Abstract
Knowledge of protein structure is essential to understand protein function. High-resolution protein structure has so far been the domain of ensemble methods. Here, we develop a simple single-molecule technique to measure spatial position of selected residues within a folded and functional protein structure in solution. Construction and mechanical unfolding of cysteine-engineered polyproteins with controlled linkage topology allows measuring intramolecular distance with angstrom precision. We demonstrate the potential of this technique by determining the position of three residues in the structure of green fluorescent protein (GFP). Our results perfectly agree with the GFP crystal structure. Mechanical triangulation can find many applications where current bulk structural methods fail.
Keywords: mechanical protein unfolding, protein stability, single molecule force spectroscopy
Determination of high-resolution protein structure has so far been the domain of ensemble methods. X-ray crystallography, although providing complete and angstrom-precise structural information (1), can only provide static pictures of a crystallized protein far from its native environment. NMR spectroscopy allows determination of protein structure in solution with atomic resolution but is still molecular-weight limited (2, 3). Still, a wide range of proteins is accessible by neither x-ray crystallography nor NMR because of insolubility, aggregation, and/or crystallization problems (4). High-resolution electron microscopy bridges the gap to supramolecular structures (5). Apart from protein structure with atomic resolution, fluorescence resonance energy transfer (FRET) (6), electron paramagnetic resonance (EPR) (7), and small-angle scattering (8), among other techniques, are widely used to obtain dynamic information about proteins at work. Yet these techniques generally provide only relative structural information with less molecular resolution. There is great need for novel assays and techniques able to report absolute and precise information about positions and intramolecular distances in a folded and functioning protein structure.
Here, we develop a simple and direct single-molecule technique that provides detailed and angstrom-precise information about the structure of a folded and functioning protein in solution. We use single-molecule force spectroscopy combined with cysteine-engineered polyproteins to determine the position of three residues within the structure of green fluorescent protein (GFP).
Results
Mechanical Triangulation: Principle. We consider a protein of unknown structure but known amino acid sequence in its native, folded conformation (compare Fig. 1a). In a first step, we focus on the folded pair distance di,j between amino acids i and j (usually of the order of several angstroms). Such distances cannot be measured directly because there is a lack of applicable scale bars or calipers. However, the linear protein sequence itself can provide the necessary scale bar. Consider that the protein could be grabbed exactly at amino acids i and j and then forced into a completely stretched conformation (Fig. 1a). The length gain ΔLi,j needed to stretch out the amino acid chain from its folded to the completely unfolded conformation is experimentally accessible in single-molecule mechanical experiments to angstrom precision (9, 10). Such length gains ΔLi,j due to mechanically induced protein unfolding and stretching are usually of the order of several nanometers. The total length of a stretched polypeptide chain Li,j is exactly predetermined. This stretched length Li,j is given by multiplying the number of amino acids between grabbing points by the length of a single stretched amino acid daa (Fig. 1a). Because in the folded conformation the two amino acids i and j already have a finite distance di,j from each other, the measured length gain ΔLi,j will be always smaller than the complete length Li,j. Hence
 |
[1] |
Fig. 1.
Principle of mechanical triangulation. (a) Grabbing a folded protein of unknown structure but known sequence exactly at amino acids i and j and forcing it into a completely stretched conformation. The folded distance di, j is given by the difference between predetermined length Li, j of the stretched amino acid chain and the recorded length gain ΔLi, j during transition. (b) Subsequent determination of pair distances dn, j and dn,i to obtain all pair distances for a certain amino acid triple i, j, and n. (c) The such-obtained pair distances allow reconstruction of the absolute spatial positions of the triangulated amino acids i, j, and n. Reconstruction of a detailed three-dimensional protein structure is straightforward by triangulating a sufficient number of pair distances.
If all distances between a set of at least three amino acids (i, j, n) are determined, triangulation known from elementary geometry can now be applied to unequivocally determine the spatial positions of those amino acids (see Fig. 1 b and c).
Cysteine-Linked Polyproteins. A currently widely used strategy for mechanical single-molecule experiments is to use polyproteins containing identical protein domains. This technique ensures that single-molecule events can be identified by a characteristic repetitive sawtooth pattern (11) due to the unfolding of the individual domains in the chain. However, the subunits of the great majority of polyproteins investigated so far were either naturally or genetically linked by their termini (12–14) because of the natural direction of translation in the ribosome. However, for our proposed experiment, control over the linkage points in polyproteins is crucial. In this study, we use pairwise introduction of cysteines into the sequence of an individual protein domain and subsequent polymerization to create almost arbitrarily linked polyproteins (see Materials and Methods). Cysteine polymerization has been successfully applied by Yang et al. (15) in a protein crystal. This approach, however, is limited to only one linkage geometry and thus is not viable for our purposes. Here, we demonstrate that arbitrary linkage can be achieved in solution.
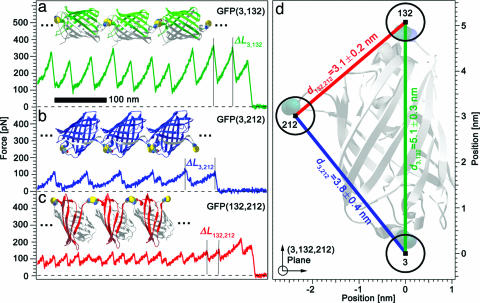
Mechanical Triangulation of GFP. To demonstrate the potential of mechanical triangulation, we chose to determine the position of amino acids 3, 132, and 212 in GFP. We engineered the three possible pairs of cysteine mutants for residues 3, 132, and 212 in GFP sequence. We polymerized the GFP variants to polyproteins that were covalently linked via disulfide bridges between the two cysteine residues. The three different GFP polyprotein chains [GFP(3, 132), GFP(3, 212), and GFP(132, 212)] are shown in Fig. 2 a–c. All mutants showed the typical bright GFP fluorescence, hence indicating native and functioning protein structure (16).
Fig. 2.
Mechanical triangulation of GFP. (a–c) Force-extension traces of single GFP(3, 132), GFP(3, 212), and GFP(132, 212) polyproteins (green, blue, and red solid lines, respectively). The gain in length ΔLi, j between peaks reflects unfolding and subsequent stretching of the number of amino acids located between linkage points in each module (colored in green, blue, and red in the structures). ΔLi, j was determined for the three polyproteins: ΔL3,132 = 41.6 ± 0.04 nm (n = 524), ΔL3,212 = 72.08 ± 0.03 (n = 500), and ΔL132,212 = 26.06 ± 0.05 nm (n = 500). (d) Intramolecular pair distances d3,132, d3,212, and d132,212 and absolute positions of residues 3, 132, and 212 in the folded GFP structure as determined from our data. Circles indicate total errors. Light gray, GFP crystal structure (PDB ID code 1EMB) (17). Backbone atoms of residues 3, 132, and 212 are shown space filling.
We recorded the force-extension response of the three differently linked GFP polyprotein chains using an atomic force microscope (see Materials and Methods). Typical traces are shown in Fig. 2 a–c for each of the GFP polyproteins (for additional data, see Fig. 4, which is published as supporting information on the PNAS web site). In the traces of Fig. 2 a–c, each peak corresponds to the forced transition of one GFP module from the folded to the completely unfolded and stretched conformation. The distance between peaks (marked ΔLi,j) in Fig. 2 a–c corresponds to the gain in length upon the transition from a folded state to the completely stretched state. We measured this length gain ΔLi,j for each GFP polyprotein as described in Materials and Methods. It is important to note that the resolution of measuring ΔLi,j in a single unfolding peak is not better than ≈2 nm. For all linkages shown, we have a statistics of N ≥ 500. Because the ΔLi,j values are normally distributed around their mean, the error of the mean is then <1 Å (see Materials and Methods). Application of Eq. 1 immediately yields the three pair distances between residues 3, 132, and 212 in the folded GFP state and allows assignment of spatial coordinates to the three amino acids in the thereby constituted plane. The result is shown in Fig. 2d.
For comparison, Fig. 2d displays additionally the high-resolution crystal structure of GFP obtained from x-ray diffraction experiments [Protein Data Bank (PDB) ID code 1EMB] (17). The crystal structure is oriented such that the carbon α atoms (displayed space-filling) in the backbone of amino acids 3, 132, and 212 fall in the figure plane. The result from mechanical triangulation almost perfectly matches the GFP crystal structure.
Discussion
To determine the absolute position of an amino acid in space using mechanical triangulation, only three measured pair distances are necessary. A more detailed structure of amino acid backbone positions by increasing the number of cysteine pair mutants will be straightforward. It will probably be not practicable to reconstruct full atomic structures by mechanical triangulation. However, for each additional position of a residue only three additional pair mutants are necessary. Therefore, up to five or six positions within a protein structure can certainly be monitored with angstrom resolution. Because solution conditions can be changed easily during a force experiment, also structural changes of enzymes can be monitored. In addition, mechanical triangulation is free from orientational issues like FRET or EPR.
Our method relies on precise knowledge of the length of an unfolded amino acid residue. Therefore we used two different calibration proteins with known high-resolution structure (DdFLN, PDB ID code 1WLH, and Ig27, PDB ID code 1TIT) to calibrate the factor daa in Eq. 1 (see Materials and Methods). The obtained factor was daa = 0.365 nm for both proteins. Such an independent agreement is especially important because a priori we cannot exclude that some amino acids may be unfolded already at low forces as has been observed with the N–C-terminally linked domain Ig27 (13).
Attention has to be paid to the fact that the linking amino acids may be contained in a part of the protein that undergoes a partial unfolding transition before the major unfolding event. In this case, Eq. 1 would be no longer valid because the measured length gain ΔLi,j would correspond to a smaller structure than the fully folded. In such a case partial unfolding of the protein may be prevented by changing the pulling direction: in an earlier study we reported that in the N–C-terminally linked GFP the N-terminal α-helix comprising amino acids 1–10 detaches at low forces (14) visible in a small hump at forces of ≈35 pN. In two of our cysteine mutants [GFP(3, 132) and GFP(3, 212)], one linkage point is amino acid 3 which is located in exactly this N-terminal α-helix. Unexpectedly, these mutants where force is applied perpendicularly to the α-helix did not show any signs of premature unfolding (see Figs. 2 and 4). Thus, the determined position of amino acid 3 matches consistently the corresponding position in the completely folded GFP crystal structure (Fig. 2b).
It is also important to note that the structure we determine by mechanical triangulation could be deformed by the load we apply. Such deformations can be estimated from protein stiffness data (18, 19) and will be generally very small (<1 Å at the typical loads). In turn, deviations of triangulation results from a known crystal structure can give valuable information about the elastic properties of certain parts within a folded protein.
An important aspect of the cysteine polymerization method presented here concerns the physics of protein stability. Earlier studies have indicated that changing the direction of load application may lower unfolding forces as compared with the N–C-terminal linkage (20–22). Surprisingly, all three GFP polyprotein mutants in Fig. 2 readily show equal or higher forces (up to three times) than the N–C-terminally linked GFP. The mutant GFP(3, 212) exhibits a previously undescribed metastable intermediate state absent in all other pulling geometries. We suggest that systematic variation of linkage geometry will open new possibilities of exploring the highly dimensional energy landscapes of proteins in an unprecedented way.
Conclusion
We demonstrated that detailed and absolute structural information can be obtained from a single protein molecule using mechanical triangulation. Our results demonstrate that angstrom-precise structural information can be obtained from single protein molecules in solution.
One key issue in protein folding is still the need for novel assays and techniques to determine the structure of insoluble folding intermediates and misfolded proteins (23, 24). We are convinced that mechanical triangulation can contribute significantly in addressing such problems.
Materials and Methods
Construction of GFP Polyproteins. Pairwise point mutation of wild-type GFP residues Lys-3–Cys, Glu-132–Cys, and Asn-212–Cys as well as wild-type Ig27 residues Glu-3–Cys and Glu-88–Cys from human cardiac titin was performed by using the QuikChange multisite-directed mutagenesis kit (Stratagene). Purification of the His6-tagged proteins was performed by using Ni-NTA affinity chromatography at 4°C. All GFP cysteine mutants showed the typical bright GFP fluorescence, hence indicating the presence of the native and functioning GFP structure. Polymerization of GFP pair-cysteine mutants was performed in PBS buffer (pH 7.4) at high protein concentrations of ≈5 mg/ml (≈0.2 mM). After 80 h of sample incubation at 37°C, sawtooth patterns in force spectroscopy experiments confirmed the presence of long protein polymers. Samples then were stored for several days at 4°C to quench further polymerization. The simple disulfide bond polymerization strategy cannot control the orientation of the monomers within the polymer. However, polarity of inclusion does not affect the length measurements.
Single-Protein Force Spectroscopy. All single-molecule force measurements were performed on a custom-built atomic force microscope. Gold-coated cantilevers (BioLevers, Olympus, Tokyo) with spring constant and resonance frequency of 30 pN/nm and 8.5 kHz (type A) were used. For the measurements, the above-described protein solutions were centrifuged for 15 min at maximum speed (15,000 × g) in a tabletop centrifuge to spin down potential larger protein aggregates. Without any further treatment, ≈10 μl of the corresponding supernatant were then applied on a clean glass surface and incubated for 60 min at room temperature. All force curves were collected at pulling speeds of 3.6 μm/s. All experiments were conducted at room temperature. See Fig. 3a for a scheme of the experimental setup.
Fig. 3.
Instrumentation schematics. (a) Schematics of an atomic force microscope. (b) Blue solid line represents a sample force-extension trace obtained on a polyprotein consisting of Ig27 domains from human cardiac titin that are covalently linked by cysteines at positions 3 and 88 in protein sequence. We used PolyIg27(3, 88) for calibration of our system (see Materials and Methods). The length gain ΔL3,88 is measured by fitting the worm-like chain model to the data, represented by black solid lines.
Measurement of Contour Lengths. For quantitative analysis of the length gain ΔLi,j due to unfolding of a cysteine-linked GFP module the force-extension traces were fit to the interpolation formula of the worm-like chain model, F(x) = (kBT/p)[0.25(1 – x/L)–2 – 0.25 + x/L] as introduced by Bustamante et al. (25). L denotes the contour length of the stretched protein, p is the persistence length, kB is Boltzmann's constant, T is the temperature in kelvin, and x is the distance between attachment points of the protein (extension or end-to-end distance). A value of p = 0.5 nm for the persistence length p was used for fitting the data collected on GFP(3, 212) and GFP(132, 212) polyproteins in the force regime 50–150 pN. A value of p = 0.35 nm was used for fitting the data collected on GFP(3, 132) polyproteins in the higher force regime 150–300 pN. To compare measurements at different persistence lengths, data needs to be corrected by a correction factor γ to account for the deviations of a real polypeptide chain from ideal worm-like chain elasticity. We determined L0.5 = γ·L0.35. The factor γ was obtained by comparing fits with p = 0.5 nm in the range 50–150 pN and fits with p = 0.35 nm in the range 150–300 pN to 70 force-extension traces pulled on polypeptide chains. We find γ = 0.966 ± 0.0009. The average length increase ΔLi,j at a persistence length p = 0.5 nm for each unfolding of a single cysteine-linked GFP module is 〈ΔL3,132 〉 = 41.6 ± 0.04 nm (N = 524), 〈ΔL3,212 〉 = 72.08 ± 0.03 (N = 500), and 〈ΔL 132,212 〉 = 26.06 ± 0.05 nm (N = 500). To facilitate data analysis, we exploited an advantage inherent in sawtooth pattern curves from polyproteins. To determine the distance between peaks, it suffices to fit the distance between the first and the last peak in a sawtooth pattern and divide this distance by the number of peaks in between (see Fig. 3b). This process is equivalent to averaging. Errors in the text are errors of the mean value as given by standard deviation divided by the square root of the number of events (N). All fits and calculations were performed with igor pro 4.01 (WaveMetrics, Lake Oswego, OR).
Calibration Factor daa. To calculate folded distances using Eq. 1, exact knowledge of the calibration factor daa is a crucial prerequisite. daa is the contour length of a single amino acid residue in the worm-like chain model used for our analysis. For this study, we decided to use a simple experimental approach for calibration. To this end, we used two different proteins of known structure. First, we used Ig-like domains of the actin crosslinker section DdFLN(1–5) from dictyostelium discoideum, containing exactly 100 aa per domain (26). The average contour length increase 〈ΔL1,100 〉 due the unfolding of a single DdFLN domain is 〈ΔL1,100 〉 = 32.5 ± 0.1 nm (14, 27). Taking into account an N–C-terminal distance d1,100 of 4 ± 0.1 nm of the folded domain from the structure (PDB ID code 1WLH) (28) we arrive at a total unfolded contour length of L1,100 = 36.5 ± 0.2 nm. By using Eq. 1, we derive a length daa of 0.365 ± 0.002 nm per unfolded and stretched amino acid residue from the measurements with DdFLN domains. To confirm this value, we constructed polyproteins of the domain Ig27 from human cardiac titin that are linked by residues 3 and 88 using the polymerization strategy described above. The average length gain when mechanically unfolding single Ig27 domains linked by residues 3 and 88 is 〈ΔL3,88 〉 = 27.62 ± 0.04 nm (N = 379) (compare Fig. 3b). The folded distance between carbon-α atoms of amino acids 3 and 88 in the Ig27 structure (PDB ID code 1TIT) is d3,88 = 3.52 ± 0.1 nm (29). Applying Eq. 1 yields then daa = 0.366 ± 0.002 nm. The calibration factors daa obtained by using two different proteins and their solution NMR structures (PDB ID codes 1TIT and 1WLH) are therefore in very good agreement and applied here for mechanical triangulation of GFP.
Calculation of Folded Distances d. The distance di,j between linking residues in the folded state of a GFP module was calculated by using Eq. 1. For the mutants GFP(3, 132) and GFP(3, 212), the chromophore of GFP has to be taken into account because it is located between linkage points. The chromophore, formed by cyclization of amino acids Ser-65, Tyr-66, and Gly-67, accounts only for a backbone length equivalent to 2 instead of 3 aa (16). Thus, the folded distance di,j for GFP(3, 132) and GFP(3, 212) was calculated by using di,j = (j – i – 1)·daa – ΔLi,j.
Supplementary Material
Acknowledgments
We thank Hermann E. Gaub for useful comments on the manuscript and Claudia Schmidt and T. A. Bornschlögl for technical assistance. This work was supported by a Deutsche Forschungsgemeinschaft SFB413 grant.
Author contributions: H.D. and M.R. designed research, performed research, analyzed data, and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PDB, Protein Data Bank.
References
- 1.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 2.Wider, G. & Wuthrich, K. (1999) Curr. Opin. Struct. Biol. 9, 594–601. [DOI] [PubMed] [Google Scholar]
- 3.Ishima, R. & Torchia, D. A. (2000) Nat. Struct. Biol. 7, 740–743. [DOI] [PubMed] [Google Scholar]
- 4.Liu, H. L. & Hsu, J. P. (2005) Proteomics 5, 2056–2068. [DOI] [PubMed] [Google Scholar]
- 5.Kurner, J., Frangakis, A. S. & Baumeister, W. (2005) Science 307, 436–438. [DOI] [PubMed] [Google Scholar]
- 6.Ha, T. (2001) Methods 25, 78–86. [DOI] [PubMed] [Google Scholar]
- 7.Hubbell, W. L., Cafiso, D. S. & Altenbach, C. (2000) Nat. Struct. Biol. 7, 735–739. [DOI] [PubMed] [Google Scholar]
- 8.Doniach, S. (2001) Chem. Rev. 101, 1763–1778. [DOI] [PubMed] [Google Scholar]
- 9.Carrion-Vazquez, M., Marszalek, P. E., Oberhauser, A. F. & Fernandez, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11288–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rief, M., Gautel, M., Schemmel, A. & Gaub, H. E. (1998) Biophys. J. 75, 3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, T. E., Marszalek, P. E. & Fernandez, J. M. (2000) Nat. Struct. Biol. 7, 719–724. [DOI] [PubMed] [Google Scholar]
- 12.Rief, M., M. Gautel, F. Oesterhelt, Fernandez, J. M. & Gaub, H. E. (1997) Science 276, 1109–1112. [DOI] [PubMed] [Google Scholar]
- 13.Marszalek, P. E., Lu, H., Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Schulten, K. & Fernandez, J. M. (1999) Nature 402, 100–103. [DOI] [PubMed] [Google Scholar]
- 14.Dietz, H. & Rief, M. (2004) Proc. Natl. Acad. Sci. USA 101, 16192–16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang, G., Cecconi, C., Baase, W. A., Vetter, I. R., Breyer, W. A., Haack, J. A., Matthews, B. W., Dahlquist, F. W. & Bustamante, C. (2000) Proc. Natl. Acad. Sci. USA 97, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmer, M. (2002) Chem. Rev. 102, 759–781. [DOI] [PubMed] [Google Scholar]
- 17.Brejc, K., Sixma, T. K., Kitts, P. A., Kain, S. R., Tsien, R. Y., Ormo, M. & Remington, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 2306–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaccai, G. (2000) Science 288, 1604–1607. [DOI] [PubMed] [Google Scholar]
- 19.Schlierf, M. & Rief, M. (2005) J. Mol. Biol. 354, 497–503. [DOI] [PubMed] [Google Scholar]
- 20.Matouschek, A. & Bustamante, C. (2003) Nat. Struct. Biol. 10, 674–676. [DOI] [PubMed] [Google Scholar]
- 21.Brockwell, D. J., Paci, E., Zinober, R. C., Beddard, G. S., Olmsted, P. D., Smith, D. A., Perham, R. N. & Radford, S. E. (2003) Nat. Struct. Biol. 10, 731–737. [DOI] [PubMed] [Google Scholar]
- 22.Carrion-Vazquez, M., Li, H., Lu, H., Marszalek, P. E., Oberhauser, A. F. & Fernandez, J. M. (2003) Nat. Struct. Biol. 10, 738–743. [DOI] [PubMed] [Google Scholar]
- 23.Weissmann, C. (2004) Nat. Rev. Microbiol. 2, 861–871. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan, R. & Lindquist, S. L. (2005) Nature 435, 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. (1994) Science 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- 26.Fucini, P., Koppel, B., Schleicher, M., Lustig, A., Holak, T. A., Muller, R., Stewart, M. & Noegel, A. A. (1999) J. Mol. Biol. 291, 1017–1023. [DOI] [PubMed] [Google Scholar]
- 27.Schwaiger, I., Kardinal, A., Schleicher, M., Noegel, A. A. & Rief, M. (2004) Nat. Struct. Mol. Biol. 11, 81–85. [DOI] [PubMed] [Google Scholar]
- 28.Popowicz, G. M., Muller, R., Noegel, A. A., Schleicher, M., Huber, R. & Holak, T. A. (2004) J. Mol. Biol. 342, 1637–1646. [DOI] [PubMed] [Google Scholar]
- 29.Improta, S., Politou, A. S. & Pastore, A. (1996) Structure (London) 4, 323–337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.