Abstract
Foamy viruses (FVs) or spumaviruses are retroviruses that have been developed as vectors, but their integration patterns have not been described. We have performed a large-scale analysis of FV integration sites in unselected human fibroblasts (n = 1,008) and human CD34+ hematopoietic cells (n = 1,821) by using a bacterial shuttle vector and a comparable analysis of lentiviral vector integration sites in CD34+ cells (n = 1,331). FV vectors had a distinct integration profile relative to other types of retroviruses. They did not integrate preferentially within genes, despite a modest preference for integration near transcription start sites and a significant preference for CpG islands. The genomewide distribution of FV vector proviruses was nonrandom, with both clusters and gaps. Transcriptional profiling showed that gene expression had little influence on integration site selection. Our findings suggest that FV vectors may have desirable integration properties for gene therapy applications.
Keywords: gene therapy, insertional mutagenesis, retroviral integration
Integrating viral vectors can provide the long-term transgene expression required in many gene therapy applications, especially when proliferating cells such as hematopoietic stem cells are transduced. However, integration also results in insertional mutagenesis that may activate oncogenes, as seen in X-linked severe combined immune deficiency patients that developed leukemia after retroviral vectors integrated near the LMO2 protooncogene (1). Large-scale integration site analyses can help define the spectrum of insertional mutagenesis expected of a viral vector, and studies of murine leukemia virus (MLV), HIV, and avian sarcoma virus (ASV) have demonstrated distinct integration patterns (2-6). The relationship of integration to transcription is especially important, as it may determine the likelihood of oncogene activation. The two most commonly used retroviral vectors differ in this respect, with HIV vectors integrating preferentially throughout transcription units, and MLV vectors integrating preferentially near transcription start sites (2, 3).
Foamy viruses (FVs), or spumaviruses, comprise a class of retroviruses distinct from oncoviruses such as MLV and lentiviruses such as HIV. FVs are found in several mammalian species (7) but are not endemic in human populations (8), and the prototype “human FV” isolate (9) is now thought to be a chimpanzee virus [SFVcpz(hu)] (10, 11). FVs have several distinguishing properties, including a transcriptional transactivator expressed from an internal promoter (12-14), a DNA-based genome that forms by reverse transcription in virion-producing cells (15, 16), and an essential cis-acting region overlapping the pol and env genes (17-19).
FV vectors have been developed (19-22) and shown to have a broad host range (21, 23, 24), large packaging capacity (25), stable virions that can be concentrated by ultracentrifugation (26), and prolonged survival of the preintegration complex in quiescent cells (27). They are especially effective at transducing hematopoietic stem cells of mice and humans (26, 28-31), making them promising vectors for the treatment of hematologic diseases. Although FV vectors integrate into the host genome of transduced cells, little is known about where integration occurs. Previous studies mapped 12 FV vector integrations to both introns and intergenic regions in human hematopoietic cells (29) and 7 FV integrations in satellite DNA and ribosomal DNA in a murine fibroblast cell line (32).
Importantly, wild-type FVs are not pathogenic in their natural hosts (33) or in humans exposed to primate FVs (34). Despite their prevalence in well studied captive primate populations used for research, FVs have not been associated with the development of malignancies. Although this lack of pathogenicity is reassuring and contrasts with the biology of MLV, it could be caused by cytotoxic effects of the wild-type virus rather than differences in integration site selection. Depending on where proviruses integrate, replication-incompetent FV vectors that contain strong internal promoters and enhancers may activate protooncogenes. To determine where FV proviruses integrate, we have performed a large-scale analysis of FV vector integration sites in normal human cells and compared our results with those obtained with other integrating vectors.
Results
We used a shuttle vector system to rescue and sequence provirus integration sites (Fig. 1). The FV vector ΔΦPFmcsNO contains a GFP gene under the control of a phosphoglycerate kinase (PGK) promoter that allowed us to measure transduction frequencies, and plasmid sequences consisting of a Tn5 promoter, neomycin resistance gene (neo), and p15A replication origin that function in bacteria. Normal human fibroblasts and human CD34+ peripheral blood cells were infected with ΔΦPFmcsNO and cultured without selection to allow time for integration to occur. Genomic DNA was then isolated from infected cells, digested with specific restriction enzymes to release the bacterial plasmid and flanking genomic sequences from each provirus, circularized, and rescued in Escherichia coli. We sequenced a total of 4,963 rescued plasmids. Different restriction enzymes were used to avoid potential biases in recognition site distribution, and several criteria were used to exclude sequence reads that might lead to inaccuracies, including those with deleted LTRs, poor alignments, and low blast scores (see Table 2, which is published as supporting information on the PNAS web site). A total of 638 plasmids contained junction sequences that were identical to other plasmid junctions, and these were considered to be duplicate recoveries of the same provirus. With these criteria, 1,008 and 1,821 unique integrants were localized to build 35 of the human genome sequence in fibroblasts and CD34+ cells, respectively. Five integrants (0.18%) mapped to the 43-kb ribosomal DNA repeats, which are not localized in the genome sequence but are present at an estimated 400 copies in a diploid human genome (35), and comprise ≈0.29% of human DNA.
Fig. 1.
FV shuttle vector rescue strategy. The shuttle vector ΔΦPFmcsNO is based on the ΔΦ (deleted foamy) backbone (25) and is shown with its PGK promoter, GFP transgene, multiple cloning site, neo gene driven by a Tn5-derived bacterial promoter, p15A bacterial replication origin, and U3-deleted LTRs (ΔLTR). To recover provirus junctions, genomic DNA from infected cells was digested with multiple cloning site enzyme(s) that have compatible sticky ends (e.g., PciI and BspHI). Fragments containing the Tn5 promoter, neo gene, p15A origin, 3′ LTR, and flanking chromosomal DNA were circularized by ligation and transferred to bacteria, where they replicate and confer kanamycin resistance. Unwanted fragments containing 5′ vector sequences or bacterial plasmid DNA were destroyed by digestion with the rare-cutting endonuclease I-SceI and the methylation-sensitive enzyme DpnI, respectively. Bacterial colonies were picked and their LTR/chromosome junctions were sequenced with a primer in the LTR.
To provide an appropriate data set for comparison, we also rescued and sequenced proviruses from CD34+ cells infected with HIV-1-based vector HIV-PYmcsNO, which expresses yellow fluorescent protein from the PGK promoter and contains the same shuttle vector sequences as ΔΦPFmcsNO. A total of 1,331 HIV vector integrants were localized to the human genome sequence by using the same criteria as for FV vectors (see Table 2).
The chromosomal features of the FV vector integration sites are shown in Table 1. As a control, we generated a set of 10,000 sequences localized to random positions in the human genome that were processed the same way as our provirus sequence reads (see Materials and Methods). We also compared our results to HIV, MLV, ASV, and adeno-associated virus (AAV; a parvovirus that has also been developed as an integrating vector) integration sites identified previously in other experiments. The data we obtained for HIV vectors with the shuttle vector approach showed remarkably similar integration frequencies to those obtained by other investigators with PCR-based approaches (HIVshuttle vs. HIVpcr; Table 1). This finding supports the conclusion that differences seen between FV and other vectors were not caused by the method used to isolate junctions and suggests that neither method imposes a large bias to the data. This comparison also demonstrates that integration preferences are similar in CD34+ cells and transformed cell lines, at least for HIV vectors.
Table 1. Genomic features of integration sites.
| Percent of integrations
|
|||||||
|---|---|---|---|---|---|---|---|
| Repeat elements‡
|
|||||||
| Virus or vector | Unique sites | RefSeq genes* | CpG islands† | LINE | SINE | DNA | LTR |
| FVFibro§ | 1,008 | 24.0 | 5.85 | 15.7 | 12.3 | 3.57 | 9.42 |
| FVCD34¶ | 1,821 | 31.1 | 12.1 | 14.6 | 16.6 | 2.91 | 6.48 |
| FVAll | 2,829 | 28.6 | 9.93 | 15.0 | 15.1 | 3.14 | 7.52 |
| Random | 10,000 | 34.1 | 2.56 | 20.0 | 13.5 | 3.20 | 8.78 |
| HIVshuttle∥ | 1,331 | 69.1 | 1.50 | 17.1 | 18.2 | 3.83 | 3.83 |
| HIVpcr** | 1,757 | 65.2 | 1.48 | 18.1 | 15.4 | 3.36 | 3.87 |
| MLV†† | 644 | 43.3 | 17.7 | 10.4 | 13.2 | 2.64 | 6.06 |
| ASV‡‡ | 480 | 42.9 | 2.71 | 20.6 | 12.7 | 2.92 | 7.29 |
| AAV§§ | 670 | 38.8 | 4.03 | 18.51 | 13.58 | 3.13 | 5.67 |
Sites in transcribed regions of RefSeq genes.
Sites inside of or within 1 kb of CpG islands.
Repetitive elements as defined at the University of California, Santa Cruz genome browser.
Sites analyzed by our criteria from published sequences (2, 3, 5) in primary lung fibroblasts, peripheral blood mononuclear cells, Sup T1 cells, H9 and HeLa cells: Genbank accession numbers CL528773-CL529239, CL529240-CL529767, BH610086-BH609398, AY516881-AY517244, and AY517245-AY517469, respectively.
Sites analyzed by our criteria from published sequences (3) in HeLa cells: Genbank accession numbers AY515855-AY516880.
Sites analyzed by our criteria from published data (4, 5) in HeLa cells and 293T-TVA cells: Genbank accession numbers AY653309-AY653534 and CL528303-CL528772, respectively.
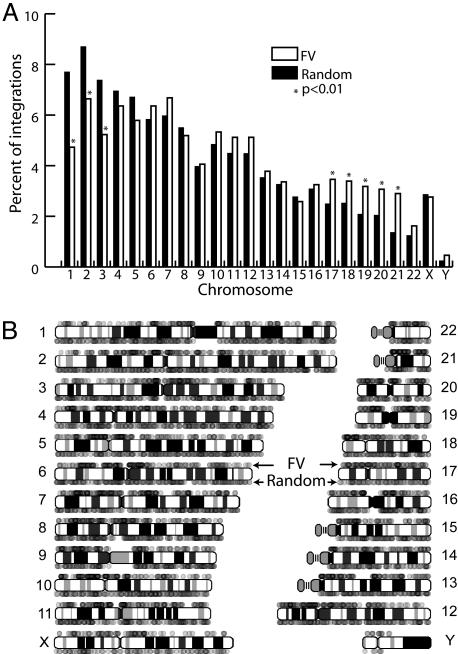
Compared with the random control set, FV vectors preferentially integrated in or near CpG islands, and there were fewer integrants than expected in genes and long interspersed nuclear element (LINE) repeats. In contrast, the three other types of retroviruses all integrated preferentially in genes. MLV vectors were similar to FV vectors regarding their preference for CpG islands and reduced integration in LINE repeats. FV vector proviruses were found in all human chromosomes (Fig. 2A). Relative to the set of random sites, the three largest chromosomes had fewer proviruses than expected, whereas several of the smallest chromosomes (17-21) had more proviruses than expected. The cytogenetic distributions of both FV vector and random sites are shown in Fig. 2B. The FV sites exhibited both clustering and gaps, which were quantified by determining the distances between neighboring integration sites (Fig. 3A). Compared with random sites, FV vectors were more likely to integrate near other integration sites, with almost 1% of neighboring integrants lying within 1 kb of each other, as opposed to 0.11% of neighboring random sites. This result can be appreciated another way by examining the number of hotspots containing three or more integrants in different-sized regions (Fig. 3B). Interestingly, neighboring FV vector integration sites were also more likely to be farther apart from each other than random sites, especially for gaps of >104 kb (Fig. 3A). Three of these large gaps were centromeric regions where integrations could not be localized for both FV and random sites, whereas there were eight large noncentromeric gaps found only in the FV data set. The largest noncentromeric gap for FV integrants was 17 Mb and can easily be identified on the cytogenetic map of chromosome 13 (Fig. 2B).
Fig. 2.
Chromosomal distribution of integration sites. (A) FV vector integration sites from both CD34+ cells and fibroblasts (n = 2,829) are plotted as a percentage of all integrants in each chromosome and compared with a set of random sites (n = 10,000). Asterisks mark chromosomes with significantly different integration frequencies (P < 0.01). (B) The same FV vector integration sites are represented as individual dots positioned above each human chromosome and compared with 2,829 random sites placed below the chromosomes. Dots are 20% opaque to display multiple overlapping integrants.
Fig. 3.
Clusters and gaps of FV vector integration sites. (A) The distances between adjacent unique FV vector integration sites were determined and binned by size, with the percent of proviruses within each bin plotted. (B) The number of FV vector integration hotspots (defined as three integrations) was determined for each sequence window size from 2.5 to 100 kb. Three size-matched sets of 2,829 random sites were also plotted with standard deviations as controls. Asterisks mark significant differences (P < 0.01).
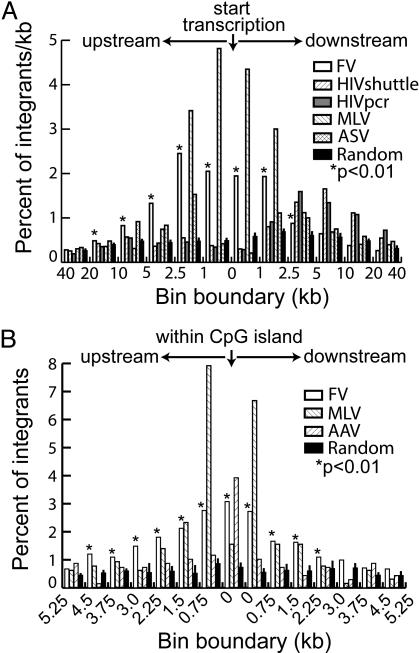
Additional analyses were performed to assess the relationship of integration to transcription. Although FV vectors did not preferentially integrate in genes, there was a modest preference for integrating near transcription start sites (Fig. 4A). When compared with other retroviruses, the FV vector preference was similar to MLV in that it was greatest within 2.5 kb upstream or downstream of start sites (3), but of a lesser magnitude (10.6% of all FV integrants as compared with 18.7% for MLV). In contrast, HIV integration sites were preferentially located throughout transcription units and not at start sites, and ASV had more modest preferences in relation to transcription units, in agreement with prior analyses (2, 4, 5).
Fig. 4.
Integration and transcription units. (A) The positions of FV (n = 2,829), HIV sites isolated by shuttle vector rescue (HIVshuttle, n = 1,331), HIV sites isolated by PCR (HIVpcr, n = 1,757), MLV (n = 644), and ASV (n = 480) integration sites were mapped relative to RefSeq gene transcription start sites, binned into different size sequence windows, and plotted as the percent of all integrations per kb. (B) The positions of FV (n = 2,829), MLV (n = 644), and AAV (n = 670) integration sites were mapped relative to those of CpG islands (GC content ≥50%, length >200 bp, ratio of observed to expected number of CpG dinucleotides >0.6). Integration sites in CpG islands and 14 0.75-kb windows flanking each island (the average length of a CpG island is 764 bp) were binned and plotted as the percent of all sites. A set of random sites (n = 10,000) were included as controls, and asterisks mark significant differences (FV vs. random; P < 0.01). See Table 1 for details on non-FV vector integration sites.
CpG islands are sequences rich in CpG dinucleotides that are frequently found near promoter regions and may regulate transcription (36, 37). As shown in Table 1, FV, MLV, and AAV vectors all integrated preferentially within 1 kb of CpG islands. However, on further examination there were distinct differences for all three vector types (Fig. 4B). FV integration preferences were greatest within CpG islands and steadily decreased in sequences extending outward. MLV integrated preferentially in sequences flanking CpG islands, but less so within islands, and AAV vectors integrated preferentially within islands but not in flanking regions. FV vectors also integrated more frequently near CpG islands in CD34+ cells than in fibroblasts (see Table 1; P < 0.01).
To determine whether restriction enzyme site distributions influenced these analyses, we plotted the locations of recognition sites for each restriction enzyme or enzyme combination used relative to Reference Sequence (RefSeq) genes (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/RefSeq) and CpG islands (Figs. 6 and 7, which are published as supporting information on the PNAS web site). Although enzyme-specific distributions were observed, they did not account for the distribution of FV vector integrants relative to these genomic features. However, in the case of published MLV data (3), the low number of integrants present within CpG islands may have been caused by a lack of MseI sites (Fig. 8, which is published as supporting information on the PNAS web site).
Gene expression arrays were used to determine whether integration occurred preferentially in expressed genes. We ranked 13,069 RefSeq genes by expression level in uninfected and infected CD34+ cells and identified those where FV vectors integrated (Fig. 5). The average expression rank of genes where FV vectors integrated within the transcribed region was 1.077- to 1.083-fold above that of genes where random sites were located in both infected and uninfected cells. The average expression rank of genes where integrations occurred within 10 kb of transcription starts was 1.061- to 1.066-fold above that of random sites. These results suggest that gene expression did not have a major impact on FV vector integration. This finding was also confirmed by comparing the expression levels of genes where FV vectors integrated in fibroblasts and CD34+ cells, which showed little effect of cell-type-specific expression on integration (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 5.
Transcriptional profiling of FV vector integration sites in CD34+ cells. RNA samples from uninfected and FV vector-infected CD34+ cells were hybridized to the human U133A plus 2.0 gene chip array (Affymetrix), and the expression levels of RefSeq genes with unique probe sets were ranked and plotted (gray dots, opacity 50%). RefSeq genes containing FV vector integrations are plotted as red circles. The average expression ranks are shown for all genes containing an FV vector integrant (n = 423; blue square), genes containing an FV vector integrant within 10 kb of the transcription start site (n = 389; green square), genes containing computer-generated random sites (n = 2,567; yellow square), and all RefSeq genes analyzed (n = 13,069; white circle).
Discussion
In this study we used a shuttle vector system to rescue integrated FV vector proviruses as bacterial plasmids, and robotic processes were used to purify plasmid DNA and sequence junctions. Bacterial rescue of retroviral shuttle vectors is a well established technique (38, 39), and here we have used this efficient and scalable method to recover thousands of proviruses from normal human cells without selection. There are certain advantages to this approach as compared with more commonly used PCR-based methods. The length of junction sequences recovered is not limited by the amplification steps of PCR, and the sequence obtained from plasmids is of high quality, so long sequence reads are generated. The average match length in our FV vector experiments was 337 bp, whereas the PCR methods used to sequence MLV and HIV integration junctions (2, 3) generated average match lengths of 107 and 236 bp, respectively when aligned by our criteria. This increase in sequence length allows a more accurate localization of junction sequences, especially those containing repeats, and additional sequence can be obtained from recovered plasmids if necessary. The major disadvantage of our approach is that the vector must include a bacterial replication origin and selectable marker. Although these would not typically be included in a gene therapy vector, they could be built into vectors when size permits. The similar results obtained for HIV integration sites generated by plasmid rescue or PCR validates both methods and strongly supports the conclusion that FV vectors have distinct integration preferences as compared with other integrating vectors.
FV vectors were found to integrate in a nonrandom distribution, with preferences for CpG islands and transcription start sites, and fewer integrations than expected in long interspersed nuclear elements. Although some of these patterns were similar to other integrating viruses that have been studied, in particular MLV and AAV, the combination of properties was unique to FV vectors. Thus there may be both common and distinct mechanisms in the life cycles of viruses before or during integration. Clustering of FV integrants was observed, and because of the large number of integrants localized, we could identify several chromosomal regions where FV vectors did not integrate. The basis for these gaps is not clear, as they contain genes expressed in both fibroblasts and CD34+ cells (data not shown) and thus are not simply inactive chromatin. The same regions also contained integrants of other retroviruses studied in different cell lines and HIV vector integrants isolated by shuttle vector rescue in CD34+ cells (data not shown).
A key issue addressed by our study is the potential for insertional mutagenesis by FV vectors and how this compares with other integrating vectors that might be used in gene therapy. Unlike MLV or HIV, FV vectors did not show an overall preference for integrating within genes (Table 1), and their preferences for integrating near transcription start sites and CpG islands were lower than those of MLV (Fig. 4), which may decrease the risk of mutating and/or activating cellular genes. We examined more directly the possibility of protooncogene activation and identified 126 FV vector integrants (4.4%) within protooncogene transcripts or 50 kb upstream from their start site (Table 3, which is published as supporting information on the PNAS web site). This percentage was slightly higher (P < 0.01) than the average obtained from three sets of 2,829 randomly generated sites (3.2% ± standard deviation of 0.5%), but lower (P < 0.01) than the HIV (shuttle 7.0%, PCR 6.7%) or MLV (7.0%) data sets.
One of four hotspots containing four or more FV vector integrants within 50 kb (Fig. 10, which is published as supporting information on the PNAS web site), was 470 kb from the EVI1 locus on chromosome 3, originally identified as a common MLV integration site in myeloid tumors of mice (40). It is not clear whether these integrants are close enough to influence EVI1 expression, although in mice an MLV vector provirus located >500 kb away from the start site increased Evi1 transcription (41). Interestingly, in nonhuman primates several MLV vector integrations were also observed near EVI1, but in the adjacent MDS1 gene that partially overlaps some EVI1 transcripts in humans (6). The closest FV vector integrant to the LMO2 protooncogene activated by MLV vector insertions in leukemias that developed in the X-linked severe combined immune deficiency trial (1) was ≈700 kb away.
Although our findings suggest that FV vectors may have a safer spectrum of insertional mutagenesis compared with MLV or HIV vectors, it is important to note that the integration preferences described generally varied by 2-fold or less, which would not provide a significant improvement in safety. Other factors are likely to play a more important role in determining the oncogenic risk of vectors, including differences in enhancer activity, potential insulating effects of proviral sequences, the number of integrations that occur in a clinical trial, and the cell types in which they occur. The lack of pathogenicity of wild-type FV is encouraging in this regard, but does not take into account potential effects of internal promoters and transgenes included in vectors. Future experiments will be required to address these factors and improve the safety of integrating viral vectors.
Materials and Methods
Vector Production. The FV vector plasmid pΔΦPFmcsNO was constructed by using standard molecular biology techniques by replacing the murine stem cell virus promoter of pΔΦMscvF (25) with a murine PGK promoter and adding bacterial plasmid sequences including a Tn5-driven neo gene and p15A replication origin (42), and a multiple cloning site containing I-SceI, BamHI, AvrII, PciI, NsiI, NdeI, and Bsp120I recognition sites. The HIV-1-based shuttle vector HIV-PYmcsNO also contains the same Tn5-driven neo gene, p15A replication origin, and multiple cloning site and was made from pRRL.sinCPPT.Pgk.EYFP.wpre (kindly provided by Luigi Naldini, Fondazione San Raffaele del Monte Tabor, Milan), which contains a yellow fluorescent protein in place of the GFP in pRRLsin.hPGK.EGFP.Wpre vector (43). FV vector preparations were produced by calcium phosphate transfection as described (25, 44) except that 12 μg of pΔΦPFmcsNO, 12 μg of pCiGSΔΨ (28), 1.6 μg of pCiPS, and 0.75 μg of pCiES in a total volume of 800 μl were used for each 10-cm tissue culture dish. HIV vector preparations were produced as described (45). Stocks were titered by determining the number of GFP-transducing units on normal human fibroblasts or human HT-1080 fibrosarcoma cells.
Cell Culture and Infections. Normal human fibroblasts obtained from the Coriell Institute for Medical Research (Camden, NJ; repository GM05387) and HT-1080 cells (46) were cultured in DMEM with 10% heat-inactivated (56°C for 30 min) FBS (HyClone, Logan, UT), 1.25 μg/ml amphotericin, 100 units/ml of penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 atmosphere. For fibroblast infections, two 10-cm dishes were each seeded with 5 × 105 cells on day 1, vector ΔΦPFmcsNO was added at a multiplicity of infection of 0.5 on day 2, the cells were expanded to four 10-cm dishes on day 5, and DNA was isolated on day 8.
Human CD34+ cells were collected from three male volunteers and isolated by magnetic beads (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions and stored in liquid nitrogen until use. Cells were thawed on day 1 and cultured in transduction medium (DMEM with 20% FBS, 100 ng/ml Flt-3 ligand, 100 ng/ml stem cell factor, and 100 ng/ml thrombopoietin). For infections, 5 × 105 CD34+ cells thawed 1 day previously were added to each well of a 6-well plate pretreated with 50 μg/ml CH-296 fibronectin fragment (Takara, New York), and exposed to vector for 10 h at multiplicity of infection (MOI) 1.5 (ΔΦPFmcsNO) or 18 h at MOI 5 (HIV-PYmcsNO). Cells were then washed, resuspended in transduction medium supplemented with 20 ng/ml IL-3 and 20 ng/ml IL-6, and plated in a 6-well plate. RNA was isolated for gene expression analysis 48 h later, and DNA was isolated 4 days later. All cytokines were obtained from PeproTech (Rocky Hill, NJ) and were of (recombinant) human origin.
Provirus Rescue and Sequencing of Junctions. DNA from infected fibroblasts or CD34+ cells was isolated by using a Puregene isolation kit (Gentra Systems) following the manufacturer's instructions, followed by extraction with phenol and chloroform and ethanol precipitation. Twenty micrograms of DNA was digested for 3 h with 40 units of either NdeI, NsiI, AvrII, and SpeI, or BspHI and PciI, and the DNA fragments were extracted with phenol and chloroform, precipitated with ethanol, and resuspended in 355 μl of H2O. For ligations, 5 μl of T4 DNA ligase (400 units/μl; New England Biolabs) and 40 μl of 10× ligation buffer were added, incubated overnight at 15°C, heat-inactivated at 65°C for 10 min, and cooled to room temperature. Twenty units of I-SceI was added and incubated at 37°C for 1 h to eliminate ligated fragments containing sequences upstream of the multiple cloning site. The reaction was then adjusted to 50 mM NaCl and digested with 40 units of methylation-sensitive DpnI for 30 min at 37°C to eliminate bacterial plasmids that may have been present in the vector stock. The DNA was then extracted with phenol and chloroform, precipitated with ethanol, and resuspended in 10 μl of 1 mM Tris (pH 8.0) buffer with 0.1 mM EDTA. DH10B cells (47) were transformed by electroporation with 1 μl (≈1-2 μg of DNA) at a time, and transformed bacteria were grown on agar containing 50 μg/ml kanamycin. Kanamycin-resistant colonies were picked by using a Qpix2 robot (Genetix, Boston), and plasmid DNA containing rescued proviruses was purified and sequenced as described (48) by using FV sequencing primer 5′-AAACCGACTTGATTCGAGAACC or HIV sequencing primer 5′-TCTCTCTGGTTAGACCAGATCTGAGC located in the respective LTRs. Sequencing reactions were electrophoresed on ABI prism 3700 (Applied Biosystems capillary sequencers and analyzed by using phred/phrap/consed software tools (49-51).
Microarray Analysis of Gene Expression. RNA from fibroblasts and CD34+ cells was isolated with the RNeasy kit and Qiashredder column (Qiagen, Valencia, CA) following the manufacturer's instructions. Five micrograms of total RNA was labeled and 15 μg of cRNA was used per HG-U133 plus 2.0 array (Affymetrix, Santa Clara, CA) per the manufacturer's instructions. The subset of probes that identify specific RefSeq transcripts (13,069) were used in our analysis. Probe sets that hybridized with more than one gene were excluded. Expression levels were determined by using Affymetrix gcos 1.1.1 software, and when multiple probe sets reflected the expression level of a single RefSeq gene, the average expression level was used in the rankings.
Analysis of Integration Sites. DNA sequences were processed with perl programs, and sequences containing an LTR junction were truncated at 500 bp, or at the first occurrence of a restriction site used during rescue. The resulting junction sequences were aligned to build 35 of the human genome, the vector plasmid sequence, and human ribosomal DNA by using a stand-alone version of blat (52) that generates a blast alignment score. The input script was as follows: blat chromosome_file query_file -out=blast8 -ooc=11.ooc output_file. Alignments were sorted by blast score, and those with the five highest scores were saved for further processing. To ensure that sequences represented true junctions and were accurately localized, additional screening criteria were used as described in Table 2. The average match length was 337 bp for all FV sequences and 301 bp for all HIV sequences. perl programs were also used to compare localized integration sites to various chromosomal features by using tables available from the University of California, Santa Cruz database (53) and to determine the positions of restriction enzyme sites in the human genome. Additional published retroviral integration sites (2-5) and AAV vector integration sites (54) were processed as above for comparison (GenBank accession numbers in Table 1).
We also produced a randomly localized set of genomic positions by generating random numbers between 1 and 5,941,037,819 (the size of the build 35 diploid male genome with chromosomes laid end to end) with the perl “rand” function and converting to chromosomal positions by splitting the numeric range of the diploid genome into separate chromosomes. These chromosomal positions were used to extract 337 bp of sequence from build 35 of the human genome at each randomly determined position, and the resulting files were aligned with the genome by using blat as described above. A set of 10,000 positions localized by this method was used as a control data set (“random integrants”). To analyze clustering and hotspots, we used similar sets of 2,829 random genomic positions as size-matched controls.
To identify FV vector integrants in protooncogenes or near their promoters we searched the Sanger Institute Cancer Gene Census Table (www.sanger.ac.uk/genetics/CGP/Census), which contains 331 unique potential oncogenes with corresponding RefSeq transcripts (as of April 8, 2005), and the Mouse Retrovirus Tagged Cancer Gene Database (http://rtcgd.ncifcrf.gov), which contains 363 potential oncogenes with corresponding RefSeq transcripts (as of November 5, 2005). perl programs were used to identify RefSeq genes that contained FV vector integration sites or random sites within their transcribed regions or within 50 kb upstream of the transcript start site.
Statistical Analysis. Statistical significance was determined by using the χ2 test. P values < 0.01 were considered significant.
Supplementary Material
Acknowledgments
We thank Maynard Olson and the University of Washington Genome Center for helpful advice and sequencing, Erik Olson and Tulin Okbinoglu for help with stock production, the Center for Expression Arrays (University of Washington) for microarray analysis, Shelly Heimfeld and the Fred Hutchinson Cell Processing Core for isolation of human CD34+ cells, and Keiko Akagi and Neal Copeland for providing a table of human genes homologous to potential mouse oncogenes from the Retrovirus Tagged Cancer Gene Database. This work was supported by grants from the National Institutes of Health, the Child Health Research Center at Children's Hospital, Seattle, and the University of Washington Center for AIDS Research.
Author contributions: G.D.T., D.G.M., H.-P.K., R.K., and D.W.R. designed research; G.D.T., D.G.M., M.A.J., and J.M.A. performed research; G.D.T., D.G.M., M.A.J., H.-P.K., and D.W.R. analyzed data; and G.D.T. and D.W.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FV, foamy virus; MLV, murine leukemia virus; ASV, avian sarcoma virus; AAV, adeno-associated virus; PGK, phosphoglycerate kinase; RefSeq, Reference Sequence.
References
- 1.Hacein-Bey-Abina, S., von Kalle, C., Schmidt, M., Le Deist, F., Wulffraat, N., McIntyre, E., Radford, I., Villeval, J. L., Fraser, C. C., Cavazzana-Calvo, M. & Fischer, A. (2003) N. Engl. J. Med. 348, 255-256. [DOI] [PubMed] [Google Scholar]
- 2.Schroder, A. R., Shinn, P., Chen, H., Berry, C., Ecker, J. R. & Bushman, F. (2002) Cell 110, 521-529. [DOI] [PubMed] [Google Scholar]
- 3.Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 4.Narezkina, A., Taganov, K. D., Litwin, S., Stoyanova, R., Hayashi, J., Seeger, C., Skalka, A. M. & Katz, R. A. (2004) J. Virol. 78, 11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell, R. S., Beitzel, B. F., Schroder, A. R., Shinn, P., Chen, H., Berry, C. C., Ecker, J. R. & Bushman, F. D. (2004) PLoS Biol. 2, E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hematti, P., Hong, B. K., Ferguson, C., Adler, R., Hanawa, H., Sellers, S., Holt, I. E., Eckfeldt, C. E., Sharma, Y., Schmidt, M., et al. (2004) PLoS Biol. 2, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooks, J. J. & Gibbs, C. J., Jr. (1975) Bacteriol. Rev. 39, 169-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweizer, M., Turek, R., Hahn, H., Schliephake, A., Netzer, K. O., Eder, G., Reinhardt, M., Rethwilm, A. & Neumann Haefelin, D. (1995) AIDS Res. Hum. Retroviruses 11, 161-170. [DOI] [PubMed] [Google Scholar]
- 9.Achong, B. G., Mansell, P. W., Epstein, M. A. & Clifford, P. (1971) J. Natl. Cancer Inst. 46, 299-307. [PubMed] [Google Scholar]
- 10.Schweizer, M. & Neumann-Haefelin, D. (1995) Virology 207, 577-582. [DOI] [PubMed] [Google Scholar]
- 11.Herchenroder, O., Renne, R., Loncar, D., Cobb, E. K., Murthy, K. K., Schneider, J., Mergia, A. & Luciw, P. A. (1994) Virology 201, 187-199. [DOI] [PubMed] [Google Scholar]
- 12.Rethwilm, A., Erlwein, O., Baunach, G., Maurer, B. & ter Meulen, V. (1991) Proc. Natl. Acad. Sci. USA 88, 941-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatesh, L. K., Theodorakis, P. A. & Chinnadurai, G. (1991) Nucleic Acids Res. 19, 3661-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lochelt, M., Muranyi, W. & Flugel, R. M. (1993) Proc. Natl. Acad. Sci. USA 90, 7317-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moebes, A., Enssle, J., Bieniasz, P. D., Heinkelein, M., Lindemann, D., Bock, M., McClure, M. O. & Rethwilm, A. (1997) J. Virol. 71, 7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu, S. F., Baldwin, D. N., Gwynn, S. R., Yendapalli, S. & Linial, M. L. (1996) Science 271, 1579-1582. [DOI] [PubMed] [Google Scholar]
- 17.Erlwein, O., Bieniasz, P. D. & McClure, M. O. (1998) J. Virol. 72, 5510-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinkelein, M., Schmidt, M., Fischer, N., Moebes, A., Lindemann, D., Enssle, J. & Rethwilm, A. (1998) J. Virol. 72, 6307-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, M., Chari, S., Yanchis, T. & Mergia, A. (1998) J. Virol. 72, 3451-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, M. & Rethwilm, A. (1995) Virology 210, 167-178. [DOI] [PubMed] [Google Scholar]
- 21.Russell, D. W. & Miller, A. D. (1996) J. Virol. 70, 217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieniasz, P. D., Erlwein, O., Aguzzi, A., Rethwilm, A. & McClure, M. O. (1997) Virology 235, 65-72. [DOI] [PubMed] [Google Scholar]
- 23.Mergia, A. & Heinkelein, M. (2003) Curr. Top. Microbiol. Immunol. 277, 131-159. [DOI] [PubMed] [Google Scholar]
- 24.Hill, C. L., Bieniasz, P. D. & McClure, M. O. (1999) J. Gen. Virol. 80, 2003-2009. [DOI] [PubMed] [Google Scholar]
- 25.Trobridge, G., Josephson, N., Vassilopoulos, G., Mac, J. & Russell, D. W. (2002) Mol. Ther. 6, 321-328. [DOI] [PubMed] [Google Scholar]
- 26.Vassilopoulos, G., Trobridge, G., Josephson, N. C. & Russell, D. W. (2001) Blood 98, 604-609. [DOI] [PubMed] [Google Scholar]
- 27.Trobridge, G. & Russell, D. W. (2004) J. Virol. 78, 2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josephson, N. C., Trobridge, G. & Russell, D. W. (2004) Hum. Gene Ther. 15, 87-92. [DOI] [PubMed] [Google Scholar]
- 29.Josephson, N. C., Vassilopoulos, G., Trobridge, G. D., Priestley, G. V., Wood, B. L., Papayannopoulou, T. & Russell, D. W. (2002) Proc. Natl. Acad. Sci. USA 99, 8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leurs, C., Jansen, M., Pollok, K. E., Heinkelein, M., Schmidt, M., Wissler, M., Lindemann, D., Von Kalle, C., Rethwilm, A., Williams, D. A. & Hanenberg, H. (2003) Hum. Gene Ther. 14, 509-519. [DOI] [PubMed] [Google Scholar]
- 31.Zucali, J. R., Ciccarone, T., Kelley, V., Park, J., Johnson, C. M. & Mergia, A. (2002) 302, 229-235. [DOI] [PubMed]
- 32.Neves, M., Peries, J. & Saib, A. (1998) Res. Virol. 149, 393-401. [DOI] [PubMed] [Google Scholar]
- 33.Falcone, V., Schweizer, M. & Neumann-Haefelin, D. (2003) Curr. Top. Microbiol. Immunol. 277, 161-180. [DOI] [PubMed] [Google Scholar]
- 34.Heneine, W., Schweizer, M., Sandstrom, P. & Folks, T. (2003) Curr. Top. Microbiol. Immunol. 277, 181-196. [DOI] [PubMed] [Google Scholar]
- 35.Bross, K. & Krone, W. (1972) Humangenetik 14, 137-141. [DOI] [PubMed] [Google Scholar]
- 36.Bird, A. P. (1986) Nature 321, 209-213. [DOI] [PubMed] [Google Scholar]
- 37.Larsen, F., Gundersen, G., Lopez, R. & Prydz, H. (1992) Genomics 13, 1095-1107. [DOI] [PubMed] [Google Scholar]
- 38.Berger, S. A. & Bernstein, A. (1985) Mol. Cell. Biol. 5, 305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cepko, C. L., Roberts, B. E. & Mulligan, R. C. (1984) Cell 37, 1053-1062. [DOI] [PubMed] [Google Scholar]
- 40.Mucenski, M. L., Taylor, B. A., Ihle, J. N., Hartley, J. W., Morse, H. C., 3rd, Jenkins, N. A. & Copeland, N. G. (1988) Mol. Cell. Biol. 8, 301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kustikova, O., Fehse, B., Modlich, U., Yang, M., Dullmann, J., Kamino, K., von Neuhoff, N., Schlegelberger, B., Li, Z. & Baum, C. (2005) Science 308, 1171-1174. [DOI] [PubMed] [Google Scholar]
- 42.Rutledge, E. A. & Russell, D. W. (1997) J. Virol. 71, 8429-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Follenzi, A., Ailles, L. E., Bakovic, S., Geuna, M. & Naldini, L. (2000) Nat. Genet. 25, 217-222. [DOI] [PubMed] [Google Scholar]
- 44.Trobridge, G., Vassilopoulos, G., Josephson, N. & Russell, D. W. (2002) Methods Enzymol. 346, 628-648. [DOI] [PubMed] [Google Scholar]
- 45.Horn, P. A., Morris, J. C., Bukovsky, A. A., Andrews, R. G., Naldini, L., Kurre, P. & Kiem, H. P. (2002) Gene Ther. 9, 1464-1471. [DOI] [PubMed] [Google Scholar]
- 46.Rasheed, S., Nelson Rees, W. A., Toth, E. M., Arnstein, P. & Gardner, M. B. (1974) Cancer 33, 1027-1033. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan, D., Jessee, J. & Bloom, F. R. (1991) Methods Enzymol. 204, 63-113. [DOI] [PubMed] [Google Scholar]
- 48.International Human Genome Sequencing Consortium (2004) Nature 431, 931-945. [DOI] [PubMed] [Google Scholar]
- 49.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 50.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 51.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 52.Kent, W. J. (2002) Genome Res. 12, 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karolchik, D., Hinrichs, A. S., Furey, T. S., Roskin, K. M., Sugnet, C. W., Haussler, D. & Kent, W. J. (2004) Nucleic Acids Res. 32, D493-D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller, D. G., Trobridge, G. D., Petek, L. M., Jacobs, M. A., Kaul, R. & Russell, D. W. (2005) J. Virol. 79, 11434-11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.