Abstract
The active transport of the plant hormone auxin plays a major role in the initiation of organs at the shoot apex. Polar localized membrane proteins of the PIN1 family facilitate this transport, and recent observations suggest that auxin maxima created by these proteins are at the basis of organ initiation. This hypothesis is based on the visual, qualitative characterization of the complex distribution patterns of the PIN1 protein in Arabidopsis. To take these analyses further, we investigated the properties of the patterns using computational modeling. The simulations reveal previously undescribed properties of PIN1 distribution. In particular, they suggest an important role for the meristem summit in the distribution of auxin. We confirm these predictions by further experimentation and propose a detailed model for the dynamics of auxin fluxes at the shoot apex.
Keywords: auxin, modeling, shoot meristem
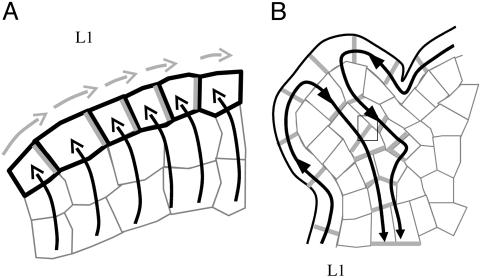
There is strong evidence that active auxin transport, generated by influx and efflux carriers, creates patterns of auxin distribution at the shoot apex. This distribution is, in turn, interpreted in terms of differential growth and cell differentiation (1-3). In Arabidopsis, AUX1, a putative influx transporter (4), is mainly located in the surface layer (L1) of the shoot apical meristem (2) (Fig. 1A). Interestingly, the protein seems to be homogeneously distributed in plasma membranes of the individual cells. Therefore, it has been proposed that AUX1 helps to restrict auxin to these layers, although additional mechanisms may be required (5). The efflux facilitator PIN1 also is localized in the surface layers of the meristem, but in contrast to AUX1 it is often localized on certain anticlinal sides of the cells only. Because neighboring cells often show coherent PIN1 positioning, it was proposed that PIN1 is responsible for directed hormone flows within the meristem L1 layer (Fig. 1 A). In particular, careful immunological studies have revealed that the membranes carrying PIN1 are preferentially oriented toward the incipient primordia, suggesting auxin transport toward the young organs (2, 3).
Fig. 1.
Models for auxin transport in the shoot apical meristem. (A) The putative auxin influx carrier AUX1, represented in black, is homogeneously distributed on the cell membranes of the surface layer of the meristem, whereas the putative auxin efflux carrier PIN1, represented in gray, seems to have a polarized localization. As proposed by ref. 2, AUX1 would help to concentrate auxin in the surface layer (black arrows), and PIN1 would direct auxin fluxes (gray arrows) within these layers. Note that additional mechanisms responsible for auxin influx into the L1 layer have been proposed (5). (B) In the provascular tissues of young primordia, PIN1 is oriented downward, evacuating auxin from the meristem surface (black arrows) to deeper tissues. Consequently, the primordia act as auxin sinks.
Together, the observations so far suggest a dynamic scenario where auxin is transported to the meristem from basally localized tissues via the L1 layer. At the meristem surface, auxin is redistributed and accumulates at particular sites where it will induce the initiation of new organs. This accumulation subsequently leads to the activation of transport in the provascular tissues causing an inward directed flow (Fig. 1B). The young organ is thus transformed into an auxin sink, which depletes its surroundings from auxin and prevents the formation of new primordia in its vicinity.
Although this scenario is relatively straightforward, the previous observations leave a number of questions open. First, it is not clear at all why auxin should start to accumulate at the site where a primordium will be initiated. Second, the immunolabelings reveal a very complex distribution of PIN1 proteins (Fig. 2). As a result, the interpretation of these patterns in terms of cell-cell interaction networks and, more specifically, in terms of auxin distribution remains extremely difficult.
Fig. 2.
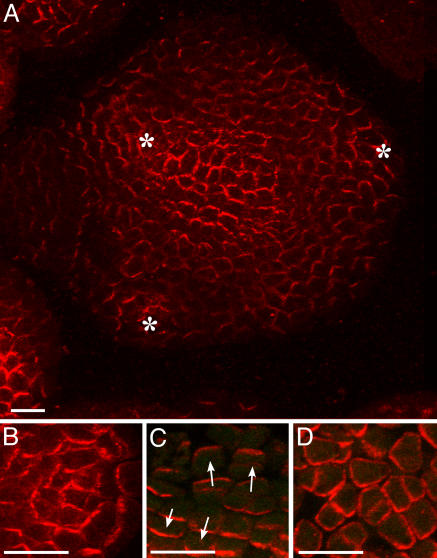
PIN1 immunolocalization in Arabidopsis shoot apical meristems (6). (A) Global view of an anti-PIN1 immunolabeling on a meristem cross section. PIN1 is localized on the membrane and polarized in most cells. Patterns are complex. Asterisks, young primordia. (Bar, 20 μm.) (B) In the peripheral zone of the meristem, concentric PIN1 orientations around young primordia are observed. The patterns suggest that the cells orient toward a single central cell of the primordium. (C) In boundaries between the meristem and the primordium, cell polarities in opposing directions are observed (arrows). (D) At the meristem summit, PIN1 localization is variable and does not seem to show any particular organization. (Scale bars for B-D,10 μm.)
To address these questions, we developed computational modeling tools that allowed us to uncover previously undescribed properties of the cell-cell interaction network and to predict auxin fluxes in the shoot apical meristems directly based on microscopical observations.
Results
Sections of shoot apical meristem were labeled with anti-PIN antibody (Ab). To interpret the complex labeling patterns in term of putative auxin distribution, we simulated the hormone fluxes on digitized meristems (Figs. 3 A-G and 4 A and B; also see Materials and Methods).
Fig. 3.
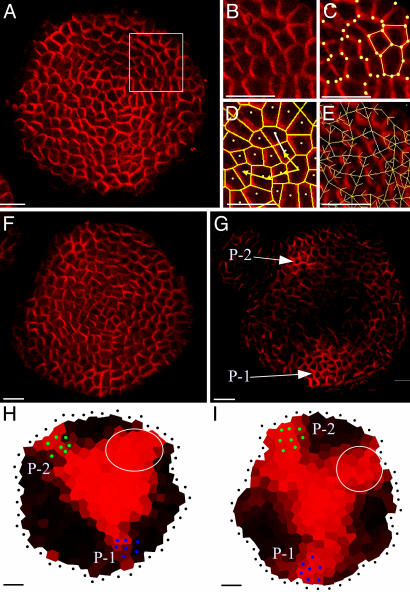
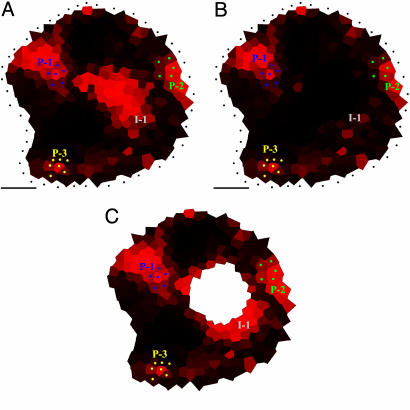
From PIN1 immunolabeling to the simulation of auxin fluxes. (A) A transverse section showing PIN labeling. [Reprinted with permission from ref. 7 (Copyright 2005, Elsevier).] The rectangle indicates the detail shown in B. merrysim (see Supporting Text) is used to capture the cell shapes and the PIN1 localization in each cell. (C) All cell vertices (spots) are manually positioned. The vertices of each cell are subsequently grouped. (D) Cells are manually connected to each other if and only if there is a PIN1 labeling on the membrane between them (arrows). The connection is oriented in the way of supposed PIN1-mediated efflux. (E) The result is a network of cell interactions. (F and G) Anti-PIN1 immunolabeling on two successive transverse sections of another meristem. In G, the labeling of the provascular strands at the level of P1 and P2 can be clearly distinguished (arrows). At these positions, called the primordium centers, auxin will be evacuated in the simulations. (H and I) Results of the simulated auxin fluxes in meristems shown in A and F. The position of the primordium centers visible on the original images are marked by green and blue dots. Virtual auxin is injected via the black dots surrounding the meristems. The quantity of virtual auxin per cell is proportional to the red intensity. Auxin accumulates where young primordia are being formed, but also at the meristem summit. Moreover, the auxin maximum at the meristem summit protrudes toward the initium I-1 (gray circle). (Scale bars, 20 μm.)
Fig. 4.
Effect of stronger active transport in primordia. (A) Reference simulation with uniform active transport. (B) Simulation with a 10-fold increase of active transport in the cells of the different primordia (colored dots).
Simulation of Auxin Fluxes. The auxin transport through the network of interconnected cells was modeled by using the following set of hypotheses:
Auxin passively diffuses via all walls (edges of the individual cells in the graph) and is actively transported via oriented connections only (1-4, 8-11). We only consider net auxin flux from cell to cell, without taking into account the molecular mechanisms involved (5, 12-14). To keep a tractable model at the tissue level, we decided to model this transport process using a simplified system, where we do not represent the compartment corresponding to the intercellular space. The net balance of auxin in a cell thus is considered to be the result of a direct exchange between cells through two processes: diffusion from cell to cell and polarized active transport due to the presence of PIN1 molecules on certain membranes of cells.
Auxin is restricted to the L1 layer and enters the meristem from the meristem border via the efflux facilitator (2) or, alternatively, auxin produced by every cell within the meristem.
Auxin is evacuated via the L1 cells that are in contact with provascular strands characterized by PIN1 labeling in deeper layers (1, 2) (Fig. 3G). Longitudinal sections show that these provascular strands are approximately three cells wide (data not shown). Therefore, a circular area of three cells wide is designated to evacuate auxin at the position of each provascular strand on the images. They are defined here as “Primordia” (P-1, P-2,...,P-1 being the nearest to the meristem summit) and behave as auxin sinks.
The simulation algorithm continues to distribute the virtual auxin in the system until the auxin distribution gets stationary. This hypothesis is to take into account that the establishment of auxin distribution is a fast process, much faster than growth and cell proliferation (2). Therefore, in a normally growing meristem, auxin distribution is likely to be near the equilibrium at all times.
Cells cannot accumulate auxin indefinitely. We modeled this constraint by using a saturation level, above which the cells no longer accept auxin influx. Simulations tests showed that this scenario was not very different from the situation where, at high level of accumulation, auxin diffusion overcomes active transport (see Supporting Text, which is published as supporting information on the PNAS web site).
Auxin is degraded at a constant rate in each cell. Situations with different degradation levels were tested, including no degradation at all.
Ten meristems that were precisely sectioned in a plane transverse to the stem were immunolabeled (6) by using the PIN Ab. Subsequently, the corresponding images were used to extract connection maps. When the six rules mentioned above were applied to these maps (for technical details, see Supporting Text and Figs. 9, 10, 11, 12, 13, 14 and Tables 1 and 2, which are published as supporting information on the PNAS web site), virtual auxin accumulated at the sites where young primordia were being formed (Fig. 3 H and I). This property of the PIN1 network could be expected from visual inspection of the immunolabelings. However, the simulations also showed a strong accumulation of virtual auxin in a domain covering the meristem summit, a property not obvious from visual inspection only. In all meristems tested, the central zone of accumulation also locally extended further to the periphery. Interestingly, this peripheral protrusion corresponded precisely to the site where the organ founder cells of the next primordium (called here initium-1 or I-1) were expected. More specifically, the divergence angle between the last formed primordium and this initium oscillated between 105° and 145° with a mean at 130°. Extensive tests showed that the patterns were robust, relatively insensitive to even major changes in the parameters (see Fig. 4 and Supporting Text).
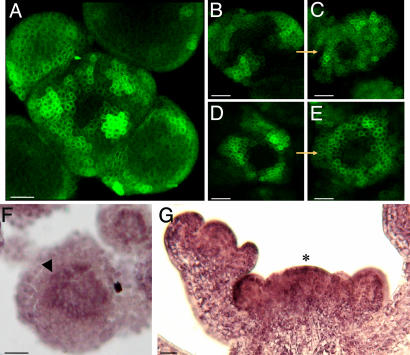
Auxin at the Meristem Summit. An unexpected simulation result was that the meristem summit accumulated auxin, suggesting a role for this domain in hormone distribution. Because previous studies only indicated a minor role for the meristem summit in this respect (15), we next tested this prediction in planta. We first analyzed plants expressing GFP under control of pDR5, a synthetic promoter that is sensitive to auxin and that has been used to estimate relative hormone threshold levels in different tissues (3). As expected, GFP was strongly expressed in the future organ primordia, even at very early stages of initiation, i.e., at the level of I-1, just next to the meristem summit (Fig. 5A) (3). As a consequence, this pattern fully coincided with those predicted by the simulations. However, in contrast to what could be expected from the simulations, GFP was not, or very weakly, expressed in the meristem summit. Therefore, either this domain contained little or no auxin, or pDR5 was insensitive to auxin in the meristem summit. To distinguish between these two possibilities, we treated young in vitro grown plants (18) expressing pDR5::GFP with auxin in absence or presence of the auxin transport inhibitor NPA. The presence of 10-5 M auxin and 10-5 M NPA caused an important increase in the amount of pDR5::GFP expressing cells. However, the meristem summit never showed any increase in GFP activity, even in meristems where the entire periphery had activated the marker (Fig. 5 B-E). We concluded that, as judged by pDR5 activity, the central domain of the meristem was auxin-insensitive. The observed insensitivity did not provide any information on the actual amount of auxin present in this domain. To address this issue, we used a monoclonal Ab directed against auxin to define local differences in auxin concentrations (19). The labeling showed a weak, but consistent, pattern, with an obvious maximum at the meristem summit (Fig. 5 F and G).
Fig. 5.
Localization of auxin in Arabidopsis shoot apical meristems. (A-E) Spatial pattern of pDR5::GFP expression in shoot apical meristems under different conditions. (A) Untreated meristem. (B and C) Treatment of a meristem with 10-5 M IAA during 22 h. 10-5 M NPA (auxin transport inhibitor) was added to keep auxin in the meristem (B, t = 0 h; C, t = 22 h). (D and E) Treatment of a meristem with 10-5 M of the synthetic auxin 2,4 D during 22 h (D, t = 0 h; E, t = 22 h). The pDR5::GFP-expressing domain covers a larger part of the periphery after the treatment with IAA-NPA or 2,4 D but the summit of the meristem remains unlabeled. (F and G) Immunolocalization of IAA in shoot apical meristems (16, 17). The presence of labeling is characterized by a purple/brown signal. (F) Cross section of a wild-type meristem; showing labeling at the meristem summit (arrowhead). (G) Longitudinal section of a wild-type meristem also showing labeling at the meristem summit (asterisk). (Scale bars, 20 μm.)
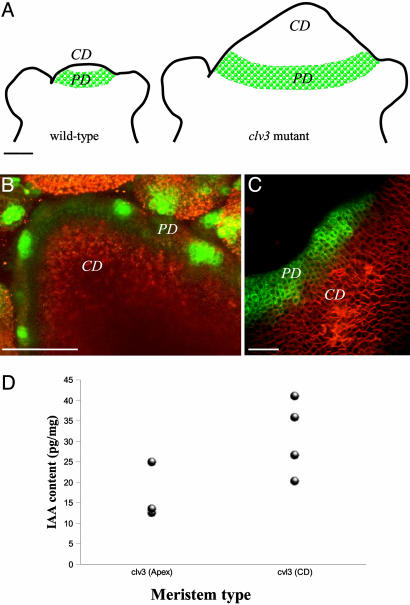
To provide additional evidence that auxin did accumulate in the central part of the meristem, we extended our analysis to gas chromatography and mass spectrometry (GCMS) (20, 21). Because a normal wild-type meristem was too small to perform this type of analysis, we decided to use the clavata3 (clv3) mutant. This mutant lacks a signaling peptide (CLV3) that is required to keep the central part of the meristem within certain size limits (22, 23). If this peptide is absent, the central domain continues to grow, until it is several millimeters wide (Fig. 6A). To confirm that the central domain of the clv3 meristem behaved like a normal wild-type meristem summit with regard to auxin sensitivity, we crossed the pDR5::GFP marker into the mutant background. In the enlarged dome of the mutant, we could only observe GFP fluorescence at the very periphery, close to the site of organ initiation (Fig. 6 B and C). The rest of the enlarged meristem did not express DR5-GFP. This result confirmed that the auxin-insensitive part of the meristem corresponded to the domain that is under control of the CLV3 pathway. This domain is believed to be equivalent to the so-called “central zone” required for meristem maintenance (23) To determine whether the clv3 summit contained auxin or not, we next performed GCMS. For this purpose we measured the auxin contents in apices containing the SAM and young flower buds of clv3 mutants. In addition, samples containing only cells coming from the enlarged meristematic summit of the clv3 mutant were taken. The results (Fig. 6D) showed that the samples enriched in central zone cells contained active IAA and were even enriched in hormone. Thus, the hypothesis that the central domain of the meristem is insensitive to auxin, but contains free IAA, as suggested by the computer simulations and the auxin immunolabeling, was further confirmed by using the IAA quantification in the clv3 mutant. Several lines of evidence suggest that PIN1 is auxin inducible (24), which might seem in contradiction with our observation that PIN is expressed in the auxin-insensitive center of the meristem. There are two possible explanations for this apparent contradiction. First, PIN expression also might depend on other parameters than auxin, and, second, the meristem summit could be partially sensitive to auxin, via a pathway that does not involve the auxin-responsive elements present in DR5.
Fig. 6.
Quantification of IAA in the central part of the clv3 meristems. (A) Schematic descriptions of wild-type and clv3 meristems illustrating the enlarged central zone in clv3 (CD, central domain). The green area represents the periphery domain (PD) where pDR5::GFP can be expressed. (B and C) Pattern of pDR5::GFP expression in clv3 meristems. (B) Global view of a full projection showing that pDR5 activity is limited to the meristem periphery, with several maxima where the next primordia will be formed. (C) Detail of a meristem. (Bars in A-C, 50 μm.) (D) Results of IAA quantification with GCMS in clv3 meristems. Samples included the young apex (CD + PD + young primordia) or the CD only. For each class, the quantification was performed on four different samples (four dots), each sample containing several meristems. The quantification shows that the central domain of clv3 meristems concentrates significantly (at 1%) more IAA than the overall apex.
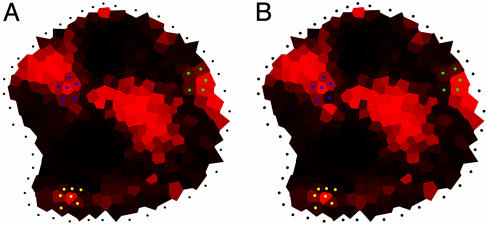
Further Simulation to Test the Role of Auxin at the Summit. What could be the function of IAA in the central domain of the meristem? To address this question, we performed additional simulations. These simulations were based on the same rules as before, but in addition the model was instructed to degrade auxin at the meristem summit. In all meristems tested, this additional instruction not only removed the auxin maximum from the meristem summit but also the maximum at the level of the I-1 initium (Fig. 7 A and B). By contrast, the maxima around the formed primordia were maintained. The results, therefore, suggest that the meristem summit plays an essential role in the creation of novel auxin maxima at the site of the organ primordium founder cells.
Fig. 7.
Testing the importance of auxin accumulation at the meristem summit. (A) Simulation of auxin distribution using the standard parameter set (i.e., there are no special instructions for the meristem summit, and auxin is evacuated only via the primordia P-1, P-2, and P-3). (B) Simulation of auxin distribution in the same meristem, but this time the auxin arriving at the summit is immediately degraded. As a result, the maximum at the initium I-1 has disappeared. (C) Simulation of auxin distribution in the same meristem, but this time, the meristem summit was removed. We defined this summit using the auxin accumulation zone. The initium I-1 is still present.
Discussion
Together, the simulations and subsequent experiments lead to a model in which auxin coming from the periphery is transported into the central zone of the meristem, which is insensitive to the organ-promoting effect of the hormone. At a certain level of accumulation, auxin can no longer freely enter the meristem summit, and because new auxin is arriving constantly, the hormone will accumulate at the site where the fluxes toward the summit are the most abundant. In a way, this scenario would be analogous to a “traffic jam” at the entry of the meristem. Our simulations predict that this site corresponds precisely to the I-1 area, i.e., the zone where the interprimordium distance is the largest (Fig. 8). At this stage, we have only considered the spiralled phyllotactic patterns observed in Arabidopsis. It will certainly be of interest to test our hypothesis that the model is also compatible with other types of phyllotaxis. For this purpose, more extensive simulation efforts using dynamic models will be required.
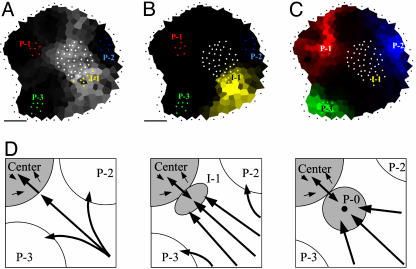
Fig. 8.
Auxin fluxes and primordium initiation. (A-C) Auxin pathways inferred from a simulation (see Supporting Text). The color intensity in each cell is proportional to the contribution of this cell to auxin accumulation in the chosen zone (black: no contribution). The different zones are indicated as groups of colored dots. (A) Auxin reaches the summit (gray dots) via corridors between primordia. The most important flux is between P-2 and P-3. I-1 is located at the limit of the summit and the most important flux toward the summit. (B) The initium I-1 (yellow dots) is mainly filled by auxin coming from the periphery. PIN patterns suggest that the center contributes little. (C) All three primordia receive auxin from the periphery. P-1 (red dots) and P-2 (blue dots) also receive some auxin from the center in contrast to P-3 (green dots). (D) Model for the formation of an auxin maximum preceding creation of a primordium. As the distance between P-2 and P-3 increases, more auxin arrives at the meristem center in this sector. Because the center can only absorb a limited amount of auxin, this situation will lead to the formation of an auxin maximum (I-1). Eventually, this maximum will be transformed into a primordium (P-0) where the provascular system behaves as an auxin sink (black dot at the center of the primordium). (Bars, 20 μm.)
The results might seem in contradiction with elegant experiments where the tomato meristem summit was ablated by using a laser (15). In this case, no modification in organ positioning was observed, at least for a period of up to four to five plastochrones, suggesting that the meristem center did not play an important role in organ positioning. To clarify this issue, we performed additional simulations, where all cells from the meristem center were removed (Fig. 7C). Interestingly, this extra hypothesis did not have an effect on the accumulation of auxin at I-1 in the model. In this context, it should be noted that an ablated meristem center is analogous to a center that no longer accepts auxin. As a consequence, it also would cause an accumulation of auxin at the site where the fluxes are most abundant. Our results are, therefore, fully compatible with the experimental evidence and provide an alternative explanation.
In this study, we have considered the molecular mechanism of auxin flux as a black box, which simply results in a net flux from cell to cell. Hereby, we assume that the PIN-labeled membranes indicate the direction of active transport. Although we show that this approach can lead to testable hypotheses, it might be of interest to include certain processes or parameters that have remained inaccessible for our simulations. For example, it could be useful to consider chemical parameters such as pH-dependent effects that influence permeability of auxin or to include more precise information on auxin concentrations. For this purpose, it will be essential to develop the biological, mathematical, and computer tools required to obtain and analyze quantitative information on these parameters.
In conclusion, our results reveal a robust network of cell interactions that is sufficient to generate auxin distribution patterns consistent with the observed organ positions (25). In addition, they suggest a role for the meristem summit in organ positioning. The next, challenging step will be to understand how the PIN1 proteins themselves are oriented. In this context, two major hypotheses have been proposed. In the first one, the patterns of cell polarity are due to the organization of local gradients of auxin concentrations. This hypothesis was originally used in ref. 12 for designing a computational model of leaf venation formation and was used recently to model various types of leaf venation patterns (26). The phyllotaxis model developed by Jönsson et al. (14) is based on a similar hypothesis. In the second hypothesis, the orientation of PIN1 pumps results from a biochemical interpretation of mechanical stresses in the meristem surface. Such a mechanism would provide a possible molecular foundation for mechanical-based models (27, 28). By any means, it will not only be important to identify cellular mechanisms leading to polar localization of PIN1, but we also need to understand how these mechanisms are coordinated at the level of the whole meristem.
Materials and Methods
Immunolabeling of PIN1 Protein. After embedding, the meristems were sectioned perpendicular to the main stems with a thickness of 12-15 μm. After labeling with anti-PIN1, the physical sections were viewed in the confocal microscope to obtain an optimal image of the labeling patterns. In some cases, a single physical section was sufficient to cover the entire dome of the meristem. In other cases, the patterns of two successive sections were combined to cover the dome.
Anti-PIN1. Based on the sequence of AtPIN1 (gene At1g73590), one potentially antigenic peptide sequence (GPTPRPSNYEEDGGPA) was selected in the large intracytosolic loop domain of AtPIN1 and used to produce Abs (made by Eurogentec, Seraing, Belgium). This Ab recognizes PIN1, because no labeling is seen at the surface of the meristem in the pin1 mutant. More detailed characterization of the Ab will be presented elsewhere. After immunostaining, the sections were viewed in a Leica confocal microscope to guarantee an optimal representation of the labeling patterns.
GCMS. For GCMS, the plant tissue was collected in a 1.5-ml microcentrifuge tube and immediately frozen in liquid nitrogen. Then, 0.5 ml of cold 0.05 M phosphate buffer (pH 7.0) containing 0.02% sodium diethyldithiocarbamic acid (antioxidant) was added to the tube, together with 13C6-IAA (Cambridge Isotope Laboratories, Cambridge, MA) internal standard (50 pg/mg tissue) and a 3-mm tungsten-carbide bead. The sample was homogenized at 30 Hz in a vibration mill (MM 301, Retsch, Haan, Germany) for 3 min and then extracted under continuous shaking for 15 min at +4°C. After extraction, the pH was adjusted to 2.7 with 1 M HCl. Purification was performed by using solid-phase extraction on a 50-mg BondElut-C18 column (Varian). The column was conditioned with 1 ml of methanol, followed by 1 ml of 1% acetic acid. After application of the sample, the column was washed with 1 ml of 10% methanol in 1% acetic acid. The column was eluted with 1 ml of methanol, and the sample then was evaporated to dryness. Then, 0.2 ml of 2-propanol and 0.5 ml of dichloromethane was added to the sample, followed by 5 μl of 2 M trimethylsilyl-diazomethane in hexane (Sigma-Aldrich). The sample was incubated in room temperature for 30 min, and excess diazomethane then was destroyed by adding 5 μl of 2 M acetic acid in hexane. After evaporation to dryness, the sample was trimethylsilylated and analyzed by selected-reaction-monitoring GCMS as described.
Modeling Tools. To interpret the labeling patterns in terms of putative auxin distribution, we developed a method relying on the simulation of auxin fluxes on digitized meristems.
Briefly, the method involves the following steps (Fig. 3 A-G). First, the membranes of the individual cells are identified on the images of immunolabeled sections. This information is used to reconstruct a graph where the nodes represent the cells and every cell is connected to its neighbors. These connections are used to simulate auxin diffusion from cell to cell. A second type of connections is used to simulate active auxin transport. For this purpose, the cells also are connected via the membranes carrying PIN1 labeling. The latter connections are oriented (represented as arrows in Fig. 3 D and E) to take into account the direction of PIN1-mediated efflux. By using these maps of interconnected cells, we simulated auxin transport by applying a set of rules based on observations and hypotheses mostly taken from the literature (for a detailed description see Supporting Text).
To test the robustness of the auxin distribution patterns, we performed a range of tests in which only one parameter was modified at the time (specified in Supporting Text). For each test, the nonvarying parameters were set to values intermediate between those having extreme effects on the simulation (see Tables 1 and 2). The results showed that the patterns were qualitatively insensitive to major changes in diffusion and transport rates. At constant transport strength, the results were qualitatively equivalent for a 13-fold increase in diffusion rates. Conversely, at constant diffusion rate, the results were qualitatively equivalent for a 5-fold increase in transport strength. The patterns, therefore, should be considered as robust.
In a minority of the cells, the immunolabeling was not clear enough to assert the polarity or even the presence of the PIN-protein. Therefore, we classified the different connections into four categories with decreasing confidence level: (i) strong signal, (ii) strong but unpolarized signal, (iii) weak but polarized signal, and (iv) weak and unpolarized signal. We next performed the simulations removing the connections ii-iv. Because the resulting patterns were not significantly different, we only considered the labeled membranes with the highest confidence level (for details, see Supporting Text).
An aspect that was not taken into account was the relative level of immunolabeling. Because there is no experimental evidence of how this level translates into transport rates, we restricted ourselves to recording only the presence/absence of PIN1 on cell walls.
Simulations showed that even an increase of transport rates by a factor of 10 in the young primordia (where PIN1 labeling was the strongest) did not significantly change the final outcome of the simulations (see Fig. 4).
Supplementary Material
Acknowledgments
We thank Przemyslaw Prusinkiewicz and Cris Kuhlemeier for critical reading of the text. We also thank Jean-Louis Giavitto (University of Evry, Evry, France) and Olivier Michel (University of Evry, Evry, France) for providing the MGS language. We thank Jiri Friml (University of Tübingen, Tübingen, Germany) for providing the DR5::GFP line. P.B.d.R. was supported by a fellowship provided by Institut National de la Recherche Agronomique. J.T. was supported by an Action Concertée Incitative (ACI) from the French Ministry of Research and by the Marie Curie program of the European Union. I.B.-C. was supported by the French Ministry of Research.
Author contributions: P.B.d.R., I.B.-C., C.G., and J.T. designed research; P.B.d.R., I.B.-C., H.M., and N.C. performed research; K.L. contributed new reagents/analytic tools; P.B.d.R., I.B.-C., C.G., and J.T. analyzed data; and P.B.d.R., I.B.-C., C.G., and J.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: GCMS, gas chromatography and MS.
References
- 1.Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P. & Traas, J. (2000) Development (Cambridge, U.K.) 127, 5157-5165. [DOI] [PubMed] [Google Scholar]
- 2.Reinhardt, D., Pesce, E.-R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J. & Kuhlemeier, C. (2003) Nature 426, 255-260. [DOI] [PubMed] [Google Scholar]
- 3.Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G. & Friml, J. (2003) Cell 115, 591-602. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., Walker, A. R., Schulz, B. & Feldmann, K. A. (1996) Science 273, 948-950. [DOI] [PubMed] [Google Scholar]
- 5.Kramer, E. M. (2004) Trends Plant Sci. 9, 578-582. [DOI] [PubMed] [Google Scholar]
- 6.Vitha, S., Baluska, F., Mews, M. & Volkmann, D. (1997) J. Histochem. Cytochem. 45, 89-95. [DOI] [PubMed] [Google Scholar]
- 7.Traas, J. & Bohn Courseau, I. (2005) Curr. Opin. Plant Biol. 8, 587-592. [DOI] [PubMed] [Google Scholar]
- 8.Vogler, H. & Kuhlemeier, C. (2003) Curr. Opin. Plant Biol. 6, 51-56. [DOI] [PubMed] [Google Scholar]
- 9.Galweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A. & Palme, K. (1998) Science 282, 2226-2230. [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt, D., Mandel, T. & Kuhlemeier, C. (2000) Plant Cell 12, 507-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carraro, N., Peaucelle, A., Laufs, P. & Traas, J. (2006) Plant Mol. Biol., in press. [DOI] [PubMed]
- 12.Mitchison, G. J. (1980) Proc. R. Soc. London Ser. B 207, 79-109. [Google Scholar]
- 13.Goldsmith, M. H. M., Goldsmith, T. H. & Martin, M. H. (1981) Proc. Natl. Acad. Sci. USA 78, 976-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jönsson, H., Heisler, M. G., Shapiro, B. E., Meyerowitz, E. M. & Mjolsness, E. (2006) Proc. Natl. Acad. Sci. USA 103, 1633-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinhardt, D., Frenz, M., Mandel, T. & Kuhlemeier, C. (2003) Development (Cambridge, U.K.) 130, 4073-4083. [DOI] [PubMed] [Google Scholar]
- 16.Thomas, C., Bronner, R., Molinier, J., Prinsen, E., van Onckelen, H. & Hahne, G. (2002) Planta 215, 577-583. [DOI] [PubMed] [Google Scholar]
- 17.Moctezuma, E. & Lewis, J. F. (1999) Planta 209, 180-186. [DOI] [PubMed] [Google Scholar]
- 18.Grandjean, O., Vernoux, T., Laufs, P., Belcram, K., Mizukami, Y. & Traas, J. (2004) Plant Cell 16, 74-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avsian-Kretchmer, O., Cheng, J.-C., Chen, L., Moctezuma, E. & Sung, Z. R. (2002) Plant Physiol. 130, 199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ljung, K., Rishikesh, P., Bhalerao, R. P. & Sandberg, G. (2001) Plant J. 28, 465-474. [DOI] [PubMed] [Google Scholar]
- 21.Edlund, A., Eklöf, S., Sundberg, B., Moritz, T. & Sandberg, G. (1995) Plant Physiol. 108, 1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark, S. E. (2001) Nat. Rev. Mol. Cell. Biol. 2, 276-284. [DOI] [PubMed] [Google Scholar]
- 23.Laux, T. (2003) Cell 113, 281-283. [DOI] [PubMed] [Google Scholar]
- 24.Vieten, A., Vanneste, S., Wisniewska, J., Benková, E., Benjamins, R., Beeckman, T., Luschnig, C. & Friml, J. (2005) Development (Cambridge, U.K.) 132, 4521-4531. [DOI] [PubMed] [Google Scholar]
- 25.Lyndon, R. (1998) The Shoot Apical Meristem (Cambridge Univ. Press, Cambridge, U.K.).
- 26.Runions, A., Fuhrer, M., Lane, B., Federl, P., Rolland-Lagan, A.-G. & Prusinkiewicz, P. (2005) ACM Trans. Graph. 24, 702-711. [Google Scholar]
- 27.Green, P., Steel, C. & Rennich, S. (1998) in Symmetry in Plants, eds. Jean, R. & Barabé, R. (World Scientific, Toh Tuck Link, Singapore), pp. 359-392.
- 28.Shipman, P. D. & Newell, A. C. (2005) J. Theor. Biol. 236, 154-197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.