., et al. (2002) Science 298, 995-1000. [DOI] [PubMed] [Google Scholar]
- 62.Yasin, B., Harwig, S. S., Lehrer, R. I. & Wagar, E. A. (1996) Infect. Immun. 64, 709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehrer, R. I., Daher, K., Ganz, T. & Selsted, M. E. (1985) J. Virol. 54, 467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bastian, A. & Schafer, H. (2001) Regul. Pept. 101, 157-161. [DOI] [PubMed] [Google Scholar]
- 65.Gropp, R., Frye, M., Wagner, T. O. & Bargon, J. (1999) Hum. Gene Ther. 10, 957-964. [DOI] [PubMed] [Google Scholar]
- 66.Kim, C., Gajendran, N., Mittrucker, H. W., Weiwad, M., Song, Y. H., Hurwitz, R., Wilmanns, M., Fischer, G. & Kaufmann, S. H. (2005) Proc. Natl. Acad. Sci. USA 102, 4830-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiethoff, C. M., Wodrich, H., Gerace, L. & Nemerow, G. R. (2005) J. Virol. 79, 1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards, R. M., Lowy, D. R., Schiller, J. T. & Day, P. M. (2006) Proc. Natl. Acad. Sci. USA 103, 1522-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kämper, N., Day, P. M., Nowak, T., Selinka, H.-C., Florin, L., Bolschev, J., Hilbig, L., Schiller, J. T. & Sapp, M. (2006) J. Virol. 80, 759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]