Abstract
The Bet protein of foamy viruses (FVs) is an auxiliary protein encoded by the 3′ end of the viral genome. Although its function during the viral replication cycle is still unknown, Bet seems to play a key role in the establishment and/or maintenance of viral persistence, representing the predominant viral protein detected during chronic infection. To clarify the function of this viral protein, the subcellular distribution of Bet from the prototypic human foamy virus (HFV) was examined. We report here that this protein is distributed in both the cytoplasm and the nucleus of HFV-infected or Bet-transfected cells. The nuclear targeting results from the presence of a bipartite nuclear localization signal at the C-terminal region, sufficient to direct heterologous reporter proteins to the nucleus. Since HFV Bet spreads between cells, we show here that the secreted protein targets the nuclei of recipient cells. HFV Bet follows an unconventional route to exit the cell since its secretion is not affected by brefeldin A, a drug which disrupts the trafficking between the endoplasmic reticulum and the Golgi complex. Finally, these inter- and intracellular movements were also observed for the equine foamy virus Bet protein, strongly suggesting that these remarkable features are conserved among FVs.
Foamy viruses (FVs) are complex retroviruses encoding auxiliary proteins from the 3′ end of their genome in addition to the structural and enzymatic gag, pol, and env genes. In the case of human foamy virus (HFV), the prototype of FVs, two additional open reading frames (ORFs) have been described, ORF1 and ORF2 (for a review, see references 14 and 20). Tas (transactivator of spumaviruses, originally called Bel1), a 36-kDa nuclear phosphoprotein, which is encoded by ORF1, transactivates viral gene expression by direct binding to the viral DNA on specific sequences in the long terminal repeat (LTR) and in the internal promoter (10). The latter sequence is located upstream from the viral auxiliary genes, at the 3′ end of the env gene, and directs their expression early in the acute infection and during the chronic state (18, 19, 21). Although the product of ORF2, named Bel2, has been reported (2, 7), its existence has never been confirmed. A multispliced viral mRNA which fuses the env gene, deleted of its membrane-spanning domain, to the bet gene, leading to the gp160 kDa Env-Bet protein, has been described (6, 16).
The auxiliary Bet protein of the prototypic HFV is a 482-amino-acid (aa) protein generated by alternative splicing that fuses the first 88 residues of Tas to 394 aa of ORF2 (Fig. 1A). Highly expressed in acute-infection and chronically infected cells, Bet was also shown to be secreted by producing cells and internalized by naïve ones (6). Although the functions of Bet in the replication cycle remain unknown, its integrity is absolutely required for efficient replication of the feline foamy virus (1) and for that of HFV to a much lesser extent (2, 34). Besides its involvement in acute infection, Bet was shown to be implicated in the establishment and/or maintenance of viral persistence. Indeed, in chronically infected cells, a Tas-defective genome, namely ΔHFV, negatively interferes with the replication of the parental virus by the production of Bet (24, 26). Moreover, ΔHFV represents the predominant viral form detected in the blood and organs of chronically infected rabbits (25). In line with these observations, HFV Bet-expressing cell lines were shown to be resistant to productive HFV infection (3). In these settings, the effect of Bet seems to take place during the early steps of infection, after virus entry but before provirus establishment.
FIG. 1.
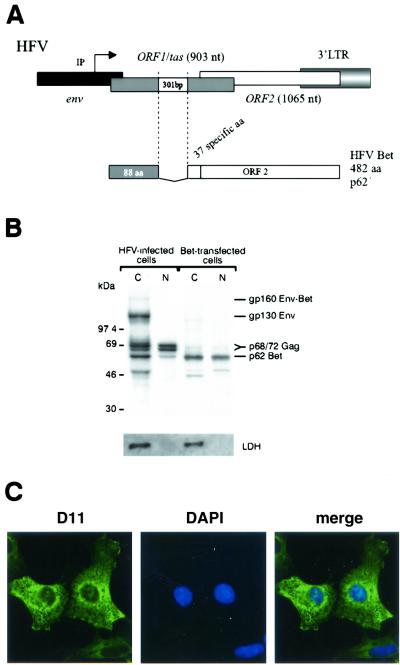
Intracellular trafficking of the HFV Bet protein. (A) Schematic representation of the 3′ end of the HFV provirus. A 301-bp splice generates the Bet mRNA. IP, internal promoter; 88 aa, 88 first residues shared with Tas. (B) Immunoprecipitation of nuclear (N) and cytoplasmic (C) fractions of radiolabeled HFV-infected U373-MG cells using rabbit polyclonal anti-HFV antibodies. Bet was detected in both fractions, while the viral glycoproteins Env and Env-Bet were strictly cytoplasmic. Immunoprecipitation using the monoclonal anti-Bet antibody (D11) of nuclear and cytoplasmic fractions of radiolabeled Cos6 cells transfected with a Bet-expressing vector (pSGHFV-Bet) confirms the nucleocytoplasmic distribution of Bet. The efficiency of the subcellular fractionation is assessed by Western blotting using a monoclonal antibody directed against LDH. (C) Confocal section of Cos6 cells transfected with pSGHFV-Bet using D11 reveals Bet in both the nucleus and the cytoplasm, 48 h posttransfection. The nuclei were revealed with DAPI.
To get insights into the functions of FV Bet, we studied the inter- and intracellular trafficking of Bet from two FVs, the prototypic HFV and the distantly related equine foamy virus (EFV) (32).
MATERIALS AND METHODS
Plasmids and mutagenesis.
Constructs were derived from either pSGHFV-Bet (previously described in reference 6), pEGFP-C1 (Clontech), or pCH110 (Amersham). All the primers used in our experiments were sequenced before their use. All mutations were performed with the Quickchange site-directed mutagenesis kit (Stratagene), and the coding sequences were verified by DNA sequence analysis using the ThermoSequenase kit (United States Biochemicals). The sizes of mutated proteins were verified by Western blotting.
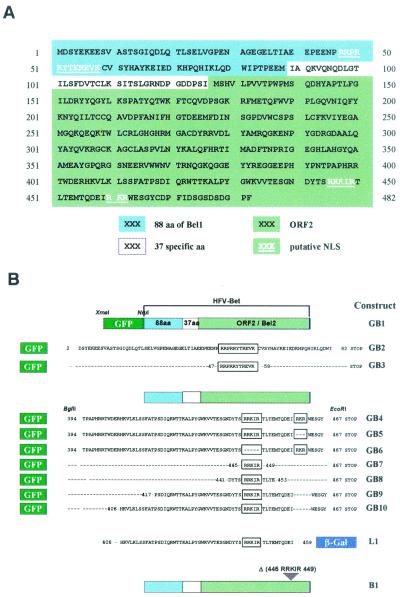
First, an XmaI site was introduced by PCR at the 5′ end of the Bet genomic clone in pSGHFV-Bet. The GFP sequence was introduced into the XmaI-NruI sites of the mutated pSGHFV-Bet (mutant GB1). This plasmid was digested by BamHI and BglII, allowing the fusion of green fluorescent protein (GFP) with the first 82 residues of Bet (mutant GB2). The RRPRRYTKREVK sequence resulting was ligated to the C terminus of GFP (mutant GB3). The Bet sequence from aa 394 to 467 was introduced by PCR into the BglII-EcoRI sites of pEGFP-C1 (mutant GB4). Basic sequences of Bet were then removed by PCR, deleting residues 445 to 449 (mutant GB6) or residues 460 to 462 (mutant GB5). From the GB5 mutant, GB10 (deletion of aa 394 to 405) and GB9 (deletion of aa 394 to 416) were generated. The RRKIR sequence was then ligated to the C terminus of GFP (mutant GB7). The DYTDRRKIRTLTE sequence was fused to GFP by a deletion performed on GB5 (mutant GB8). Deletion was performed in pSGHFV-Bet, leading to a recombinant vector encoding Bel2.
Bet residues 406 to 454 were amplified by PCR and ligated to the N-terminal end of lacZ gene in pCH110 (mutant L1). Finally, sequences coding for residues 444 to 449 were removed from pSGHFV-Bet (mutant B1). Primers used for the generation of recombinant plasmids are available on request.
To produce the EFV Bet expressing vector, the 3′ end EFV genome (nucleotides [nt] 9430 to 11885) was amplified from a λEMBL3 clone harboring a full-length EFV genome using the following primers: GGA ATT CAG GAT ATT ATC ATG GCT A and CCC AAG CTT ATG GTT CTC GAA TAA AGC GGT (the HinDIII site is underlined). This PCR product was subsequently cloned into the pSG5M plasmid (provided by Libin Ma, Leiden, The Netherlands) in EcoRI and HindIII, leading to the pEFV-AUX vector. The 297-bp intron was deleted from the pEFV-AUX by using primers TCC CCA GCT CAT CTG ACT and CAC TTT GTA AGC TGA AGA, leading to an EFV Bet-expressing vector, pSGEFV-Bet.
The Env-Bet-expressing vector (p1EB) was previously described (6).
RT-PCR experiments.
For reverse transcriptase (RT)-PCR analysis, total cellular RNAs from EFV-infected cells were extracted with an RNA extraction kit (Bioprobe Systems). RT-PCR experiments were performed with the Access RT-PCR system (Promega). Briefly, 500 ng of total RNA was used as a template for the synthesis of the first-strand cDNA for 45 min at 48°C in the presence of avian myeloblastosis virus RT. After denaturation at 94°C, the synthesis of the second strand and enzymatic amplifications were carried out with Tfl DNA polymerase for 40 cycles of 94°C for 45 s, 63°C for 45 s, and 72°C for 1 min. The primers used were GGC TAG CAG CTC TTG GAC CC (nt 9443 to 9462) and GTC AAA TTC CTG AAG AGC TGA ATC (nt 10140 to 10163). PCR products subcloned into the pGEM-Easy vector (Promega) were sequenced with the ThermoSequenase kit (United States Biochemicals).
Cells and virus.
Mycoplasma-free HFV stocks were produced on U373-MG cells, a human neural cell line, maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with l-glutamine (2 mM), penicillin (100 μg/ml), streptomycin (50 μg/ml), HEPES (240 mM), and 5% heat-inactivated fetal calf serum (FCS). Cos6 and BHK21 cells were maintained in the same medium. Virus stocks were subjected to titer determination by the LTR-GFP reporter BHK21 (FAG) cells (31). Mycoplasma-free EFV stocks were produced on ED cells, a horse fibroblast cell line, maintained in RPMI medium with 8% FCS.
Intercellular trafficking was studied by using 5 × 105 Cos6 cells seeded in lower and upper chambers of a Transwell-Clear-based culture system (0.4-μm pore size; Costar).
Transfection experiments.
A total of 5 × 105 cells were transfected with 1 μg of recombinant plasmid and the Lipofectin reagent (Gibco-BRL) as specified by the manufacturer. Forty-eight hours posttransfection, cells were fixed for immunofluorescence or lysed in lysis buffer for protein analysis. All plasmids used for transfection were purified on anion-exchange resin columns (Qiagen).
Immunofluorescence analysis.
Forty-eight hours posttransfection, cells were fixed with 4% paraformaldehyde at 4°C for 5 min and permeabilized with 100% methanol at 4°C for 10 min. Antibodies used were a rabbit anti-HFV antiserum (7), used at a 1/600 dilution in phosphate-buffered saline-Tween 20 (0.1%)-bovine serum albumin (0.2%), containing a monoclonal antibody against the HFV Bet protein, D11 (used at a 1/400 dilution), a polyclonal antibody against the β-galactosidase (used at a 1/100 dilution; Euromedex), and a rabbit anti-EFV serum (used at a 1/400 dilution). The latter was obtained after infection of male New Zealand rabbits by a single intravenous dose of EFV stock. Anti-immunoglobulin G-fluorescein isothiocyanate (FITC)-coupled antibodies (Biosys) were used as second fluorescent conjugates and when used alone gave no staining on infected or transfected cells. Nuclei were revealed with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) at 250 ng/ml.
For confocal analysis, cells were embedded in Mowiol (Calbiochem) and examined by using a Bio-Rad MRC-1024 confocal imaging system (Bio-Rad Microscience Ltd.) and an inverted Diaphot 300 Nikon microscope. For fluorescein, a krypton/argon ion laser (Ion Laser Technology Inc.) operating with the 488-nm line was used. Images of FITC and DAPI were pseudocolored in green for FITC and blue for DAPI.
Protein analysis.
For radioimmunoprecipitation, 107 cells were labeled with [35S]methionine-cysteine (50 μCi/ml; 1,245 Ci of specific activity per mmol; Dupont NEN) for 16 h in minimal essential medium (MEM; Gibco-BRL) lacking methionine-cysteine and supplemented with 5% FCS. Cells were lysed in 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate (SDS), 1 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. Nuclei were separated from the lysate by centrifugation at 12,000 × g for 5 min at 4°C and lysed in the same buffer containing 1 M NaCl. Immunoprecipitation of viral proteins with specific antisera was performed as previously described (7).
For the study of HFV Bet internalization, supernatant of radiolabeled Cos6 cells was filtered with a 0.45-μm-pore-size filter (Corning).
For the pulse-chase experiment, cells were radiolabeled with [35S]methionine-cysteine for 2 h and then chased for 4 or 8 h in cold medium. Treatment with 10 μg of brefeldin A (BFA) (Roche) per ml was performed during the entire pulse-chase period. Chase media and cell lysates were immunoprecipitated with polyclonal anti-HFV antibodies and analyzed by SDS-PAGE.
For Western blot analysis, cells were heat disrupted at 100°C for 5 min in Laemmli sample buffer. About 50 μg of protein extracts were resolved by SDS-PAGE and transferred by electroblotting onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). To detect lactate dehydrogenase (LDH), we used a goat anti-LDH antibody (Sigma) (used at a 1/2,000 dilution in phosphate-buffered saline-Tween) and then a peroxidase-conjugated anti-goat antibody (Biosys), used at a 1/3,000 dilution. HFV Bet was detected with monoclonal antibody D11 used at 1/400 dilution and then a peroxidase-conjugated anti-mouse antibody (Biosys), used at a 1/8,000 dilution. EFV Bet was detected with rabbit anti-EFV antibodies, described above, used at a 1/600 dilution, and then with a peroxidase-conjugated anti-rabbit antibody (Biosys), diluted 1/8,000. Western blot results were revealed by enhanced chemiluminescence (Amersham).
RESULTS
Subcellular localization of the HFV Bet protein.
To determine the subcellular distribution of Bet, nuclear and cytoplasmic fractions were prepared from radiolabeled U373-MG cells infected with HFV at a multiplicity of infection of 0.1 and immunoprecipitated with rabbit anti-HFV antibodies (Fig. 1B). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis revealed that glycoproteins gp130 and gp160, corresponding to the Env precursor and to the Env-Bet fusion protein, respectively, were detected solely in the cytoplasm. On the other hand, the Gag doublet at 71 and 68 kDa was located in both the cytoplasm and the nucleus of infected cells, consistent to what has been previously reported (5, 28). In these settings, Bet was detected in both nuclear and cytoplasmic compartments. A similar distribution was observed by immunoprecipitation using the anti-Bet monoclonal antibody, D11, in simian Cos6 cells transfected with a Bet-expressing vector, pSGHFV-Bet (Fig. 1B). The subcellular distribution of Bet, after transfection, is not dependent on the cell type since similar results were obtained with hamster BHK21 and human U373MG cells (data not shown). The efficiency of the subcellular fractionation was controlled by Western blotting using a monoclonal antibody directed against LDH, a soluble protein strictly confined to the cytoplasm.
These results were confirmed by immunofluorescence since Bet was detected both in the cytoplasms and in the nuclei of pSGHFV-Bet transfected Cos6 cells by confocal analysis (Fig. 1C).
Secretion of HFV Bet is not dependent on the classical endoplasmic reticulum (ER)-Golgi secretory pathway.
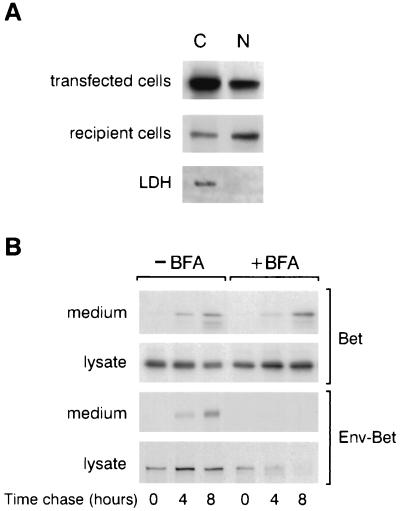
Since HFV Bet was previously shown to be secreted and internalized by surrounding cells (6), we sought to determine whether Bet targets the nuclei of recipients cells. For that purpose, Cos6 cells were transfected with pSGHFV-Bet and labeled overnight with a [35S]methionine-cysteine-containing medium, 48 h posttransfection. The next day, the cell-free supernatant was added to 106 naïve Cos6 cells at 37°C for 4 h. Cytoplasmic or nuclear fractions from transfected or recipient cells were then immunoprecipitated with rabbit anti-HFV antiserum. As shown in Fig. 2A, Bet was detected in the nuclei of transfected and recipients cells.
FIG. 2.
(A) HFV Bet targets the nuclei of recipient cells. Cos6 cells were transfected with pSGHFV-Bet, and 48 h later, cells were labeled overnight and cell-free supernatant was incubated with naïve Cos6 cells. Nuclear and cytoplasmic fractions of transfected and recipient cells were immunoprecipitated with rabbit anti-HFV antiserum and analyzed by SDS-PAGE. (B) HFV Bet secretion is not dependent on the classical ER-Golgi pathway. Aliquots of pulse-chase media and cell lysates immunoprecipitated with polyclonal anti-HFV antibodies reveal that, although Env-Bet secretion is inhibited upon treatment with BFA (10 μg/ml), secretion of Bet is not affected in these settings.
Such remarkable intercellular movements have already been described for other viral proteins (29). Since, in these cases, exocytosis does not require the classical ER-to-Golgi pathway, we wondered whether Bet could be secreted by an alternative route. Indeed, in contrast to HFV Env-Bet, HFV Bet lacks a signal sequence targeting it to the ER-Golgi pathway. To test this issue, secretion of Bet in the presence of BFA, a drug which disrupts early-to-intermediate events in the classical secretory pathway (17), was analyzed. Cos6 cells were transfected with pSGHFV-Bet or with the vector coding for the Env-Bet fusion protein (p1EB [6]), and 2 days later, cells were radiolabeled with [35S]methionine-cysteine for 2 h in the presence or absence of 10 μg of BFA per ml and then chased for 4 and 8 h with the same drug treatment. Chase media and cellular lysates were immunoprecipitated with polyclonal anti-HFV antibodies and analyzed by SDS-PAGE. Under treatment with BFA, the stability of Env-Bet was drastically altered, its secretion being prevented, whereas its biosynthesis was not affected (Fig. 2B, bottom). Conversely, biosynthesis and secretion of HFV Bet were not affected by BFA (Fig. 2B, top).
These observations demonstrated that secretion of HFV Bet occurred via a nonclassical, Golgi apparatus-independent mechanism.
NLS at the C terminus of HFV Bet.
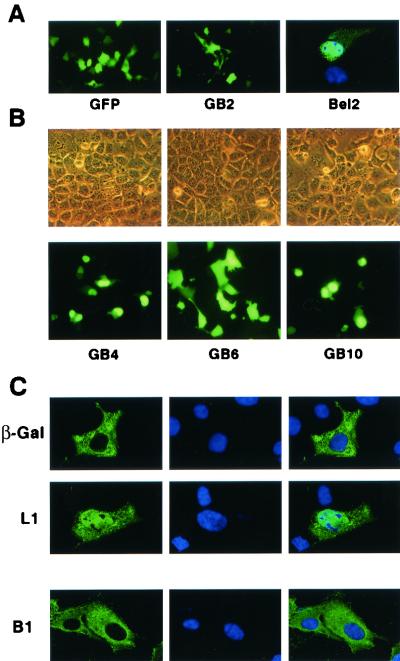
While small proteins are thought to passively diffuse between the cytoplasm and the nucleus, larger molecules, like HFV Bet protein (62 kDa), have to harbor specific signals allowing their active translocation through the nuclear pore complex (8). Analysis of the primary sequence of HFV Bet, based on sequence homology with most documented basic nuclear localization signals (NLSs) (PSORTII software), revealed two such putative sequences: one monopartite NLS at the N terminus (47RRPRRYTKREVK58) and one bipartite motif at the C terminus (445RRKIRX10RKR462) (Fig. 3A). To assess the functionality of these motifs, the corresponding sequences, flanked with various lengths of their amino acid environment, were fused to the GFP reporter gene (Fig. 3B) and fluorescence was directly monitored on live transfected Cos6 cells. The parental GFP localized in both the cytoplasms and the nuclei of transfected cells, according to its molecular mass (27 kDa; Fig. 4A). This nucleocytoplasmic distribution was affected neither by fusion with the minimal sequence 47RRPRRYTKREVK58 (construct GB3) nor by the addition of neighboring amino acids (Bet residues 2 to 82, construct GB2; Fig. 4A). Thus, the basic sequence located at the N terminus of Bet did not meet the criteria of a classical NLS.
FIG. 3.
(A) HFV Bet primary structure. Computer analysis, performed by the PSORTII software, revealed the presence of two putative NLSs: a monopartite NLS at the N terminus of Bet (aa 47 to 58, RRPRRYTKREVK) and a bipartite NLS at the C terminus (aa 445 to 462, RRKIRX10RKR). (B) Schematic representation of the derivative GFP and β-galactosidase Bet fusions.
FIG. 4.
Characterization of an NLS at the C terminus of Bet. (A) Direct fluorescence analysis of live Cos6 cells transfected with the GB2 construct reveals that Bet residues 2 to 82 do not affect the subcellular distribution of the GFP. Confocal section of Cos6 cells transfected with the Bel2 construct shows that this protein is located mainly in the nucleus. The nuclei were revealed with DAPI. (B) Phase-contrast (top) and corresponding fluorescence photomicrographs (bottom) of live transfected Cos6 cells. Although the wild-type GFP is equally distributed in the nucleus and the cytoplasm, fusion with the C-terminal domain of Bet from residues 394 to 467 allows the nuclear targeting of the corresponding fusion protein (GB4). Deletion of the 445RRKIR449 sequence totally abolishes the nuclear pattern (GB6). The sequence spanning aa 406 to 459 constitutes a functional NLS (GB10). (C) Confocal section of the L1 and B1 mutants. The wild-type β-galactosidase is strictly localized in the cytoplasm, whereas mutant L1 is also detected in the nuclei of transfected cells. Deletion of residues 445 to 449 (RRKIR) on HFV Bet (mutant B1) reveals that this protein is strictly confined in the cytoplasm. The nuclei were revealed with DAPI.
Conversely, the product of ORF2 (Bel2, the C-terminal domain of Bet) localized mainly in the nuclei of transfected cells, confirming the presence of an NLS in this region (Fig. 4A). Fusion of Bet residues 394 to 467 (containing the predicted bipartite NLS) to the GFP reporter targeted the fusion protein to the nucleus (construct GB4; Fig. 4B). Nuclear targeting of the GB4 fusion was not affected by deletion of the distal part of the predicted bipartite NLS (460RKR462, mutant GB5) (data not shown), while deletion of the proximal basic stretch (445RRKIR449, mutant GB6; Fig. 4B) abolished its nuclear translocation. Thus, the sequence delineated by basic NLS homology, spanning aa 445 to 462, seemed to represent a monopartite NLS rather than a bipartite one. To test this hypothesis, the minimal 445RRKIR449 sequence alone or flanked with neighboring amino acids (441DYTSRRKIRTLTE453 [the basic amino acid cluster is in bold]) were fused to GFP (constructs GB7 and GB8, respectively). Strikingly, none of these constructs gave rise to a nuclear staining (data not shown). To clarify this point, constructs GB9 (GFP harboring Bet residues 417 to 459) and GB10 (GFP harboring Bet residues 406 to 459) were generated. Indeed, the sequence spanning aa 406 to 416 harbors a basic stretch (406HKVLK410), which, together with aa 445 to 449, could constitute a bipartite NLS. Although GB9 was uniformly distributed in the nucleus and the cytoplasm, as was the parental GFP (data not shown), GB10 was strictly confined to the nucleus, demonstrating that residues from positions 406 to 459 were necessary to direct the reporter protein to the nucleus (Fig. 4B).
Since GFP passively diffuses through the nuclear pore, we could not exclude that the characterized signal was only a nuclear retention signal. To test this hypothesis, a fusion protein between Bet residues 406 to 459 and the β-galactosidase reporter was generated (mutant L1). Expression of the wild-type β-galactosidase led to a strict cytoplasmic staining due to its molecular mass (i.e., 115 kDa; Fig. 4C). Conversely, the L1 mutant was detected in the cytoplasms but also in the nuclei of transfected cells (Fig. 4C), demonstrating that the Bet sequence between aa 406 to 459 constituted an effective NLS.
Finally, the involvement of this bipartite NLS in the Bet context was studied. A mutant, which harbored a deletion of the distal basic stretch (residues 445 to 449, mutant B1) was generated, and its subcellular localization was studied by indirect immunofluorescence by use of rabbit anti-HFV polyclonal antibodies. Confocal analysis revealed that mutant B1 was strictly localized in the cytoplasm (Fig. 4C). This result confirmed the involvement of residues 445 to 449 in the nuclear targeting of HFV Bet, as previously described with the GFP and β-galactosidase reporters.
Inter- and intracellular trafficking of EFV Bet protein.
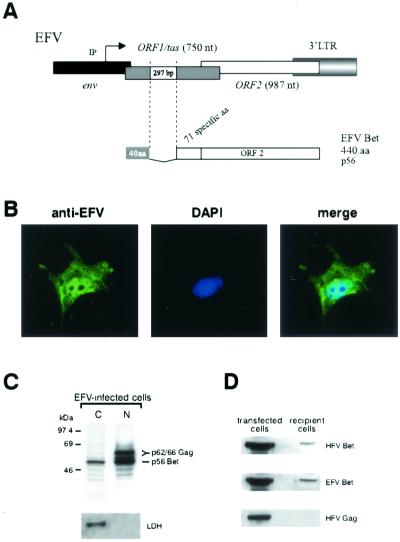
To assess whether the nucleocytoplasmic distribution of HFV Bet is encountered for other FV Bet proteins, the subcellular localization of EFV Bet was investigated. For that purpose, an EFV Bet-expressing vector was generated. The splice sites leading to the synthesis of the EFV Bet mRNA were identified by RT-PCR on total RNAs extracted from EFV-infected BHK21 cells, revealing that the splice donor and acceptor sites are located at positions 9562 and 9859, respectively. This splice event fuses the first 40 aa of EFV Tas to the entire ORF2 (329 aa), leading to a Bet protein of 440 aa (Fig. 5A). Concomitantly, the 3′ end of EFV was amplified by PCR on a λEMBL3 clone harboring a full-length EFV genome, and the 297-bp Bet intron was subsequently deleted by PCR, leading to the pSGEFV-Bet vector. Expression of pSGEFV-Bet was tested by Western blotting, revealing that EFV Bet migrated at 56 kDa (data not shown). Indirect immunofluorescence, using polyclonal anti-EFV antibodies, showed that EFV Bet was detected in both the nuclei and the cytoplasms of pSGEFV-Bet-transfected Cos6 cells (Fig. 5B). We further confirmed the nucleocytoplasmic distribution of EFV Bet on radiolabeled EFV-infected BHK21 cells. Nuclear and cytoplasmic fractions, immunoprecipitated with rabbit anti-EFV antibodies, showed that EFV Bet was detected in both the nucleus and the cytoplasm of infected cells (Fig. 5C). Note that, as described for HFV Gag, the EFV Gag doublet was detected mainly in the nucleus.
FIG. 5.
Inter- and intracellular trafficking of EFV Bet protein.(A) Schematic representation of the 3′ end of the EFV provirus. A 297-bp splice generates the Bet mRNA. 40 aa, 40 first residues shared with Tas. (B) Confocal analysis of pSGEFV-Bet-transfected Cos6 cells using anti-EFV polyclonal antibodies confirms the nucleocytoplasmic distribution of EFV Bet. (C) Immunoprecipitation of nuclear (N) and cytoplasmic (C) fractions of radiolabeled EFV-infected BHK21 cells. EFV Bet is detected in both fractions. Western blot analysis using anti-LDH antibodies attests to the efficiency of the subcellular fractionation. (D) Western blot analysis of Cos6 cells, seeded in the lower chamber and transfected with EFV or HFV Bet-expressing vectors, and of upper na&ıuml;ve cells reveals that EFV Bet exhibits the same intercellular movement as HFV Bet. EFV Bet and HFV Bet were detected with anti-EFV and anti-HFV polyclonal antibodies, respectively. As a control, an HFV Gag-expressing vector was transfected in Cos6 cells in the lower chamber, showing the characteristic Gag precursor at 71 kDa only in transfected cells.
To directly test the intercellular spreading of EFV Bet, a Transwell-Clear-based culture system, in which Cos6 cells were seeded in the lower chamber and transfected with pSGHFV-Bet or pSGEFV-Bet, was developed. Two days after transfection, Cos6 cells from the upper and lower chambers were harvested and protein extracts were analyzed by Western blotting with rabbit anti-HFV or anti-EFV antibodies, revealing that, like HFV Bet, EFV Bet was secreted by transfected cells and internalized by the upper recipient ones (Fig. 5D). As a control, Cos6 cells were transfected with an HFV Gag-expressing construct since HFV Gag does not leave the cell without the expression of the homologous envelope glycoprotein (22). As seen in Fig. 5D, the HFV Gag precursor was detected at 71 kDa in transfected cells but not in recipient cells from the upper chamber, demonstrating that our observation is not due to nonspecific cell lysis.
Taken together, our results showed that EFV Bet shared the same intra- and intercellular trafficking as HFV Bet.
DISCUSSION
In this report, we provide evidence that HFV Bet is located in both the nucleus and the cytoplasm of Bet-expressing or recipient cells. The nuclear targeting requires a nuclear localization signal at the C terminus of the protein. The basic sequence 47RRPRRYTKREVK58, highlighted in the N-terminal region by sequence homology with known basic NLS, does not constitute such a signal since it fails to direct the GFP to the nucleus. This is consistent with previous reports showing that the N-terminal region of Bet, shared with Tas, is not involved in nuclear localization of the viral transactivator or in its transactivation properties (11, 15, 33). Conversely, analysis of the localization of a GFP chimera harboring the C-terminal region of Bet and derived deletion mutants suggested the presence of a sequence which meets the criteria of a bipartite NLS. Indeed, a fusion protein between the β-galactosidase and Bet residues 406 to 459 (mutant L1) allowed the nuclear targeting of this high-molecular-weight protein, demonstrating that this sequence represents an effective NLS. Note that sequence homology with basic NLS revealed that EFV Bet harbors a basic stretch in ORF2 (277PGKKERV283) which could constitute an NLS.
EFV Bet exhibits these inter-and intracellular movements. Given that EFV is the FV most phylogenetically distant from HFV, our results support the idea that these features are conserved among FVs. The remarkable property of spreading between cells is shared by several proteins, like Drosophila Antennapedia, human immunodeficiency virus type 1 Tat, or herpes simplex virus type 1 VP22, which lack a signal sequence but can traverse biological membranes (29). Linking these proteins to compounds, peptides, or nucleic acids lets them enter any cell type in a receptor- and transporter-independent manner. Interestingly, preliminary studies revealed that HFV Bet allowed the secretion of a Bet-GFP fusion protein (data not shown), suggesting that Bet can also transduce a heterologous protein.
Proteins lacking a signal peptide presumably exit cells through alternative, nonclassical, BFA-insensitive routes by which they are released into the medium in free forms or within vesicles. Although the precise mechanisms of these secretions are largely unknown, the cytoskeleton underlying the plasma membrane was shown to be required in certain cases (12). For instance, the weakening of the actin network has been directly implicated in the formation of secretion vesicles enclosing alkaline phosphatase (9). In the case of annexin I, an interaction with bundles of actin beneath the plasma membrane is required for efficient secretion (13). These two proteins exit the cells by ectocytosis, a term first used to describe the shedding of right-side-out the membrane vesicles from the surface of neutrophils during sublytic complement attack (30). The broad mechanism appears to be the extrusion of excess membrane blebs from the cell surface releasing extracellular vesicles. In the case of Bet, we have shown that this protein is also detected at the vicinity of the plasma membrane, where it colocalizes with the cortical actin (data not shown). Note, in that sense, that it has already been suggested that Bet could be a membrane-associated protein (23) and could participate in viral budding (1). It remains to establish whether the actin-Bet interaction is directly involved in Bet secretion and subsequently whether Bet releases cells into vesicles, following the ectocytosis pathway. In the case of HFV, an intact microtubule network is absolutely required for the early stages of viral replication (27). Our observations point to a role of the cytoskeleton in the function of Bet too.
That Bet spreads to surrounding cells and moves into the nucleus suggests that these properties are relevant to some aspects of the virus cycle. Bock et al. reported that Bet expression, prior to FV infection, blocks the replication cycle at early stages (3). The nuclear localization of Bet suggests that this protein affects viral replication at nuclear stages, in particular at the import of the preintegration complex or provirus integration steps. Finally, through its secretion property, Bet could act from a distance to limit FV propagation, one possible mechanism to establish viral persistence. Indeed, while a Tas-defective provirus (ΔHFV) is detected in chronically infected hematopoietic cell lines, viral persistence is, in that case, associated with a constant production of infectious viruses (35), suggesting that the molecular basis of viral persistence seems to depend on the cell type (1). Moreover, deleterious mutations in the Bet gene were detected in a simian foamy virus isolated from a case of persistent zoonotic infection (4). Therefore, these observations suggest that several mechanisms have been developed by foamy viruses to persist in their hosts.
Acknowledgments
We thank Michel Schmidt for confocal analysis and Robin Nancel and Elisabeth Savariau for photographic work. Members of CNRS UPR9051, especially Claudine Pique and Joelle Tobaly-Tapiero, are acknowledged for stimulating discussions and critical reading of the manuscript. We thank Hugues de Thé for continuous support.
This work was supported by ARC (grant no. 5981) and F. Lacoste.
REFERENCES
- 1.Alke, A., A. Schwantes, K. Kido, M. Flotenmeyer, R. M. Flügel, and M. Löchelt. 2001. The bet gene of feline foamy virus is required for virus replication. Virology 287:310-320. [DOI] [PubMed] [Google Scholar]
- 2.Baunach, G., B. Maurer, H. Hahn, M. Kranz, and A. Rethwilm. 1993. Functional analysis of human foamy virus accessory reading frames. J. Virol. 67:5411-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, M., M. Heinkelein, D. Lindemann, and A. Rethwilm. 1998. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology 250:194-204. [DOI] [PubMed] [Google Scholar]
- 4.Callahan, M. E., W. M. Switzer, A. L. Matthews, B. D. Roberts, W. Heneine, T. M. Folks, and P. A. Sandstrom. 1999. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol. 73:9619-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giron, M. L., S. Colas, J. Wybier, F. Rozain, and R. Emanoil-Ravier. 1997. Expression and maturation of human foamy virus Gag precursor polypeptides. J. Virol. 71:1635-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giron, M. L., H. de Thé, and A. Saïb. 1998. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J. Virol. 72:4906-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giron, M. L., F. Rozain, M. C. Debons-Guillemin, M. Canivet, J. Périès, and R. Emanoil-Ravier. 1993. Human foamy virus polypeptides: identification of env and bel gene products. J. Virol. 67:3596-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorlich, D. 1997. Nuclear protein import. Curr. Opin. Cell Biol. 9:412-419. [DOI] [PubMed] [Google Scholar]
- 9.Hale, J. E., and R. E. Wuthier. 1987. The mechanism of matrix vesicle formation. Studies on the composition of chondrocyte microvilli and on the effects of microfilament-perturbing agents on cellular vesiculation. J. Biol. Chem. 262:1916-1925. [PubMed] [Google Scholar]
- 10.He, F., W. S. Blair, J. Fukushima, and B. R. Cullen. 1996. The human foamy virus Bel-1 transcription factor is a sequence-specific DNA binding protein. J. Virol. 70:3902-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, F., J. D. Sun, E. D. Garrett, and B. R. Cullen. 1993. Functional organization of the Bel-1 trans activator of human foamy virus. J. Virol. 67:1896-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, R. C. 1999. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta 1473:172-185. [DOI] [PubMed] [Google Scholar]
- 13.Khanna, N. C., E. D. Helwig, N. W. Ikebuchi, S. Fitzpatrick, R. Bajwa, and D. M. Waisman. 1990. Purification and characterization of annexin proteins from bovine lung. Biochemistry 29:4852-4862. [DOI] [PubMed] [Google Scholar]
- 14.Lecellier, C. H., and A. Saïb. 2000. Foamy viruses: between retroviruses and pararetroviruses. Virology 271:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. W., J. Chang, K. J. Lee, and Y. C. Sung. 1994. The Bel1 protein of human foamy virus contains one positive and two negative control regions which regulate a distinct activation domain of 30 amino acids. J. Virol. 68:2708-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippincott-Schwartz, J., J. G. Donaldson, A. Schweizer, E. G. Berger, H. P. Hauri, L. C. Yuan, and R. D. Klausner. 1990. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell 60:821-836. [DOI] [PubMed] [Google Scholar]
- 18.Löchelt, M., R. M. Flügel, and M. Aboud. 1994. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J. Virol. 68:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löchelt, M., W. Muranyi, and R. M. Flügel. 1993. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc. Natl. Acad. Sci. USA 90:7317-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meiering, C. D., C. Rubio, C. May, and M. L. Linial. 2001. Cell-type-specific regulation of the two foamy virus promoters. J. Virol. 75:6547-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rethwilm, A. 1995. Regulation of foamy virus gene expression. Curr. Top. Microbiol. Immunol. 193:1-24. [DOI] [PubMed] [Google Scholar]
- 24.Saïb, A., M. H. Koken, P. van der Spek, J. Périès, and H. de Thé. 1995. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J. Virol. 69:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saïb, A., M. Neves, M. L. Giron, M. C. Guillemin, J. Valla, J. Périès, and M. Canivet. 1997. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology 228:263-268. [DOI] [PubMed] [Google Scholar]
- 26.Saïb, A., J. Périès, and H. de Thé. 1993. A defective human foamy provirus generated by pregenome splicing. EMBO J. 12:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saïb, A., F. Puvion-Dutilleul, M. Schmid, J. Périès, and H. de Thé. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarze, S. R., K. A. Hruska, and S. F. Dowdy. 2000. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 10:290-295. [DOI] [PubMed] [Google Scholar]
- 30.Stein, J. M., and J. P. Luzio. 1991. Ectocytosis caused by sublytic autologous complement attack on human neutrophils. The sorting of endogenous plasma-membrane proteins and lipids into shed vesicles. Biochem. J. 274:381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobaly-Tapiero, J., P. Bittoun, M. L. Giron, M. Neves, M. Koken, H. de Thé, and A. Saïb. 2001. Human foamy virus capsid formation requires an interaction domain in the N terminus of Gag. J. Virol. 75:4367-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobaly-Tapiero, J., P. Bittoun, M. Neves, M. C. Guillemin, C. H. Lecellier, F. Puvion-Dutilleul, B. Gicquel, S. Zientara, M. L. Giron, H. de Thé, and A. Saïb. 2000. Isolation and characterization of an equine foamy virus. J. Virol. 74:4064-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesh, L. K., and G. Chinnadurai. 1993. The carboxy-terminal transcription enhancement region of the human spumaretrovirus transactivator contains discrete determinants of the activator function. J. Virol. 67:3868-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, S. F., and M. L. Linial. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, S. F., J. Stone, and M. L. Linial. 1996. Productive persistent infection of hematopoietic cells by human foamy virus. J. Virol. 70:1250-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]