Abstract
Calcium signaling pathways control a variety of cellular events such as gene transcription, protein phosphorylation, nucleotide metabolism, and ion transport. These pathways often involve a large number of calcium-binding proteins collectively known as the calmodulin or EF-hand protein superfamily. Many EF-hand proteins undergo a large conformational change upon binding to Ca2+ and target proteins. All members of the superfamily share marked sequence homology and similar structural features required to sense Ca2+. Despite such structural similarities, the functional diversity of EF-hand calcium-binding proteins is extraordinary. Calmodulin itself can bind >300 different proteins, and the many members of the neuronal calcium sensor and S100 protein families collectively recognize a largely different set of target proteins. Recent biochemical and structural studies of many different EF-hand proteins highlight remarkable similarities and variations in conformational responses to the common ligand Ca2+ and their respective cellular targets. In this review, we examine the essence of molecular recognition activities and the mechanisms by which calmodulin superfamily proteins control a wide variety of Ca2+ signaling processes.
The Ca2+ ion is a highly versatile intracellular signal regulating many different cellular functions, including fertilization, cell cycle, apoptosis, muscle contraction, vision, and memory (1). In eukaryotic cells, cytoplasmic Ca2+ entry and outflow are governed by two sources: intracellular stores such as the endoplasmic reticulum and extracellular Ca2+ that enters the cell through various transporters on the plasma membrane (2). Ca2+ entry into the cytoplasm is tightly regulated by a variety of components of the Ca2+ signaling toolkit, which were elegantly summarized by Berridge et al. (3). The Ca2+ flux machinery, consisting of ion channels, pumps, and exchangers, gives rise to highly localized and transient Ca2+ signals that are, in turn, transduced by calcium-binding proteins acting on various enzymes and down-stream effector proteins.
A central question in the field of Ca2+ signaling is how different Ca2+ signaling systems control so many divergent cellular processes (3)? Such control is achieved at both the cellular and molecular level. The spatial and temporal variation of Ca2+ signals, known as Ca2+ waves, spikes, and puffs, are responsible for generating diverse output required for different physiological conditions (4). Also, cell-specific expression of a unique set of components from the Ca2+ signaling toolkit (3) is required for generating cell-specific responses to Ca2+ signals. At the molecular level, a variety of Ca2+ sensor proteins provide totally different physiological responses by stimulating or suppressing different intracellular signaling pathways (5–7).
The calmodulin superfamily is a major class of Ca2+ sensor proteins, which collectively play a crucial role in various cellular signaling cascades through regulation of numerous target proteins in a Ca2+-dependent manner (Table 1). It has been reported that there are nearly 600 members in this superfamily (60), all of which contain one or more Ca2+ binding motifs known as the EF-hand, first identified in parvalbumin by Kretsinger and coworkers (7). Calmodulin contains four EF-hand motifs, with highly conserved amino acid sequences in all eukaryotes. In fact, this sequence is ranked fifth in an amino acid conservation contest in proteomes after the histones H4 and H3, actin B, and ubiquitin (61). In contrast, the troponin C family, which also contains four EF-hand motifs, has two isoforms in the human (skeletal and cardiac muscles) and many isoforms in invertebrates, all diverse in amino acid sequence (6). Similarly, the neuronal calcium sensor (NCS) and S100 proteins are diverse in sequence and function. Recent advances in the structural and biochemical understanding of these Ca2+ sensor proteins unveiled two emerging themes that explain the vast multifunctionality of the calmodulin superfamily. These molecular themes are genetic polymorphism and protein conformational plasticity, and are likely to be relevant to other protein superfamilies with diverse functions.
Table 1. EF-hand Ca2+-binding proteins and their functional target proteins.
| EF-hand protein | Functional targets |
|---|---|
| Calmodulin | Myosin light chain kinases (8) |
| Calmodulin-dependent protein kinases (9) | |
| Phosphorylase kinase (10) | |
| Myristoylated protein kinase C substrates (11) | |
| G protein-coupled receptor kinases (GRKs) (12) | |
| Calcineurin (13) | |
| Adenylate cyclases (14) | |
| Glutamate decarboxylase (15) | |
| Nitric oxide synthases (16) | |
| Phosphodiesterases (17, 18) | |
| Plasma membrane Ca2+ ATPase pump (19) | |
| Cyclic-nucleotide gated ion channels (20) | |
| SK channels (21) | |
| Voltage-gated Ca2+ channels (22) | |
| Inositol 1,4,5-trisphosphate receptors (23) | |
| Ryanodine receptors (24) | |
| Troponin C | |
| Skeletal (100%) | Skeletal muscle Troponin I (25) |
| Cardiac (65%) | Cardiac muscle Troponin I (26, 27) |
| Invertebrate (37%) | Invertebrate Troponin I (28) |
| NCS family | |
| Frequenin (100%) | Pl4-kinase (29), K+ channels (30), Ca2+ channels (31) |
| Neurocalcin-δ (55%) | Nicotinic acetylcholine receptors (32) |
| Recoverin (43%) | Rhodopsin kinase (33) |
| KChIP1-4 (40%) | Shaker channels (34) |
| DREAM/calensilin | Dynorphin DRE (35), presenilin (36) |
| GCAPs (35%) | Retinal guanylate cyclases (retGCs) (37) |
| Calcineurin B (32%) | Calcineurin A-subunit (38) |
| CIB (24%) | Integrin (39), presenilins (40) |
| CaBP1 (22%) | Inositol 1,4,5-trisphosphate receptors (41), voltage-gated Ca2+ channels (42) |
| S100 family | |
| S100A1 (100%) | Titin (43), SERCA2a (44), ryanodine receptor (45) |
| S100A4 (53%) | Methionine aminopeptidase 2 (46) |
| S100A6 (48%) | Ubiquitin ligase (47) |
| S100A10 (47%) | Annexin II (48) |
| S100A11 (43%) | Annexin I (49) |
| S100A12 (40%) | RAGE (50) |
| S100B (60%) | p53 (51), NDR kinase (52), CapZ (53), retinal guanylate cylcase (54) |
| Penta EF-hand family | |
| Grancalcin (100%) | L-plastin (55) |
| Sorcin (54%) | Ca2+ channels (56) |
| ALG-2 (32%) | Alix/AIP1 (57), annexins (58) |
| Calpain (29%) | Calpastatin (59) |
Relative amino acid sequence identities within each subfamily are indicated in parentheses (%). References are provided after each target in parentheses.
EF-Hand as a Building Block of Ca2+ Sensor Proteins
The EF-hand motif consists of a simple helix-loop-helix architecture in which the interhelical loop region contains several amino acids essential to the coordination of a single Ca2+ ion. Typically, a pair of EF-hand motifs in tandem array constitutes a stable structural unit, together generating cooperativity in the binding of Ca2+ ions (62, 63). Many EF-hand proteins, such as calmodulin and members of the NCS family, consist of four EF-hand motifs. This results in two globular structural units in a single protein. We will discuss the importance of this feature to the multifunctionality of these proteins. The S100 family, on the other hand, consists of only one globular structural unit comprised of two EF-hand motifs. However, members of this family all form a stable homo- or heterodimer, which contains four EF-hand motifs in a single structural entity.
The direct interaction with Ca2+ en-ables these Ca2+ sensor proteins to change their conformation from the inactive state (P) to the intermediate state (Ca2+-P*), which is a prerequisite to the formation of an active conformation in complex with a target (Ca2+-P**-E*) required to transform the target protein from its inactive state (E) to the active state (E*)
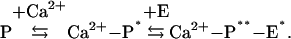 |
The first conformational transition is key to the Ca2+ sensory function and is universal to all Ca2+ sensor proteins. The second conformational change plays a critical role in the activation and recognition of specific targets.
Genetic Polymorphism in NCS and S100 Proteins
A growing number of EF-hand proteins exist as splice variants or multiple isoforms that exhibit diverse biological functions. This evolutionary diversity is best illustrated by members of the NCS (64 and 65) and S100 (63 and 65) families. A total of 24 human genes code for S100 isoforms (66), and >15 genes code for NCS isoforms and >10 splice variants (67). The S100 genes are typically 40–60% identical in sequence and the NCS genes range from 22–55% identity (Table 1). Each protein isoform binds to a distinct target protein and performs a highly specialized physiological task.
Functional Diversity of NCS and S100 Proteins
The NCS proteins, as their name implies, are expressed almost exclusively in the central nervous system. The common features of NCS proteins are an ≈200-residue chain containing four EF-hand motifs and an amino-terminal myristoylation consensus sequence (65). The best characterized NCS protein is recoverin, a calcium-myristoyl switch protein in retinal rod cells (68, 69) that controls desensitization of rhodopsin (33) by regulating rhodopsin kinase activity (70). Recoverin was also identified as the antigen in cancer-associated retinopathy (71). Other NCS proteins include neurocalcin (72), frequenin (NCS-1) (73), K+ channel-interacting proteins (KChIPs) (34), DREAM/calsenilin (35, 36) and hippocalcin (74). Frequenin also is expressed in invertebrates, including flies (73), worms (75) and yeast (29, 76). Yeast and mammalian frequenins bind and activate a particular phosphatidylinositol 4-OH kinase isoform (Pik1 gene in yeast) (29, 77) required for vesicular trafficking in the late secretory pathway (78). Mammalian frequenin (NCS-1) also regulates voltage-gated Ca2+ channels (31) and K+ channels (30). The KChIP proteins regulate the gating kinetics of Shaker K+ channels (34). DREAM/calsenilin binds to specific DNA sequence elements in the prodynorphin and c-fos genes (35) and serves as a transcriptional repressor for pain modulation (79, 80). Hence, the physiological functions of the NCS proteins are highly diverse and nonoverlapping.
The S100 proteins also have diverse physiological functions involved in regulating cell cycle control, transcription, and secretion (Table 1). S100A1 controls cardiac contractility and is associated with a number of cardiomyopathies (81). S100A2 is localized to the nucleus where it regulates transcription of various genes and acts as a tumor suppressor in breast cancer cells (82). In contrast, S100A4 (83) and S100A6 (47) are both tumor promoters. S100A7 acts extracellularly and is implicated in epidermal inflammatory diseases such as psoriasis (84). The S100A8/A9 heterodimer acts extracellularly as a chemotactic molecule in inflammation (85). S100A10 and S100A11 bind to distinct annexin proteins and regulate annexin trafficking to cell membranes (48, 49). S100A12 is expressed in phagocytes, where it regulates secretion of proinflammatory mediators (50). Perhaps the best characterized S100 protein, S100B is highly expressed in the brain, where it is implicated in various neurodegenerative diseases such as Alzheimer's disease, Down syndrome, and multiple sclerosis. S100B is multifunctional and binds to multiple target proteins, including p53, an interaction that is involved in regulating transcription (51). S100B also interacts with NDR kinase (52), CapZ (53), and retinal guanylate cyclase (86) to control neuronal excitability. In summary, the physiological functions of S100 proteins are even more wide ranging and diverse than those of the NCS family.
Structure and Target Recognition of NCS and S100 Proteins
The three-dimensional structures of various NCS proteins [recoverin (69, 87), neurocalcin (88) and frequenin (89)] reveal that their main-chain globular folds are nearly identical, which is not surprising given the sequence conservation. In all NCS proteins, four EF-hands form a highly compact, globular fold, in contrast to the dumbbell arrangement of two independent domains found in calmodulin (90) and troponin C (91). The three-dimensional structures of various S100 isoforms are also very similar to one another and contain a symmetrical dimer of EF-hands arranged in a compact globular fold (51, 92–94). If the overall main-chain conformation of the various NCS and S100 isoforms is so similar, how can one explain the functional differences? An important distinguishing structural property is the distribution of charged and hydrophobic side-chains on the protein surface. For NCS proteins, the surface properties of the C-terminal domain are quite variable, whereas the N-terminal domain is more conserved.
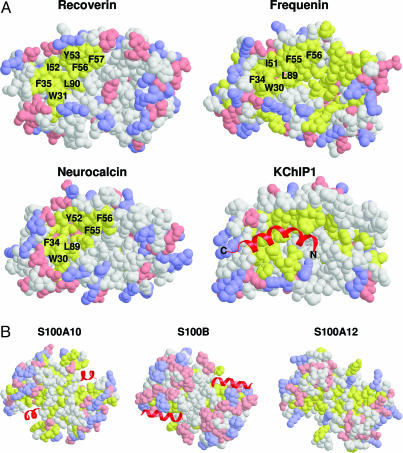
Surface representations of hydrophobicity and the charge density of the various NCS structures are shown in Fig. 1A. All NCS structures exhibit a similar exposed hydrophobic surface located on the N-terminal half of the protein (F35, W31, F56, F57, Y86, and L90 for recoverin in Fig. 2A). The exposed hydrophobic residues in this region are conserved and correspond to residues of recoverin that interact with the myristoyl group in the Ca2+-free state (69, 95, 96). Upon Ca2+ binding, these exposed residues in the hydrophobic patch have been implicated in target recognition from mutagenesis studies (97–100), and these residues form intermolecular contacts with target proteins as observed in the recent crystal structure of KChIP1 (101). The structural interaction of KChIP1 with its target helix (highlighted red in Fig. 1A) is very similar to the interaction of the N-terminal domain of CaM bound to myosin light chain kinase (MLCK) (Fig. 2C).
Fig. 1.
Space-filling structures of NCS (A) and S100 (B) proteins. Positive, negative, and hydrophobic residues on the protein surface are highlighted in blue, pink, and yellow, respectively. Ribbon diagram of bound target helices are shown in red. Conserved hydrophobic residues implicated in target recognition are labeled. Atomic coordinates are available from the Protein Data Bank: 1rec (recoverin), 1bjf (neurocalcin), 1g8i (frequenin), 1s6c (KChIP1), 1bt6 (S100A10), 1dt7 (S100B), and 1e8a (S100A12).
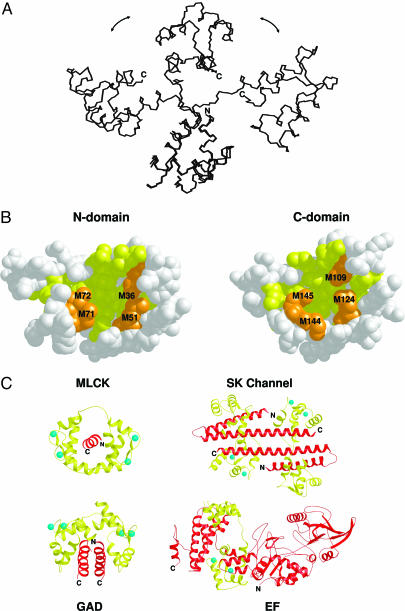
Fig. 2.
Structure and target recognition by calmodulin. (A) Superposition of selected NMR structures of apo-calmodulin (Protein Data Bank entry 1dmo). (B) Space-filling representation of hydrophobic target binding sites (yellow) flanked by surrounding methionine residues (orange). (C) Main-chain ribbon diagrams of calmodulin (yellow) bound to the target proteins myosin light chain kinase (MLCK), SK channel, GAD, and EF (red). Bound Ca2+ ions are shown in blue.
Although the hydrophobic patch on the N-terminal domain is conserved among NCS proteins, the surface properties of the C-terminal domain appear to be unique. For example, frequenin exhibits exposed hydrophobic residues (highlighted yellow in Fig. 1A) in the C-terminal domain that, together with the conserved hydrophobic crevice in the N-terminal domain, form one continuous and elongated patch. By contrast, recoverin has mostly charged residues on the surface of the C-terminal half (highlighted blue and pink), whereas neurocalcin and KChIP1 have mostly uncharged and polar residues shown in white. We propose that these different patterns of charge distribution and hydrophobicity on the surface of the C-terminal half of NCS proteins are important structural determinants for conferring target specificity.
A similar contrast in surface charge distribution is also apparent in representative structures of S100 proteins (Fig. 1B). S100B contains an abundance of negatively charged residues (Asp and Glu shaded pink) located on the protein surface balanced by a nearly equal number of positively charged surface residues (blue). By contrast, S100A10 and S100A12 have fewer charged residues on their protein surfaces and a much higher percentage of hydrophobic residues (yellow). Two target helices derived from p53 (shown dark red in Fig. 1B) are bound to the S100B homodimer in a symmetrical fashion (51). Target helices derived from annexin II bind to related binding sites on S100A10 (48). Both the p53 and annexin II target helices contain an amphipathic hydrophobic face that interacts with a complementary hydrophobic crevice on S100B and S100A10, respectively. The orientation of the p53 target helix with respect to S100B is quite different from that of the annexin target helix bound to S100A10 (Fig. 1B). The different target orientations are explained, in part, by differences in the surface contour properties at the respective binding sites. Another important factor is the distribution of charged residues on the surface near the target-binding site. The highly charged environment on S100B may exert local forces that influence the disposition of the target helix by forming electrostatic and/or hydrogen-bonding contacts. By contrast, the corresponding residues on S100A10 are mostly neutral and do not interact strongly with the target helix. The characteristic surface charge properties of the various S100 isoforms help explain the unique target recognition of each isoform that is important for specifying their diverse biological functions.
In summary, the structures of NCS and S100 proteins both contain EF-hands arranged in a compact globular fold, in contrast to the two independent domains found in calmodulin. The highly conserved and compact main chain conformation found in either NCS or S100 proteins serves as a scaffold that can be adapted with amino acid side-chain groups of different size and/or charge to alter the protein surface and facilitate unique target interactions. This type of surface remodeling appears to be an effective means of modifying target specificity, thereby increasing the repertoire of downstream effector proteins.
Conformational Plasticity in Calmodulin
In contrast to the highly specialized multiple isoforms of S100 and NCS families, the single protein, CaM, regulates numerous target proteins that are functionally and structurally diverse (Table 1). In 1970, CaM was first discovered to activate cyclic nucleotide phosphodiesterase (17, 18). Subsequently, many proteins such as Ca2+-transporting ATPase (102, 103), myosin light chain kinase (8) and phosphorylase kinase (10) were all found to be regulated by CaM. Protein kinases (104), the protein phosphatase calcineurin (13) and nitric oxide synthetase (NOS) (16) were also found to be targets of Ca2+/CaM-dependent stimulation. The list of known CaM-dependent proteins exceeds 300 in number (105) and now includes many ion transporters, such as inositol 1,4,5-trisphosphate receptor (23), ryanodine receptor (24), plasma membrane Ca2+ pump (19), and L-type Ca2+ channel (22), all negatively or positively regulated by Ca2+/CaM. Amazingly, some infectious bacteria enzymes, such as adenylyl cyclase from Bacillus anthracis and Bordetella pertussis, require host CaM for its toxic enzymatic activity of converting ATP to cyclic-ADP (106).
Now a major question is how CaM binds and regulates all these proteins. Crystallographic and NMR studies on various CaM target proteins have shed light on this unusual property of CaM. The first structure determined for CaM in complex with a target protein showed a remarkable conformational change in CaM's two EF-hand domains upon binding to a peptide derived from myosin light chain kinase (MLCK) (107, 108). This structure revealed two important properties. First, the central domain linker (residues 78–81) is highly flexible and can be bent dramatically upon binding to the target protein (Fig. 2A; refs. 109–111). The flexibility of the domain linker permits the orientation of the two domains of CaM to change independently to accommodate the structural nature of the target protein. Secondly, two hydrophobic anchoring residues from the smooth muscle MCLK peptide (Trp-800 and Leu-813) bind simultaneously to the hydrophobic pocket in N- and C-terminal domains, which is extremely rich in methionine residues (four Met residues in each pocket, see Fig. 2B). CaM contains nine methionines corresponding to 6% of the entire sequence, which is significantly higher than the average of known proteomes (1%). Conservative mutations of these methionines to leucines at different sites significantly altered CaM's activity to stimulate cAMP phosphodiesterase (112). The vital role of methionines in interactions with target proteins is evident in a number of new structures reported for CaM in complex with CaMKII (113), CaM kinase kinase (114), NO synthatase (115), glutamate decarboxylase (Fig. 2C; ref. 15), MARCKS (116), Ca2+-activated K+ channel (Fig. 2C; ref. 21), and anthrax adenylyl cyclase (Fig. 2C; ref. 14). These structures reinforce the significance of the aforementioned structural properties of CaM by clearly illustrating that CaM can adopt largely different, global conformations depending on the structural entity that CaM binds. In addition to plasticity of the protein fold, the amino acid side chains that interact with target proteins, in particular the Met residues, are remarkably flexible. Indeed, the hydrophobic pockets of CaM are so flexible that they can accommodate a variety of amino acid side chains, such as those of Trp, Phe, Ile, Leu, Val, and Lys. This structural plasticity at both the level of the individual side chains and the orientation of entire domains is crucial for CaM to the recognition of numerous target proteins for regulation by CaM.
Cellular Level of Control
The exact and precise recruitment of the Ca2+ signaling toolkit proteins under a specific cellular condition depends on differential gene transcription of its components. In the case of CaM alone, the temporal and spatial expression of its gene is tightly regulated in most, if not all, cell types. In many vertebrates, three bona fide CaM genes (CaM I, CaM II, and CaM III), which are collectively transcribed into at least eight mRNAs, eventually translated into an identical CaM protein consisting of 148 amino acids. These proteins are highly expressed in the brain and the testis (117). Interestingly, recent data using highly polarized cells, such as neurons, (118) indicate that CaM mRNAs are targeted to different intracellular compartments and that the translocation of mRNA(s), instead of the protein, provides a means to enrich the CaM protein in a specific site(s) in the cell. Overall, CaM seems to be a limiting factor in cell function (119). Furthermore, the CaM protein has been shown to undergo posttranslational modifications including acetylation, trimethylation, carboxylmethylation, proteolytic cleavage, and phosphorylation (120–122). These chemical modifications may modulate CaM's biological activity as a Ca2+ sensor protein; however, the physiological roles of such posttranslational events remain to be fully explored.
Similarly, the CaM target proteins identified thus far are all transcriptionally regulated. For instance, the inositol 1,4,5-trisphosphate receptor (IP3R) possesses three isoforms in mammals: Type I IP3R is highly expressed in cerebellar Purkinje cells of the central nervous system, whereas type II is predominantly found in cardiac ventricular myocytes (123). These isoforms not only have different subcellular distribution, which is dynamically regulated dependent on physiological conditions, but also generate different splice variants with different biophysical properties in terms of ligand binding (124). Moreover, these CaM targets are often posttranslationally modified, which may affect their ability to interact with CaM. For example, phosphorylation of the catalytic subunit of CaM kinase modulates its CaM sensitivity (104). Clearly, the CaM–target interaction is modulated by a number of cellular mechanisms involving temporal and spatial subcellular localization of CaM itself and the target protein as well as structural modifications of the CaM-binding site within the target protein.
Like CaM, the various NCS proteins are variably expressed in many different cell types, and NCS splice variants exhibit tissue-specific expression. The NCS proteins, recoverin and guanylate cyclase-activating proteins, are found almost exclusively in retinal rod and cone cells (125, 126), whereas many other NCS proteins, such as neurocalcin and frequenin, are present only in the brain and spinal cord. Mammalian frequenin (NCS-1) is expressed throughout the central nervous system (127). By contrast, hippocalcin is localized in hippocampal pyramidal neurons, and VILIP-3 is found only in cerebellar Purkinje and granule cells. In addition, hippocalcin, neurocalcin-δ, and recoverin each possess a calcium-myristoyl switch that controls subcellular membrane localization and capacity to interact with membrane-bound targets (68, 69). In contrast, the KChIP proteins lack myristoylation and generally do not possess a functional myristoyl switch. Instead, splice variants of KChIPs are differentially expressed in various sensory neurons and cardiac cells. The DREAM protein (KChIP3) is found mostly in the cerebellar granular cortex (128), whereas KChIP2 and related splice variants are found only in cardiac myocytes (129). Hence, the tissue-specific expression of NCS proteins and their membrane localization controlled by N-terminal myristoylation help confer the distinctive physiological roles of NCS proteins generally.
S100 proteins also exhibit tissue-specific expression patterns and different subcellular localization. S100 proteins in the cytosol act as intracellular calcium sensors and/or calcium buffers. S100A1 is expressed exclusively in cardiac cells, where it has been implicated in a number of cardiomyopathies (81). S100A2, S100A6, and S100B are all present in the nucleus, where they control gene expression and are implicated in various forms of cancer. S100B is expressed also throughout the central nervous system, where it is linked to a number of neurodegenerative disorders. Many S100 proteins (such as S100B, S100A4, S100A7, S100A8, and S100A9) are also extracellularly targeted, where they interact with the extracellular domain of the receptor for advanced glycation end products. Extracellular S100 proteins also display an unusually high affinity for Zn2+ or Cu2+, which may serve a unique regulatory role in the extracellular space. In summary, the cellular localization and tissue-specific expression of S100 proteins help explain their functional diversity.
Summary and Perspectives
In this review, we summarized our current knowledge on the diversity of proteins that function as Ca2+ sensors in eukaryotic cells. The NCS and S100 protein families has undergone extensive rounds of evolutionary mutation to equip each isoform with a specific function at a specific location in the cell. In contrast, the evolutionary history of the single multifunctional Ca2+ sensor protein, CaM, is strikingly different. Presumably through a gene duplication event, CaM has become equipped with two EF-hand domains that are connected by a magical flexible linker that enables this multifunctional protein to change its conformation as needed. This event presumably happened at a relatively early stage of evolution, and the molecular architecture has been kept since then as the amino acid sequence of CaM is remarkably conserved from yeast to human. It is interesting to note that all known NCS and S100 family proteins adopt a single globular domain architecture, unlike the two independent domains of CaM. Protein conformational plasticity of CaM thus emerged as a means of achieving functional diversity rather than employing the more traditional approach of genetic polymorphism.
In human and other eukaryotic proteomes, signaling module domains such as SH2, SH3, and PDZ domains can be linked with a kinase or a receptor module to introduce regulatory mechanisms (130, 131). Generally, the insertion of protein domains enables the protein to acquire additional functions one at a time. The uniqueness of CaM's modularity is that by simply connecting two EF-hand modules via a flexible linker, an exponential increase in the number of target interactions was achieved. Is this conformational plasticity unique to CaM, or is there any other protein(s) that might use the same principle for its multifunctionality? Finally, with regard to NCS and S100 proteins, how could each of the protein isoforms coevolve with their respective target proteins to achieve such specificity and exclusivity? Further biochemical and structural studies will be needed to address those key questions.
Acknowledgments
We thank several of our colleagues for their helpful discussions and Kyoko Yap, Jane Gooding, Peter Stathopulos, and Chris Marshall for their helpful comments on the manuscript. This work was supported by the Canadian Institutes of Health Research (M.I.) and the National Institutes of Health (J.B.A.). M.I. holds the Canada Research Chair of Cancer Structural Biology.
Author contributions: M.I. and J.B.A. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: KChIP, K+ channel-interacting protein; NCS, neuronal calcium sensor.
References
- 1.Carafoli, E. (2005) FEBS J. 272, 1073–1089. [DOI] [PubMed] [Google Scholar]
- 2.Bootman, M. D. & Berridge, M. J. (1995) Cell 83, 675–678. [DOI] [PubMed] [Google Scholar]
- 3.Berridge, M. J., Bootman, M. D. & Roderick, H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529. [DOI] [PubMed] [Google Scholar]
- 4.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21. [DOI] [PubMed] [Google Scholar]
- 5.Haiech, J., Moulhaye, S. B. & Kilhoffer, M. C. (2004) Biochim. Biophys. Acta 1742, 179–183. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki, H., Nakayama, S. & Kretsinger, R. H. (1998) Biometals 11, 277–295. [DOI] [PubMed] [Google Scholar]
- 7.Kretsinger, R. H. & Nockolds, C. E. (1973) J. Biol. Chem. 248, 3313–3326. [PubMed] [Google Scholar]
- 8.Yazawa, M. & Yagi, K. (1977) J. Biochem. (Tokyo) 82, 287–289. [DOI] [PubMed] [Google Scholar]
- 9.Soderling, T. R. (1999) Trends Biochem. Sci. 24, 232–236. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, P., Burchell, A., Foulkes, J. G. & Cohen, P. T. (1978) FEBS Lett. 92, 287–293. [DOI] [PubMed] [Google Scholar]
- 11.Matsubara, M., Nakatsu, T., Kato, H. & Taniguchi, H. (2004) EMBO J. 23, 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sallese, M., Iacovelli, L., Cumashi, A., Capobianco, L., Cuomo, L. & De Blasi, A. (2000) Biochim. Biophys. Acta 1498, 112–121. [DOI] [PubMed] [Google Scholar]
- 13.Klee, C. B., Ren, H. & Wang, X. (1998) J. Biol. Chem. 273, 13367–13370. [DOI] [PubMed] [Google Scholar]
- 14.Drum, C. L., Yan, S. Z., Bard, J., Shen, Y. Q., Lu, D., Soelaiman, S., Grabarek, Z., Bohm, A. & Tang, W. J. (2002) Nature 415, 396–402. [DOI] [PubMed] [Google Scholar]
- 15.Yap, K. L., Yuan, T., Mal, T. K., Vogel, H. J. & Ikura, M. (2003) J. Mol. Biol. 328, 193–204. [DOI] [PubMed] [Google Scholar]
- 16.Stuehr, D. J., Cho, H. J., Kwon, N. S., Weise, M. F. & Nathan, C. F. (1991) Proc. Natl. Acad. Sci. USA 88, 7773–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung, W. Y. (1970) Biochem. Biophys. Res. Commun. 38, 533–538. [DOI] [PubMed] [Google Scholar]
- 18.Kakiuchi, S. & Yamazaki, R. (1970) Biochem. Biophys. Res. Commun. 41, 1104–1110. [DOI] [PubMed] [Google Scholar]
- 19.Carafoli, E. (1997) Basic Res. Cardiol. 92, Suppl. 1, 59–61. [DOI] [PubMed] [Google Scholar]
- 20.Trudeau, M. C. & Zagotta, W. N. (2003) J. Biol. Chem. 278, 18705–18708. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher, M. A., Rivard, A. F., Bachinger, H. P. & Adelman, J. P. (2001) Nature 410, 1120–1124. [DOI] [PubMed] [Google Scholar]
- 22.Zuhlke, R. D., Pitt, G. S., Deisseroth, K., Tsien, R. W. & Reuter, H. (1999) Nature 399, 159–162. [DOI] [PubMed] [Google Scholar]
- 23.Michikawa, T., Hirota, J., Kawano, S., Hiraoka, M., Yamada, M., Furuichi, T. & Mikoshiba, K. (1999) Neuron 23, 799–808. [DOI] [PubMed] [Google Scholar]
- 24.Tang, W., Sencer, S. & Hamilton, S. L. (2002) Front. Biosci. 7, d1583–d1589. [DOI] [PubMed] [Google Scholar]
- 25.McKay, R. T., Pearlstone, J. R., Corson, D. C., Gagne, S. M., Smillie, L. B. & Sykes, B. D. (1998) Biochemistry 37, 12419–12430. [DOI] [PubMed] [Google Scholar]
- 26.Takeda, S., Yamashita, A., Maeda, K. & Maeda, Y. (2003) Nature 424, 35–41. [DOI] [PubMed] [Google Scholar]
- 27.Lindhout, D. A. & Sykes, B. D. (2003) J. Biol. Chem. 278, 27024–27034. [DOI] [PubMed] [Google Scholar]
- 28.Ruksana, R., Kuroda, K., Terami, H., Bando, T., Kitaoka, S., Takaya, T., Sakube, Y. & Kagawa, H. (2005) Genes Cells 10, 261–276. [DOI] [PubMed] [Google Scholar]
- 29.Hendricks, K. B., Wang, B. Q., Schnieders, E. A. & Thorner, J. (1999) Nat. Cell Biol. 1, 234–241. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, T. Y., Pountney, D. J., Ozaita, A., Nandi, S., Ueda, S., Rudy, B. & Coetzee, W. A. (2001) Proc. Natl. Acad. Sci. USA 98, 12808–12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss, J. L., Archer, D. A. & Burgoyne, R. D. (2000) J. Biol. Chem. 275, 40082–40087. [DOI] [PubMed] [Google Scholar]
- 32.Lin, L., Jeanclos, E. M., Treuil, M., Braunewell, K. H., Gundelfinger, E. D. & Anand, R. (2002) J. Biol. Chem. 277, 41872–41878. [DOI] [PubMed] [Google Scholar]
- 33.Makino, C. L., Dodd, R. L., Chen, J., Burns, M. E., Roca, A., Simon, M. I. & Baylor, D. A. (2004) J. Gen. Physiol. 123, 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An, W. F., Bowlby, M. R., Betty, M., Cao, J., Ling, H. P., Mendoza, G., Hinson, J. W., Mattsson, K. I., Strassle, B. W., Trimmer, J. S. & Rhodes, K. J. (2000) Nature 403, 553–556. [DOI] [PubMed] [Google Scholar]
- 35.Carrion, A. M., Link, W. A., Ledo, F., Mellstrom, B. & Naranjo, J. R. (1999) Nature 398, 80–84. [DOI] [PubMed] [Google Scholar]
- 36.Buxbaum, J. D., Choi, E. K., Luo, Y., Lilliehook, C., Crowley, A. C., Merriam, D. E. & Wasco, W. (1998) Nat. Med. 4, 1177–1181. [DOI] [PubMed] [Google Scholar]
- 37.Palczewski, K., Sokal, I. & Baehr, W. (2004) Biochem. Biophys. Res. Commun. 322, 1123–1130. [DOI] [PubMed] [Google Scholar]
- 38.Griffith, J. P., Kim, J. L., Kim, E. E., Sintchak, M. D., Thomson, J. A., Fitzgibbon, M. J., Fleming, M. A., Caron, P. R., Hsiao, K. & Navia, M. A. (1995) Cell 82, 507–522. [DOI] [PubMed] [Google Scholar]
- 39.Gentry, H. R., Singer, A. U., Betts, L., Yang, C., Ferrara, J. D., Sondek, J. & Parise, L. V. (2005) J. Biol. Chem. 280, 8407–8415. [DOI] [PubMed] [Google Scholar]
- 40.Stabler, S. M., Ostrowski, L. L., Janicki, S. M. & Monteiro, M. J. (1999) J. Cell Biol. 145, 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kasri, N. N., Holmes, A. M., Bultynck, G., Parys, J. B., Bootman, M. D., Rietdorf, K., Missiaen, L., McDonald, F., De Smedt, H., Conway, S. J., et al. (2004) EMBO J. 23, 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, A., Westernbroek, R. E., Haeseleer, F., Palczewski, K., Scheuer, T. & Catterall, W. A. (2002) Nat. Neurosci. 5, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki, R., Berri, M., Wu, Y., Trombitas, K., McNabb, M., Kellermayer, M. S., Witt, C., Labeit, D., Labeit, S., Greaser, M. & Granzier, H. (2001) Biophys. J. 81, 2297–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiewitz, R., Acklin, C., Schafer, B. W., Maco, B., Uhrik, F., Wuytack, P., Erne, C. W. & Heizmann, C. W. (2003) Biochem. Biophys. Res. Commun. 306, 550–557. [DOI] [PubMed] [Google Scholar]
- 45.Most, P., Remppis, A., Pleger, S. T., Loffler, E., Ehlermann, P., Bernotat, J., Kleuss, C., Heierhorst, J., Ruiz, P., Witt, H., et al. (2003) J. Biol. Chem. 278, 33809–33817. [DOI] [PubMed] [Google Scholar]
- 46.Endo, H., Takenaga, K., Kanno, T., Satoh, H. & Mori, S. (2002) J. Biol. Chem. 277, 26396–26402. [DOI] [PubMed] [Google Scholar]
- 47.Nowotny, M., Spiechowicz, M., Jastrzebska, B., Filipek, A., Kitagawa, K. & Kuznicki, J. (2003) J. Biol. Chem. 278, 26923–26928. [DOI] [PubMed] [Google Scholar]
- 48.Rety, S., Sopkova, J., Renouard, M., Osterloh, D., Gerke, V., Tabaries, S., Russo-Marie, F. & Lewit-Bentley, A. (1999) Nat. Struct. Biol. 6, 89–95. [DOI] [PubMed] [Google Scholar]
- 49.Rety, S., Osterloh, D., Arie, J. P., Tabaries, S., Seeman, J., Russo-Marie, F., Gerke, V. & Lewit-Bentley, A. (2000) Structure Fold Des. 8, 175–184. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann, M. A., Drury, S., Fu, C., Qu, W., Taguchi, A., Lu, Y., Avila, C., Kambham, N., Bierhaus, A., Nawroth, P., et al. (1999) Cell 97, 889–901. [DOI] [PubMed] [Google Scholar]
- 51.Rustandi, R. R., Baldisseri, D. M. & Weber, D. J. (2000) Nat. Struct. Biol. 7, 570–574. [DOI] [PubMed] [Google Scholar]
- 52.Bhattacharya, S. S., Large, E., Heizmann, C. W., Hemmings, B. & Chazin, W. J. (2003) Biochemistry 42, 14416–14426. [DOI] [PubMed] [Google Scholar]
- 53.Kilby, P. M., Van Eldik, L. J. & Roberts, G. C. (1997) Protein Sci. 6, 2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitaramayya, A. (2002) Adv. Exp. Med. Biol. 514, 389–398. [DOI] [PubMed] [Google Scholar]
- 55.Lollike, K., Johnson, A. H., Durussel, I., Borregaard, N. & Cox, J. A. (2001) J. Biol. Chem. 276, 17762–17769. [DOI] [PubMed] [Google Scholar]
- 56.Valdivia, H. H. (1998) Trends Pharmacol. Sci. 19, 479–482. [DOI] [PubMed] [Google Scholar]
- 57.Vito, P., Pellegrini, L., Guiet, C. & D'Adamio, L. (1999) J. Biol. Chem. 274, 1533–1540. [DOI] [PubMed] [Google Scholar]
- 58.Satoh, H., Nakano, Y., Shibata, H. & Maki, M. (2002) Biochim. Biophys. Acta. 1600, 61–67. [DOI] [PubMed] [Google Scholar]
- 59.Yang, H. Q., Ma, H., Takano, E., Hatanaka, M. & Maki, M. (1994) J. Biol. Chem. 269, 18977–18984. [PubMed] [Google Scholar]
- 60.Carafoli, E., Santella, L., Branca, D. & Brini, M. (2001) Crit. Rev. Biochem. Mol. Biol. 36, 107–260. [DOI] [PubMed] [Google Scholar]
- 61.Copley, R. R., Schultz, J., Ponting, C. P. & Bork, P. (1999) Curr. Opin. Struct. Biol. 9, 408–415. [DOI] [PubMed] [Google Scholar]
- 62.Ikura, M. (1996) Trends Biochem. Sci. 21, 14–17. [PubMed] [Google Scholar]
- 63.Bhattacharya, S. S., Bunick, C. G. & Chazin, W. J. (2004) Biochim. Biophys. Acta. 1742, 69–79. [DOI] [PubMed] [Google Scholar]
- 64.Burgoyne, R. D., O'Callaghan, D. W., Hasdemir, B., Haynes, L. P. & Tepikin, A. V. (2004) Trends Neurosci. 27, 203–209. [DOI] [PubMed] [Google Scholar]
- 65.Ames, J. B., Tanaka, T., Stryer, L. & Ikura, M. (1996) Curr. Opin. Struct. Biol. 6, 432–438. [DOI] [PubMed] [Google Scholar]
- 66.Marenholz, I., Heizmann, C. W. & Fritz, G. (2004) Biochem. Biophys. Res. Commun. 322, 1111–1122. [DOI] [PubMed] [Google Scholar]
- 67.Weiss, J. L. & Burgoyne, R. D. (2002) in Handbook of Cell Signaling, ed. Bradshaw, R. (Academic, San Diego), Vol. 2, pp. 79–82. [Google Scholar]
- 68.Zozulya, S. & Stryer, L. (1992) Proc. Natl. Acad. Sci. USA 89, 11569–11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ames, J. B., Ishima, R., Tanaka, T., Gordon, J. I., Stryer, L. & Ikura, M. (1997) Nature 389, 198–202. [DOI] [PubMed] [Google Scholar]
- 70.Chen, C. K., Inglese, J., Lefkowitz, R. J. & Hurley, J. B. (1995) J. Biol. Chem. 270, 18060–18066. [DOI] [PubMed] [Google Scholar]
- 71.Polans, A. S., Buczylko, J., Crabb, J. & Palczewski, K. (1991) J. Cell. Biol. 112, 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hidaka, H. & Okazaki, K. (1993) Neurosci. Res. 16, 73–77. [DOI] [PubMed] [Google Scholar]
- 73.Pongs, O., Lindemeier, J., Zhu, X. R., Theil, T., Engelkamp, D., Krah-Jentgens, Lambrecht, H. G., Kock, K. W., Schwerner, J., Rivosecchi, R., et al. (1993) Neuron 11, 15–28. [DOI] [PubMed] [Google Scholar]
- 74.Kobayashi, M., Takamatsu, K., Saitoh, S., Miura, M. & Noguchi, T. (1992) Biochem. Biophys. Res. Commun. 189, 511–517. [DOI] [PubMed] [Google Scholar]
- 75.Gomez, M., De Castro, E., Guarin, E., Sasakura, H., Kuhara, A., Mori, I., Bartfai, T., Bargmann, C. I. & Nef, P. (2001) Neuron 30, 241–248. [DOI] [PubMed] [Google Scholar]
- 76.Hamasaki-Katagiri, N., Molchanova, T., Takeda, K. & Ames, J. B. (2004) J. Biol. Chem. 279, 12744–12754. [DOI] [PubMed] [Google Scholar]
- 77.Kapp, Y., Melnikov, S., Shefler, A., Jeromin, A. & Sagi, R. (2003) J. Immunol. 171, 5320–5327. [DOI] [PubMed] [Google Scholar]
- 78.Hama, H., Schnieders, E. A., Thorner, J., Takemoto, J. Y. & DeWald, D. B. (1999) J. Biol. Chem. 274, 34294–34300. [DOI] [PubMed] [Google Scholar]
- 79.Cheng, H. Y., Pitcher, G. M., Laviolette, S. R., Whishaw, I. Q., Tong, K. I., Ikura, M., Salter, M. W. & Penninger, J. M. (2002) Cell 108, 31–43. [DOI] [PubMed] [Google Scholar]
- 80.Lilliehook, C., Bozdagi, O., Yao, J., Gomez-Ramirez, M., Zaidi, N. F., Wasco, W., Gandy, S., Santucci, A. C., Haroutunian, V., Huntley, G. W. & Buxbaum, J. D. (2003) J. Neurosci. 23, 9097–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du, X. J., Cole, T. J., Tenis, N., Gao, X. M., Kontgen, F., Kemp, B. E. & Heierhorst, J. (2002) Mol. Cell Biol. 22, 2821–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wicki, R., Franz, C., Scholl, F. A., Heizmann, C. W. & Schafer, B. W. (1997) Cell Calcium 22, 243–254. [DOI] [PubMed] [Google Scholar]
- 83.Schmidt, B., Klingelhofer, J., Grum-Schwensen, B., Christensen, A., Andresen, S., Kruse, C., Hansen, T., Ambartsumian, N., Lukanidin, E. & Grigorian, M. (2004) J. Biol. Chem. 279, 24498–24504. [DOI] [PubMed] [Google Scholar]
- 84.Broome, A. M., Ryan, D. & Eckert, R. L. (2003) J. Histochem. Cytochem. 51, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newton, R. A. & Hogg, N. (1998) J. Immunol. 160, 1427–1435. [PubMed] [Google Scholar]
- 86.Duda, T., Koch, K. W., Venkataraman, V., Lange, C., Beyermann, M. & Sharma, R. K. (2002) EMBO J. 21, 2547–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flaherty, K. M., Zozulya, S., Stryer, L. & McKay, D. B. (1993) Cell 75, 709–716. [DOI] [PubMed] [Google Scholar]
- 88.Vijay-Kumar, S. & Kumar, V. D. (1999) Nat. Struct. Biol. 6, 80–88. [DOI] [PubMed] [Google Scholar]
- 89.Bourne, Y., Dannenberg, J., Pollmann, V. V., Marchot, P. & Pongs, O. (2001) J. Biol. Chem. 276, 11949–11955. [DOI] [PubMed] [Google Scholar]
- 90.Babu, Y. S., Bugg, C. E. & Cook, W. J. (1988) J. Mol. Biol. 204, 191–204. [DOI] [PubMed] [Google Scholar]
- 91.Herzberg, O. & James, M. N. (1988) J. Mol. Biol. 203, 761–779. [DOI] [PubMed] [Google Scholar]
- 92.Potts, B. C., Smith, J., Akke, M., Macke, T. J., Okazaki, K., Hidaka, H., Case, D. A. & Chazin, W. J. (1995) Nat. Struct. Biol. 2, 790–796. [DOI] [PubMed] [Google Scholar]
- 93.Drohat, A. C., Baldisseri, D. M., Rustandi, R. R. & Weber, D. J. (1998) Biochemistry 37, 2729–2740. [DOI] [PubMed] [Google Scholar]
- 94.Wilder, P. T., Varney, K. M., Weiss, M. B., Gitti, R. K. & Weber, D. J. (2005) Biochemistry 44, 5690–5702. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka, T., Ames, J. B., Harvey, T. S., Stryer, L. & Ikura, M. (1995) Nature 376, 444–447. [DOI] [PubMed] [Google Scholar]
- 96.Ames, J. B., Hamasaki, N. & Molchanova, T. (2002) Biochemistry 41, 5776–5787. [DOI] [PubMed] [Google Scholar]
- 97.Ermilov, A. N., Olshevskaya, E. V. & Dizhoor, A. M. (2001) J. Biol. Chem. 276, 48143–48148. [DOI] [PubMed] [Google Scholar]
- 98.Krylov, D. M., Niemi, G. A., Dizhoor, A. M. & Hurley, J. B. (1999) J. Biol. Chem. 274, 10833–10839. [DOI] [PubMed] [Google Scholar]
- 99.Tachibanaki, S., Nanda, K., Sasaki, K., Ozaki, K. & Kawamura, S. (2000) J. Biol. Chem. 275, 3313–3319. [DOI] [PubMed] [Google Scholar]
- 100.Olshevskaya, E. V., Boikov, S., Ermilov, A., Krylov, D., Hurley, J. B. & Dizhoor, A. M. (1999) J. Biol. Chem. 274, 10823–10832. [DOI] [PubMed] [Google Scholar]
- 101.Zhou, W., Qian, Y., Kunjilwar, K., Pfaffinger, P. J. & Choe, S. (2004) Neuron 41, 573–586. [DOI] [PubMed] [Google Scholar]
- 102.Gopinath, R. M. & Vincenzi, F. F. (1977) Biochem. Biophys. Res. Commun. 77, 1203–1209. [DOI] [PubMed] [Google Scholar]
- 103.Jarrett, H. W. & Penniston, J. T. (1977) Biochem. Biophys. Res. Commun. 77, 1210–1216. [DOI] [PubMed] [Google Scholar]
- 104.Hook, S. S. & Means, A. R. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 471–505. [DOI] [PubMed] [Google Scholar]
- 105.Yap, K. L., Kim, J., Truong, K., Sherman, M., Yuan, T. & Ikura, M. (2000) 1, 8–14. [DOI] [PubMed]
- 106.Collier, R. J. & Young, J. A. (2003) 19, 45–70. [DOI] [PubMed]
- 107.Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu, G., Klee, C. B. & Bax, A. (1992) Science 256, 632–638. [DOI] [PubMed] [Google Scholar]
- 108.Meador, W. E., Means, A. R. & Quiocho, F. A. (1992) Science 257, 1251–1255. [DOI] [PubMed] [Google Scholar]
- 109.Persechini, A. & Kretsinger, R. H. (1988) J. Biol. Chem. 263, 12175–12178. [PubMed] [Google Scholar]
- 110.Trewhella, J. (1992) Cell Calcium 13, 377–390. [DOI] [PubMed] [Google Scholar]
- 111.Barbato, G., Ikura, M., Kay, L. E., Pastor, R. W. & Bax, A. (1992) Biochemistry 31, 5269–5278. [DOI] [PubMed] [Google Scholar]
- 112.Zhang, M., Li, M., Wang, J. H. & Vogel, H. J. (1994) J. Biol. Chem. 269, 15546–15552. [PubMed] [Google Scholar]
- 113.Meador, W. E., Means, A. R. & Quiocho, F. A. (1993) Science 262, 1718–1721. [DOI] [PubMed] [Google Scholar]
- 114.Osawa, M., Tokumitsu, H., Swindells, M. B., Kurihara, H., Furuya, T. & Ikura, M. (1999) Nat. Struct. Biol. 6, 819–824. [DOI] [PubMed] [Google Scholar]
- 115.Aoyagi, M., Arvai, A. S., Tainer, J. A. & Getzoff, E. D. (2003) EMBO J. 22, 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamauchi, E., Nakatsu, T., Matsubara, M., Kato, H. & Taniguchi, H. (2003) Nat. Struct. Biol. 10, 226–231. [DOI] [PubMed] [Google Scholar]
- 117.Kortvely, E. & Gulya, K. (2004) 74, 1065–1070. [DOI] [PubMed]
- 118.Palfi, A., Kortvely, E., Fekete, E., Kovacs, B., Varszegi, S. & Gulya, K. (2002) Life Sci. 70, 2829–2855. [DOI] [PubMed] [Google Scholar]
- 119.Persechini, A. & Stemmer, P. M. (2002) Trends Cardiovasc. Med. 12, 32–37. [DOI] [PubMed] [Google Scholar]
- 120.Benaim, G. & Villalobo, A. (2002) Eur. J. Biochem. 269, 3619–3631. [DOI] [PubMed] [Google Scholar]
- 121.Murtaugh, T. J., Rowe, P. M., Vincent, P. L., Wright, L. S. & Siegel, F. L. (1983) Methods Enzymol. 102, 158–170. [DOI] [PubMed] [Google Scholar]
- 122.Murtaugh, T. J., Wright, L. S. & Siegel, F. L. (1986) J. Neurochem. 47, 164–172. [DOI] [PubMed] [Google Scholar]
- 123.Vermassen, E., Parys, J. B. & Mauger, J. P. (2004) Biol. Cell 96, 3–17. [DOI] [PubMed] [Google Scholar]
- 124.Iwai, M., Tateishi, Y., Hattori, M., Mizutani, A., Nakamura, T., Futatsugi, A., Inoue, T., Furuichi, T., Michikawa, T. & Mikoshiba, K. (2005) J. Biol. Chem. 280, 10305–10317. [DOI] [PubMed] [Google Scholar]
- 125.Dizhoor, A. M., Ray, S., Kumar, S. Niemi, G., Spencer, M., Brolley, D., Walsh, K. A., Philipov, P. P., Hurley, J. B. & Stryer, L. (1991) Science 251, 915–918. [DOI] [PubMed] [Google Scholar]
- 126.Palczewski, K., Subbaraya, I., Gorczyca, W. A., Helekar, B. S., Ruiz, C. C., Ohguro, H., Huang, J., Zhao, X., Crabb, J. W. & Johnson, R. S. (1994) Neuron 13, 395–404. [DOI] [PubMed] [Google Scholar]
- 127.Paterlini, M., Revilla, V., Grant, A. L. & Wisden, W. (2000) Neuroscience 99, 205–216. [DOI] [PubMed] [Google Scholar]
- 128.Hammond, P. I., Craing, T. A., Kumar, R. & Brimijoin, S. (2003) Brain Res. Mol. Brain Res. 111, 104–110. [DOI] [PubMed] [Google Scholar]
- 129.Kuo, H. C., Cheng, C. F., Clark, R. B., Lin, J. J., Lin, J. L., Hoshijima, M., Nguyen-Tran, V. T., Gu, Y., Ikeda, Y., Chu, P. H., et al. (2001) J. Biol. Chem. 278, 36445–36454. [Google Scholar]
- 130.Pawson, T. (2004) Cell 116, 191–203. [DOI] [PubMed] [Google Scholar]
- 131.Pawson, T. & Scott, J. D. (2005) Trends Biochem. Sci. 30, 286–290. [DOI] [PubMed] [Google Scholar]