Abstract
Processed pseudogenes (PPGs) are cDNA sequences that were generated through reverse transcription of mature, spliced mRNAs and have subsequently been reinserted at a new genomic location. These cDNA sequences are usually no longer transcribed and are considered “dead on arrival.” Here we show that PPGs can be used to generate a map of the transcriptome. By analyzing thousands of human PPGs, we were able to discover hundreds of transcript variants so far unidentified. An experimental verification of a subset of these variants by RT-PCR indicates that most of them are still active in the human transcriptome. Furthermore, we demonstrate that PPGs can enable the identification of ancient splice variants that were expressed ancestrally but are now extinct. Our results show that the genome itself carries a “virtual cDNA library” that can readily be used to analyze both present and ancestral transcripts. Our approach can be applied to sequenced metazoan genomes to computationally annotate splicing variation even when expressed sequences are unavailable.
Keywords: alternative splicing, processed pseudogenes, pseudogenes, retropseudogenes
It is well established that alternative splicing plays a key role in generating transcript and protein diversity from a limited number of genes in mammals (1). Employing computational analyses of ESTs, numerous studies have identified a plethora of splice variants in human genes (2–7).
Although more than 6 million human ESTs were deposited in dbEST (as until January 2005), the mapping of the human transcriptome is still far from being complete. Splice variants unique to specific conditions, tissue types or developmental stages that are poorly represented in EST libraries might escape detection. A striking example comes from the IFN alpha gene family, comprised of 13 genes on chromosome 9. These genes were the first cytokines to be discovered, and their protein products are produced en masse upon viral infection in many different cell types (8). However, there is no representation for any of these genes in dbEST (R. Sorek, unpublished data), probably because no EST library was ever produced from virally infected cells. In concordance with the above example, many isoforms are missing from maps of human splice variants. Indeed, we have recently shown that up to 30% of human–mouse conserved exon-skipping events are not represented by ESTs (9). Hundreds of splice variants were also discovered by microarray-based analysis (10).
Another problem with inference of alternative splicing from ESTs stems from their partiality nature. It was suggested that different alternative splicing events in the same transcript are coordinated, e.g., the choice of an alternative first exon influences alternative splicing patterns of internal exons (11). As the average length of an EST is <500 bp, it seldom spans the entire length of a gene. This results in local predictions of alternative splicing (i.e., skipping of a specific exon) without full knowledge of the complete mature transcript. Cloning and sequencing of full-length transcripts solve this problem, but this procedure is much more expensive and time consuming than EST sequencing, so that only ≈67,000 full-length (complete coding sequence) human transcripts are currently found in GenBank.
We propose a different source for full-length transcripts information: processed pseudogenes (PPGs). PPGs (or retropseudogenes) are generated by reverse transcription of a spliced, mature mRNA, presumably by a virus or retrotransposon-encoded reverse transcriptase (12–16). Their cDNA is then incorporated back into the genome (17). Most PPGs lack promoter and regulatory elements, and are therefore not transcribed. Because pseudogenes are usually nonfunctional, they can rapidly accumulate substitutions, insertions, and deletions (18, 19). With the limitation of post pseudogenization sequence changes, PPGs represent the mature form of the spliced mRNA from which they originated (20, 21), and can therefore potentially be used for the prediction of full-length transcripts. PPGs also provide the unique opportunity to learn of the ancient transcriptome, because most of them were retroposed millions of years ago.
Results and Discussion
For the purpose of using PPGs for alternative splicing prediction, we have computationally screened the database of PPGs of Zhang et al. (22) (www.pseudogene.org) for the existence of transcripts that are not represented in EST or cDNA databases. Zhang et al. (22) compiled a data set of 8,605 PPGs by aligning TrEMBL protein sequences onto the human genome. In this database, the sequence of each PPG is linked to a TrEMBL sequence corresponding to its gene of origin. We remapped the nucleotide sequence of each PPG to the human genome (build 34) and to its gene of origin. Possible artifacts, such as putative PPGs showing significant alignment with their gene of origin only, PPGs for which the gene of origin was intronless, and PPGs that showed no alignment with their annotated genomic location, were discarded (see Methods for PPG filtering procedure). After this filtering stage, 5,697 PPGs associated with 1,933 genes remained.
Most analyses to find PPGs, including the one by Zhang et al. (22), use the protein coding sequence (CDS) of the original gene to search for pseudogenes, leaving the UTRs outside the mapped PPG. However, splicing variations can also occur within UTRs. To address this, we extended the PPG sequence by 10 kb of surrounding genomic sequence from each side (Fig. 1A). Each extended PPG (masked for repetitive elements) was then aligned to the genomic locus of its gene of origin by using the spliced alignment program sim4 (23), and the region showing significant alignment around the core CDS was regarded as the complete PPG sequence (Fig. 1A). We considered only consecutive regions of the alignment longer than 60 bp, in which there was at least one intron. Thus, 4,457 PPGs belonging to 1,567 genes were further analyzed.
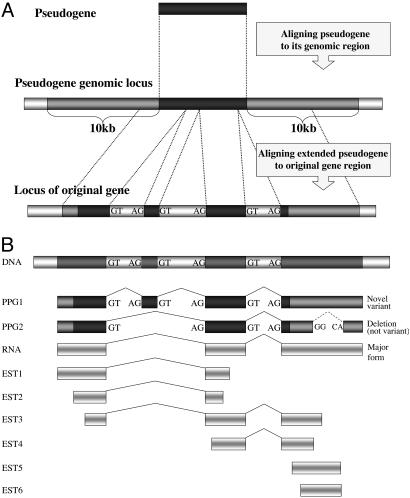
Fig. 1.
Flow of computational detection of PPG-inferred variants. (A) Annotated PPGs from www.pseudogene.org were aligned to their genomic locus and expanded by 10 kb from each their sides. The extended PPGs were then aligned to the gene of origin locus by using SIM4 (23). (B) PPGs were added to their corresponding EST/cDNA cluster, and unique splice variants were searched. To distinguish a true splicing event from insertions/deletions that occurred within the PPG sequence, only GT/AG (or GC/AG) splicing events were taken into account (see Methods).
To search for PPGs showing alternative splicing, each PPG was added to the corresponding EST cluster that was prepared by using the leads software (24) run on cDNAs and ESTs from GenBank version 139 (www.ncbi.nlm.nih.gov/GenBank). The genomic alignment of the PPG was compared to that of the ESTs, and unique variants were identified. Clearly, deletions in the PPG that occurred after the retropseudogenization might appear as alternative splicing if aligned to the genome. Therefore, we considered only splice variants that present unambiguous GT/AG or GC/AG splice sites. We also considered only PPGs that shared at least one splicing event with their gene of origin, to ensure that the pseudogene has indeed been originated from that gene. This left us with 3,776 PPGs (1,409 genes).
Of these 3,776 aligned PPGs, 992 (26%) presented a splice variant that was not indicated by any Refseq sequence at the gene locus (509 genes). Of these, 184 (19%) represented unique splice variants (in 163 genes) for which no support was detected in any of the ESTs or cDNAs in GenBank (Table 3, which is published as supporting information on the PNAS web site). These splice variants are therefore exclusively predicted by PPGs. The splicing pattern distribution of these variants is presented in Table 1.
Table 1. Types of alternative splicing events detected by PPGs.
| Splicing type* | PPGs | Novel events | Percent of events |
|---|---|---|---|
| Skipping over a known exon | 19 | 16 | 9.8 |
| Internal novel exon | 25 | 22 | 13.5 |
| Alternative donor | 26 | 22 | 13.5 |
| Alternative acceptor | 35 | 32 | 19.6 |
| Alternative promoter | 22 | 19 | 11.7 |
| Novel intron | 13 | 11 | 6.7 |
| Intron retention | 3 | 3 | 1.8 |
| Mutually exclusive exons | 1 | 1 | 0.6 |
| Other | 40 | 37 | 22.7 |
| Total splice variants | 184 | 163 | 100.0 |
| 5′ extension | 64 | 53 | 63 |
| 3′ extension | 43 | 30 | 36 |
| Both 5′ and 3′ extension | 1 | 1 | 1 |
| Total extensions | 108 | 84 | 100 |
Alternative splicing type. The type “exon skipping” is the combination of the “skipping over a known exon” and the “internal novel exon” types, therefore comprising 38 events (23% of total). We note that although “exon skipping” is the most abundant splicing event, its frequency is lower than the 38% exon-skipping fraction reported in alternative splicing inferred from ESTs (33). This difference probably stems from the PPG identification parameters of Zhang et al. (22), which did not allow long gaps in the coding sequence of the gene of origin when aligned to the PPG. Accordingly, the fraction of alternative donors and acceptors is higher than that present in EST-inferred variants (33), because these splicing patterns often affect smaller coding regions than exon skipping
In addition to splice variants, we detected 108 PPGs indicating 84 cases of 5′ or 3′ transcript extensions (Table 3). An example for such an extension is presented in Fig. 2. The average extension length was 331 bp (median, 40 bp), with the longest extension being 3,475 bp. Thus, we mapped the complete transcript sequences of 84 genes for which the UTR was only partially mapped previously. This finding underscores the advantage of PPGs, which represent full-length mature mRNAs, over ESTs that are a partial representation of the mRNA, and over putative full-length mRNAs that were accidentally primed from genomic poly(A) sequences (Fig. 2).
Fig. 2.
PPG-inferred extension of the last exon in GNA11. Shown is a snapshot from the University of California, Santa Cruz, genome browser [http://genome.ucsc.edu/cgi-bin/hgGateway (hg17)]. Boxes depict exons and thin arrowed lines are introns. The “Blat search” track shows the alignment of the PPG to its gene of origin. Index numbers of exons are marked above the track. The last exon (exon 7) in the Refseq of guanine nucleotide binding protein alpha 11 (GNA11) contains 466 bp. The PPG chr7_P29992.1 shows a 2,539-bp extension of the last Refseq exon when aligned to the GNA11 locus. No polyadenylation signal appears near the end of the Refseq's last exon, but a genomic A-rich sequence (underlined) appears immediately downstream the Refseq terminus, implying that the Refseq sequence was primed from this internal genomic poly(A). A canonical polyadenylation signal is found 12 bp upstream the PPG-inferred end (italicized and underlined), indicating that this is probably the real terminus of the GNA11 mRNA.
To test whether the PPG-inferred splice variants represent real transcript isoforms, we selected a subset of the variants (14 events) for experimental verification (Table 2 and Fig. 3A). RT-PCR was carried out by using total RNA pools from 16 human tissues, and corresponding electrophoresis bands were validated by direct sequencing (see Methods). Eight of the 14 predicted variants tested were confirmed by RT-PCR (Table 2 and Fig. 3A), indicating that a significant portion of the variants predicted by PPGs are still active in the human transcriptome.
Table 2. Variants selected for experimental validation.
| Gene name | RefSeq/mRNA* | Pseudogene† | Predicted variant | RT-PCR confirmed?‡ | Description of gene of origin |
|---|---|---|---|---|---|
| PLA2G10 | NM_003561 | chr3_O15496.1 | Skipping exon 3 | No | Phospholipase A2, group X |
| TYRO3 | NM_006293 | chr15_Q06418.1 | Novel exon 18A (54 bp) | Yes | TYRO3 protein tyrosine kinase |
| RAD52 | NM_002879 | chr2_P43351.1 | Skipping exon 10 | Yes | DNA repair protein RAD52 |
| PTPN11 | NM_002834 | chr5_Q06124.1 | Skipping exon 4 | No | Protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) |
| HAVCR1 | NM_012206 | chr12_O43656.1 | Alternative promoter | Yes | Hepatitis A virus cellular receptor 1 |
| MMRN2 | NM_024756 | chr6_Q9H8L6.1 | Skipping exons 3–4 | No | Multimerin 2 |
| RYK | NM_002958 | chr17_P34925.1 | Skipping exon 5 | Yes | RYK receptor-like tyrosine kinase |
| IMP-2 | NM_006548 | chr8_Q9Y6M1.1 | Skipping exons 10 and 13 | No | IGF-II mRNA-binding protein 2 |
| PIP5K1A/PSMD4 | NM_003557 and NM_002810 | chr10_P55036.1 | Fusion protein | Yes | Phosphatidylin ositol-4-phosphate 5-kinase, type I, alpha fused to proteasome 26S non-ATPase subunit 4 |
| ESRRA | NM_004451 | chr13_P11474.1 | Extended alt. first exon | Yes | Estrogen-related receptor alpha |
| ENDOGL1 | NM_005107 | chr18_Q9Y2C4.1 | Novel combination of splicing events (3′ truncation of exon 2 and skipping exon 3) | Yes | Endonuclease G-like 1 |
| BMPR1A | NM_004329 | chr6_P36894.1 | Alternative promoter | Yes | Bone morphogenetic protein receptor, type IA |
| CTAGE5 | NM_005930 | chr9_O15320.1 | Exon 5 truncation of 24 bp (alt. donor) | No | CTAGE family, member 5 |
| TARDBP | NM_007375 | chr2_Q13148.1 | Skipping exon 4 | No | TAR DNA binding protein |
Accession of the RNA at the gene of origin locus
Processed pseudogene accession as appears in www.pseudogene.org (22)
See Fig. 3A
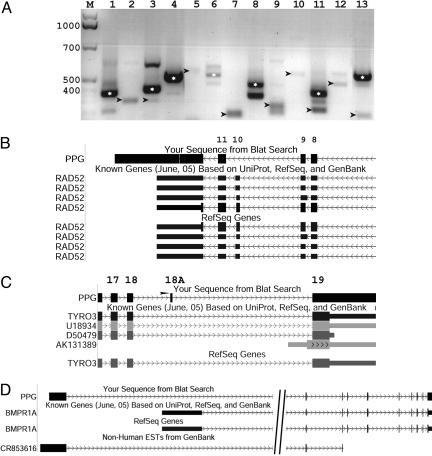
Fig. 3.
Experimental verification of selected variants that were predicted by using PPGs. (A) RT-PCR was carried out by using RNA pool from 16 different tissue types (see Methods). White point indicate the major splice form (“wild type”) for each gene (wild type does not exist in the gel when variant-specific primers were used); arrows indicate the validated isoforms. Lanes: M, 1-kb DNA size marker; 1, PLA2G10, variant not validated; 2, TYRO3, a unique exon (18A) validated; 3, RAD52, skipping on exon 10 validated; 4, PTPN11, variant not validated; 5, HAVCR1, unique promoter validated; 6, MMRN2, variant not validated; 7, RYK, skipping on exon 5 validated; 8, IMP-2, variant not validated; 9, PIP5K1A/PSMD4, transcription-mediated gene fusion validated; 10, ESRRA, 5′ extension of alternative first exon validated; 11, ENDOGL1, a unique combination of splicing events (3′ truncation of exon 2 and skipping on the following exon) validated; 12, BMPR1A, alternative promoter validated; 13, IPO7, skipping exons 21 and 22 validated [this PPG was not found in the original PPG database of Zhang et al. (22), and is presented here to demonstrate that additional variants exist in other genomic PPGs]. Two additional genes (CTAGE5 and TARDBP), for which no variant was detected, are not included in this figure. (B–D) Detailed examples of three transcript isoforms predicted by PPGs and experimentally validated. Shown are snapshots from the University of California, Santa Cruz, genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway; hg17). Boxes depict exons, and thin arrowed lines are introns. The “Blat search” track shows the alignment of the PPG to its gene of origin. Index numbers of exons are marked above the track. (B) Skipping on exon 10 in human RAD52. PPG sequence aligns to the RAD52 locus at 84.3% identity. (C) New exon (marked by arrowhead) in the TYRO3 gene. PPG sequence aligns to the TYRO3 locus at 90.3% identity. (D) PPG showing promoter (alternative first exon) in the gene BMP1RA (PPG is 98% identical with original gene). An EST (CR853616) from Pongo pygmaeus (orangutan) indicates the same variant when aligned to the human genome.
Fig. 3 B–D shows detailed examples of several PPG-inferred variants. The gene RAD52 is found on chromosome 12 and plays a key role in DNA double strand break repair (25). A PPG from chromosome 2 aligns well to RAD52 (84.3% identity), and presents skipping over exon 10, which was confirmed by RT-PCR and direct sequencing (Fig. 3B). Exon 10 contains a hydrophilic region, which is found within the C terminus of RAD52. This region was found to be involved in interactions with RAD51 and RPA (26). Therefore, eliminating this exon in the variant might have an effect on the assembly of a double strand break repair complex.
The gene TYRO3 (Fig. 3C) is found on chromosome 15 and may be involved in cell adhesion processes, particularly in the central nervous system (27). A PPG, also found on chromosome 15 aligns to TYRO3 with 90.3% identity, and presents an exon (18A) that could not be detected by using ESTs (Fig. 3C). Interestingly, exon 18A is inserted, in-frame, within the intracellular tyrosine-kinase domain, so that the resulting variant might have altered kinase activity properties.
A third example is of the gene BMPR1A (Fig. 3D), a bone morphogenetic factor receptor, which is found on chromosome 10. This gene was implicated in abnormal function to juvenile intestinal polyposis and gastrointestinal cancer (28). BMPR1A shares 98% identity with a PPG lying on chromosome 6 (Fig. 3D). Inspecting the alignment of the PPG to the BMPR1A locus shows that the PPG indicates a variant, which has an alternative promoter upstream to the first exon of the major splice form (Fig. 3D). Thus, the expression of this variant is probably regulated differently from the expression of the major splice form. Interestingly, although this promoter is not indicated by any human EST, a recently sequenced Pongo pygmaeus (orangutan) EST shows this variant when aligned to the human genome (Fig. 3D).
As indicated above, only 8 of the 14 PPG-indicated variants we tested experimentally were validated by RT-PCR. Some of the variants we were unable to validate might actually be expressed in a tissue other than the ones we tested, or at a different developmental stage. Alternatively, they could represent mRNA isoforms that existed at the time when the PPG was created, but became extinct later on in evolution. It was recently shown that alternative splicing undergoes high evolution rates, with at least 11% of human alternative cassette exons being constitutively spliced in mouse (29). Therefore, PPGs can provide us with the unique opportunity to study not only the present transcriptome but also features of the ancestral transcriptome. Indeed, PPGs were successfully used before to study evolutionary sequence changes in the RNA of the gene BC200 (30).
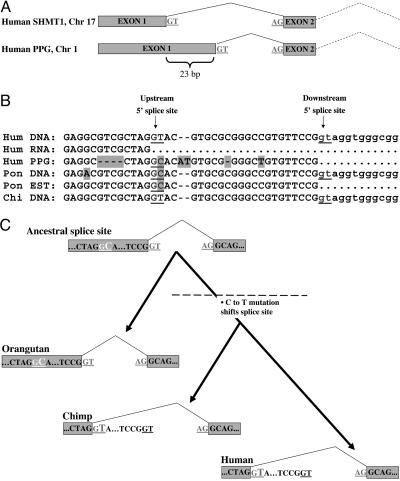
To further pursue this possibility, we searched for PPGs that might report on such evolution of alternative splicing. Fig. 4 presents an example for a PPG indicating an extinct transcript in the gene SHMT1, and describes the genomic mutations that lead to the extinction. The human PPG indicates a transcript that extends the first exon of SHMT1 by 23 nucleotides. No cDNA or EST supports this variant, but several sequences from Pongo (orangutan) support the extended variant. Does this mean that the variant is currently active in human? Inspecting the sequences suggests that this is not the case: Significantly, the Pongo sequences as well as the Pongo genomic DNA differ from the human sequence by a C instead of T at position +2 of the 5′ splice site used by the human to produce the shorter variant. The PPG also contains C at that position, indicating that the longer variant is the ancestral one. Thus, the C → T mutation in the human lineage created a 5′ splice site, which generated the shorter form. Because this mutation (as well as the PPG) exists also in the chimp genome, it probably occurred after the Pongo/human lineage separation but before the chimp/human separation (Fig. 4 B and C). This case demonstrates how PPG analysis enables the mapping of mutational changes that lead to the rapid evolution of alternative splicing, and exemplifies the power of PPGs in studying ancestral features of the transcriptome.
Fig. 4.
PPGs as a tool to study ancient transcript states. (A) The gene serine hydroxymethyltransferase 1 (SHMT1) resides on chromosome 17 in the human genome. A PPG, residing on human chromosome 1, shows a splice variant extending exon 1 by 23 bp. This exon contains the 5′ UTR of the gene. No human EST supports the long variant. (B) Alignment of genomic DNA and cDNAs from various primates at the 3′ terminus of exon 1. Upstream and downstream 5′ splice sites (5′ss) are underlined; mutations relative to human are shaded. Evidently, the GT dinucleotide at the 5′ss used by the human short form appears as a GC in the PPG. Presumably, the ancestral transcript used the downstream 5′ss, and a C → T mutation activated the upstream 5′ss, which is currently used in human. Several Pongo pygmaeus (orangutan) ESTs support the long variant (for example, CR762890). These ESTs, as well as the Pongo DNA, contain GC at the upstream splice site. On the other hand, DNA from Pan troglodytes (chimp) contains T at the upstream 5′ss, indicating that the C → T mutation occurred before the human/chimp lineage separation. Therefore, the pseudogene retroposition must also have occurred before that separation. Indeed, this pseudogene is found in the chimp genome as well (chromosome 1, not presented). (C) Evolutionary tree presenting the ancestral and current states of the transcript in human, chimp and orangutan (Pongo). Dashed line indicates the chronological point before which pseudogene retroposition must have occurred.
Following this result, we conducted a systematic search for such PPGs that might represent ancient transcripts, by looking either for SHMT1 kind of examples, or for PPGs spanning non GT/AG gaps when aligned to their gene of origin. We found 1,147 such PPG candidates. However, except for the above-described SHMT1 example, none of these PPGs had a convincing enough evidence for being an ancestral variant rather than a postpseudogenization deletion/insertion product. Because such postpseudogenization changes are relatively frequent, these results may suggest that the number of extinct splice variants that are fixed as PPGs is relatively small in the human genome. Still, genomes other than the human might contain larger portion of PPGs showing extinct splice variant, depending on their relative rates of pseudogenization and transcript evolution.
We have shown that PPGs are an extremely useful tool for identification of full-length cDNA sequences so far overlooked. We have found 992 splice variants, including 247 unique transcripts (163 splice variants plus 84 transcript extensions) using a database of 8,605 PPGs (22), but it is noteworthy that this database contains only a fraction of the complete human processed-pseudogen-ome. For example, a newly deposited database in the University of California, Santa Cruz (UCSC) version of May 2004 (hg17) contains ≈25,000 PPGs and processed genes, more than three times the data we used. Manually inspecting this database, we were able to discover more PPG-inferred variants (for example, double exon skipping in the gene IPO7; Fig. 3A, lane 13). Therefore, we conclude that additional few hundreds of unique human transcripts that are undetectable by ESTs could be inferred from PPGs.
We note that to be heritable and subsequently be fixed in the genome, PPG insertions must occur in the germ line. Consequently, all of the splice variants represented in the PPG data set must have been expressed in the germ line at the time of pseudogenization. This finding points to a limitation in our approach: the transcriptome map generated by the PPG analysis is biased toward transcripts expressed in germ line or pre-germ-line cells. Still, we have shown that many splice variants evident from PPGs are expressed in cell types other than germ-line cells (Fig. 3A), reflecting the fact that splice variants are often not confined to one tissue (alternatively, this could point to ectopic expression of germ-line splice variants; ref. 31).
Although we used PPGs to find variants in the human genome, our approach could also be applied to detect splice isoforms in other genomes. Of specific interest would be newly sequenced genomes for which very few (or zero) ESTs exist. Thus, a map of the alternatively spliced transcriptome (at least in the germ line) could be inferred from the genome sequence itself, needing neither expressed sequences nor a closely related genome. The results of such analyses should eventually bring us a few steps closer to a global understanding of the transcriptome and its evolution.
Methods
Human ESTs and cDNAs (RNAs) were obtained from NCBI GenBank version 139 (December 15, 2003; www.ncbi.nlm.nih.gov/GenBank) and aligned to the human genome build 34 (July 2003; ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/ARCHIVE) by using the leads clustering and assembly software as described (24). Briefly, the software cleans the expressed sequences from vectors and immunoglobulins, masking them for repeats and low complexity regions. It then aligns the expressed sequences to the genome, taking alternative splicing into account, and clusters overlapping expressed sequences into “clusters” that represent genes or partial genes. Clusters were separated to sense/antisense clusters by using the “Antisensor” algorithm as described in Yelin et al. (32).
PPG sequences were obtained from the database at www.pseudogene.org (22). In this database, each PPG sequence is named after the gene from which it is putatively originated by the TrEMBL accession. Each PPG was assigned to its cluster by using the TrEMBL accession.
Each PPG was aligned to the genome by using blastn (Fig. 1A). PPGs having poor or no alignment to the genome, as well as PPGs that their genomic mapping overlapped their TrEMBL locus were discarded. Also discarded were PPGs for which the TrEMBL locus (marking the putative gene of origin) was intronless.
For each PPG, a genomic piece that included the predicted PPG sequence extended by 10 kbp at both sided was generated. Regions of common repeats and low complexity were masked, and the extended PPG was aligned to the TrEMBL genomic locus by using sim4 (23) (Fig. 1A). Cases in which the PPG alignment was fragmented were divided into subPPGs, demanding each such segment to be at least 60 bp long and to include at least one putative intron. Because sim4 aligns sequences with tendency to create putative introns that support the canonical splicing pattern (i.e., GT/GC-AG) if possible, it may create alignments with poor sequence similarity close to the putative splice site. Therefore, the alignment at splice sites (regarded as the 10 bp spanning the gap) was inspected and a canonical intron was regarded as such only when there were, at most, two mismatches at both sides of the splice site or at most, one deletion/insertion at one of the sides of the splice site, as long as it was not adjacent to the splice site itself. Introns shorter than 50 bases were regarded as possible deletion products and ignored.
The new extended PPG sequences, as well as the ESTs, RNAs, and the genomic sequences of each cluster, were multiply aligned by the LEADS software into clusters (Fig. 1B). PPG sequences not sharing even one splice site with at least one of the sequences in the cluster were discarded. Clusters were then inspected for PPGs showing unique splice variants or transcript extensions.
For experimental validation, cDNA was obtained by reverse transcription of total RNA from assorted samples of the following tissue types: cervix, uterus, ovary, placenta, breast, prostate, testis, kidney, colon and intestine, pancreas, liver and spleen, brain, lung, thyroid, WBC, and the cell-lines HeLa (cervical cancer, American Type Culture Collection) and HepG2 (liver tumor, American Type Culture Collection). Each tissue type was represented by one to five samples of different origin and pathology (tumor and normal).
RNA was added with a random hexamer primer mix (Invitrogen), denaturated at 70°C for 5 min, and transferred to ice for hexamer annealing. Reverse transcription was done by Superscript II reverse transcriptase (Invitrogen), in the presence of RNAsin (Promega) at 37°C for 1 h. Reaction was terminated by enzyme deactivation on beads.
cDNA was amplified by PCR using either ReddyMix (ABgene) or HotStarTaq (Qiagen). Reaction mix was constructed of cDNA, primers (Table 4, which is published as supporting information on the PNAS web site) and the reaction master-mix, which included Taq polymerase, buffer, and dNTP mix. Some reactions required addition of Q solution (Qiagen) to enhance specificity or efficiency. The PCR consisted of 35–45 cycles of denaturation at 94°C for 45 s (preceded by an activation phase of 2 min at 95°C for HotStarTaq), annealing at a primer-specific temperature for 45 s, and extension at 72°C for 1 min. The cycle was ended by one stage of gap filling at 72°C for 10 min.
To avoid cross-reactions with the genomic locus of the PPG itself (the genomic sequence of which closely resembles the sequence of the proposed splice variant), specific primers were constructed to fully align to the target gene and to contain as many mismatches relative to the PPG as possible (Table 4).
Reaction product was separated on a 1.8–2% agarose gel in TAEx1 at ≈100–150 V, and relevant DNA bands were sent for commercial gel extraction and direct sequencing using the same primers used for PCR. Toothpick re-PCR was done in cases where the resulting band was too week for sequencing. In some cases PCR was done by using variant-specific primers (samples 2, 5–7, 9, and 10 in Fig. 2 and Table 4) so that no WT product was expected.
Supplementary Material
Acknowledgments
We thank Dan Graur, Eli Eisenberg, Dvir Dahary, Nir Tsabar, Amir Toporik, Erez Levanon, Yossef Kliger, Tal Dagan, Gad Cojocaru, and Gil Ast for insightful ideas and remarks to the manuscript. We also thank Assaf Wool, Yaron Kinar, Ami Haviv, Tomer Zekharya, Naomi Keren, and Avi Rosenberg for their technical assistance with the computational analysis. R. Sorek was supported by the Clore Israel Foundation.
Author contributions: R. Shemesh, A.N., and R. Sorek designed research; R. Shemesh, A.N., S.E., and R. Sorek performed research; R. Shemesh, A.N., and R. Sorek analyzed data; and R. Sorek wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PPG, processed pseudogene.
References
- 1.Maniatis, T. & Tasic, B. (2002) Nature 418, 236-243. [DOI] [PubMed] [Google Scholar]
- 2.Mironov, A. A., Fickett, J. W. & Gelfand, M. S. (1999) Genome Res. 9, 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J. & Bork, P. (2000) FEBS Lett. 474, 83-86. [DOI] [PubMed] [Google Scholar]
- 4.Kan, Z., Rouchka, E. C., Gish, W. R. & States, D. J. (2001) Genome Res. 11, 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan, Z., States, D. & Gish, W. (2002) Genome Res. 12, 1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 7.Modrek, B., Resch, A., Grasso, C. & Lee, C. (2001) Nucleic Acids Res. 29, 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi, T. & Takaoka, A. (2002) Curr. Opin. Immunol. 14, 111-116. [DOI] [PubMed] [Google Scholar]
- 9.Sorek, R., Shemesh, R., Cohen, Y., Basechess, O., Ast, G. & Shamir, R. (2004) Genome Res. 14, 1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141-2144. [DOI] [PubMed] [Google Scholar]
- 11.Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D. A., Hayashizaki, Y. & Gaasterland, T. (2003) Genome Res. 13, 1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanin, E. F. (1985) Annu. Rev. Genet 19, 253-272. [DOI] [PubMed] [Google Scholar]
- 13.Carlton, M. B., Colledge, W. H. & Evans, M. J. (1995) Mamm. Genome 6, 90-95. [DOI] [PubMed] [Google Scholar]
- 14.Esnault, C., Maestre, J. & Heidmann, T. (2000) Nat. Genet. 24, 363-367. [DOI] [PubMed] [Google Scholar]
- 15.Goncalves, I., Duret, L. & Mouchiroud, D. (2000) Genome Res. 10, 672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graur, D. & Li, W.-H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
- 17.Mighell, A. J., Smith, N. R., Robinson, P. A. & Markham, A. F. (2000) FEBS Lett. 468, 109-114. [DOI] [PubMed] [Google Scholar]
- 18.Graur, D., Shuali, Y. & Li, W.-H. (1989) J. Mol. Evol. 28, 279-285. [DOI] [PubMed] [Google Scholar]
- 19.Harrison, P. M. & Gerstein, M. (2002) J. Mol. Biol. 318, 1155-1174. [DOI] [PubMed] [Google Scholar]
- 20.Maestre, J., Tchenio, T., Dhellin, O. & Heidmann, T. (1995) EMBO J. 14, 6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balakirev, E. S. & Ayala, F. J. (2003) Annu. Rev. Genet. 37, 123-151. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Z., Harrison, P. M., Liu, Y. & Gerstein, M. (2003) Genome Res. 13, 2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. (1998) Genome Res. 8, 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorek, R., Ast, G. & Graur, D. (2002) Genome Res. 12, 1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Dyck, E., Stasiak, A. Z., Stasiak, A. & West, S. C. (1999) Nature 398, 728-731. [DOI] [PubMed] [Google Scholar]
- 26.New, J. H., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. (1998) Nature 391, 407-410. [DOI] [PubMed] [Google Scholar]
- 27.Lan, Z., Wu, H., Li, W., Wu, S., Lu, L., Xu, M. & Dai, W. (2000) Blood 95, 633-638. [PubMed] [Google Scholar]
- 28.Howe, J. R., Bair, J. L., Sayed, M. G., Anderson, M. E., Mitros, F. A., Petersen, G. M., Velculescu, V. E., Traverso, G. & Vogelstein, B. (2001) Nat. Genet. 28, 184-187. [DOI] [PubMed] [Google Scholar]
- 29.Pan, Q., Bakowski, M. A., Morris, Q., Zhang, W., Frey, B. J., Hughes, T. R. & Blencowe, B. J. (2005) Trends Genet. 21, 73-77. [DOI] [PubMed] [Google Scholar]
- 30.Kuryshev, V. Y., Skryabin, B. V., Kremerskothen, J., Jurka, J. & Brosius, J. (2001) J. Mol. Biol. 309, 1049-1066. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Trelles, F., Tarrio, R. & Ayala, F.J. (2005) BioEssays 27, 592-601. [DOI] [PubMed] [Google Scholar]
- 32.Yelin, R., Dahary, D., Sorek, R., Levanon, E. Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. (2003) Nat. Biotechnol. 21, 379-386. [DOI] [PubMed] [Google Scholar]
- 33.Sugnet, C. W., Kent, W. J., Ares, M., Jr., & Haussler, D. (2004) Pac. Symp. Biocomput. 66-77. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.