Abstract
The goal of this study was to elucidate the molecular mechanism by which type I IFN inhibits assembly and release of HIV-1 virions. Our study revealed that the IFN-induced ubiquitin-like protein ISG15 mimics the IFN effect and inhibits release of HIV-1 virions without having any effect on the synthesis of HIV-1 proteins in the cells. ISG15 expression specifically inhibited ubiquitination of Gag and Tsg101 and disrupted the interaction of the Gag L domain with Tsg101, but conjugation of ISG15 to Gag or Tsg101 was not detected. The inhibition of Gag-Tsg101 interaction was also detected in HIV-1 infected, IFN-treated cells. Elimination of ISG15 expression by small interfering RNA reversed the IFN-mediated inhibition of HIV-1 replication and release of virions. These results indicated a critical role for ISG15 in the IFN-mediated inhibition of late stages of HIV-1 assembly and release and pointed to a mechanism by which the innate antiviral response targets the cellular endosomal trafficking pathway used by HIV-1 to exit the cell. Identification of ISG15 as the critical component in IFN-mediated inhibition of HIV-1 release advances the understanding of the IFN-mediated inhibition of HIV-1 replication and uncovers a target for the anti HIV-1 therapy.
Keywords: assembly, interferon inhibition, ubiquitination, ISG15
Type I IFN (IFN-α and -β) signaling is an essential component of the innate antiviral response to infection. The biological response to IFN is mediated by its binding to cell type specific receptors, activation of the Jak-Stat signaling pathway and stimulation of transcription of several hundreds of IFN-induced genes (ISG) (1). Although some of the proteins encoded by these genes have a direct antiviral activity (2), one group of IFN-induced proteins are enzymes of the ubiquitin (Ub)-like pathway and the Ub-like protein ISG15 (2, 3).
ISG15 was one of the first recognized ISGs (4). ISG15 is covalently conjugated to targeted proteins through a series of steps similar to ubiquitin conjugation. The ISG15 activating enzyme is ubiquitin E1 like protein (UBE1L), and the major E2 enzyme for ubiquitin conjugation, UbcH8, also recognizes ISG15 (5). An ISG15-specific ligase E3 has not yet been identified, but the IFN-induced Ub E3-like enzymes Rsp5 and CEB1/Hc5 may function in ISG15 conjugation (6, 7). Like Ub (8), ISG15 is removed from conjugated proteins by an ISG15-specific protease, UBP43 (9). Interestingly, type I IFN stimulates expression not only of ISG15 but also UBEL1, UbcH8, and UBP43 (3, 10) and markedly increases ISG15 conjugation. ISG15 targets a large number of cellular proteins (7); however, modification by ISG15 does not typically cause substrate degradation (11).
Ub is a central cellular regulator (12), and Ub-mediated proteolysis plays a regulatory role in the immune system (13). In general, polyubiquitination targets proteins toward 26S protease-associated degradation, whereas monoubiquitination is a signal for internalization and vesicle sorting. However, whereas polyubiquitination on lysine 43 targets proteins for degradation, lysine 63 polyubiquitination is a signal for kinase activation. Many viruses modulate the Ub-proteasome pathway to alter cellular signaling and the antiviral response (14, 15). HIV-1 uses ubiquitination at two steps of its replication cycle. First, HIV-1-encoded protein Vif targets cellular cytidine deaminase APOBEC3G for Ub mediated degradation, thus preventing APOBEC3G incorporation into viral particles and deamination of the viral genome during reverse transcription (16-18). Vif itself is monoubiquitinated and this may help its recruitment to the site of viral assembly (19). Second, ubiquitination of Gag by Ub ligase Nedd4.1 is critical for assembly and release of virions from infected cells (20). HIV-1 assembly is driven by the Gag poly protein (21), and deletion of the PTAP-L domain in p6 of Gag, or inhibition of the Gag interaction with Tsg101, a protein of the endosomal sorting complex (ESCRT-I) (22), results in accumulation of HIV-1 virions at the plasma membranes.
An important role for type I IFN in the innate response to HIV-1 infection is suggested by the observation that deletion of high IFN producing dendritic pDC2 cells results in rapid progression of HIV-1 infection in vivo (23). In vitro, type I IFN inhibits HIV-1 infection both at early steps of replication (24, 25) and at the late steps of virus assembly and release (26-28). HIV-1 virions assembled in IFN-treated T cells show a low infectivity, accumulate at the plasma membrane, and have altered morphogenesis (29-31). Similar defects were seen in IFN-treated cells infected with murine leukemia virus (32). We have shown that IFN-α- and IFN-β-mediated inhibition correlates with the induction of ISG15 (33).
The goal of this study was to further elucidate the role of ISG15 in the IFN-mediated inhibition of HIV-1 replication and to determine whether ISG15 interferes with the ubiquitination steps critical for HIV-1 budding and release. We have found that ectopic ISG15 mimics the IFN effect and inhibits release of HIV-1 virions, but does not affect synthesis of HIV-1 proteins. The elimination of ISG15 expression by small interfering RNA (siRNA) rescues virus replication in IFN-treated cells. These results implicate a critical role of ISG15 in the IFN-mediated inhibition of HIV-1 budding and release.
Results
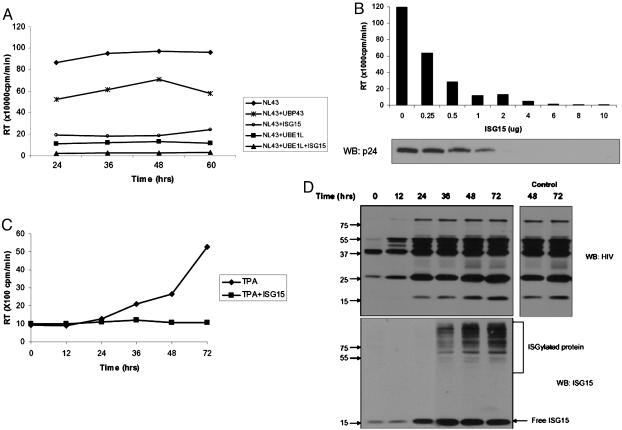
ISG15 Inhibits HIV-1 Replication. Type I IFN was shown to inhibit HIV-1 replication at the late steps of virus assembly and release (25, 29, 34), and the IFN-α-mediated inhibition of HIV-1 replication correlated with the induction of ISG15 (33, 35). Therefore, we have examined whether overexpression of ISG15 can mimic the IFN effect. Cells were transfected with HIV-1 provirus (NL43) alone or in the presence of ISG15-expressing plasmid, and the levels of released HIV-1 virions were analyzed by reverse transcriptase (RT) assay. As shown in Fig. 1A, release of HIV-1 virions into the medium was significantly inhibited in cells transfected with ISG15, and cotransfection of ISG15 with the ISG15-activating enzyme UBE1L completely blocked HIV-1 replication. The expression of the ISG15 deconjugating enzyme, UBP43, partially rescued HIV-1 released in ISG15-expressing cells (Fig. 1A). There was a direct relation between the levels of ISG15 and inhibition of HIV-1 released to the medium evaluated both by the RT activity and the levels of p24 in purified virions (Fig. 1B).
Fig. 1.
ISG 15 inhibits HIV-1 replication. (A) HIV-1 (NL43) proviral DNA and plasmids expressing ISG15, UBE1L, or UBP43 (1 μg) were cotransfected to 293T cells. Supernatants were collected at indicated times after transfection, and the levels of released virus were analyzed by RT assay. (B) 293T cells were cotransfected with plasmids expressing NL43 (2 μg) and ISG15 (0 to 10 μg), and 48 h after transfection; supernatants were collected for RT assay. The purified virions (Supporting Text) were analyzed by immune blotting with p24 antibodies. The levels of transfected DNA were kept constant by the inclusion of plasmids DNA. (C) U1.1 cells were first treated with TPA for 24 h and then infected with 301 ISG15 vector or 301 control vector that were VSV pseudotyped. At different times after TPA treatment, supernatants were collected and the virus levels determined by the RT assay. (D) Cell lysates from TPA-treated U1.1 cells, infected with 301-ISG15 viral vector (described in C) or the empty 301 vector, were analyzed at different times after TPA treatment by Western blot with HIV and ISG15 antibodies. The levels of Gag proteins in TPA-treated U1.1 cells infected with an empty 301 vector at 48 and 72 h are shown.
To determine whether ectopic ISG15 can also inhibit HIV-1 replication in HIV-1-infected cells, ISG15 was transduced by lentivirus-vesicular stomatitis virus (VSV) pseudotype to U1.1 cells, which contain a single copy of latent HIV-1 provirus. Because phorbol 12-tetradecanoate 13-acetate (TPA) treatment induces HIV-1 replication in these cells; cells were treated with TPA for 24 h before the ISG15 transduction. Levels of HIV-1 released into the medium were analyzed at different times after TPA treatment and compared to the levels of virus released from TPA-treated control cells (Fig. 1C). In TPA-treated U1.1 cells, HIV-1 virions were detected in the medium as soon as 36 h after induction, with further increase at 48 and 96 h. Release of HIV-1 virions from ISG15-transduced cells was inhibited over this time period. The immune analysis of ISG15 expression in transduced cell lysates showed the ectopic ISG15 expression at 24 h after transduction. Increased ISGylation of cellular proteins was delayed and could be detected only at 36 h after transduction. Under the conditions when release of HIV-1 virions was completely inhibited, the expression of the ectopic ISG15 did not affect synthesis of HIV-1 proteins in the transduced cells (Fig. 1D), and the levels of Gag proteins were similar to those expressed in TPA treated control U1.1 cells. These results revealed that ectopic ISG15 expression inhibited released of HIV-1 virions, but not the synthesis of viral protein in U1.1 cells.
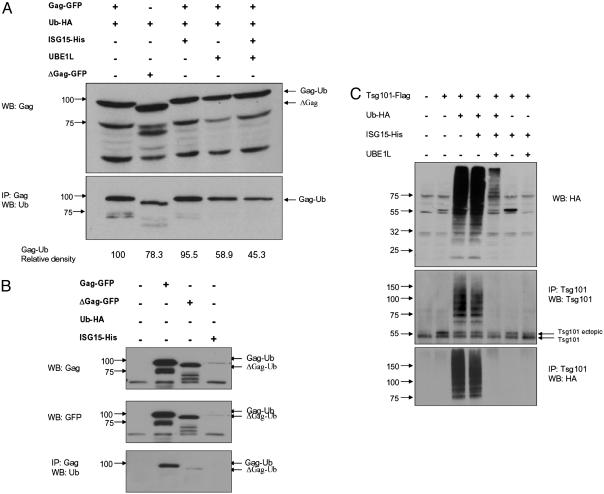
ISG15 Inhibits Ubiquitination of Gag and Tsg101. The Gag polyprotein and the PTAP motif of p6-Gag are necessary for the release of HIV-1 particles from infected cells, and the link between the ubiquitination of Gag and exocytosis of viral particles has been established (36). Because ISG15, like Ub, is conjugated to cellular proteins through lysine residues, and the ISG15 conjugation pathway has been found to intersect the ubiquitination pathway, we examined whether ectopic ISG15 modulates ubiquitination of Gag. Cells were cotransfected with a Gag expression plasmid or its p6 deletion mutant and the Ub-expressing plasmid in the presence and absence of ISG15 and UBE1L plasmids (Fig. 2A). Gag ubiquitination occurs in the absence of ectopic Ub by the endogenous Ub pathway (Fig. 2B). Expression of Ub did not down-regulate Gag levels, and even the p6 deletion mutant of Gag was ubiquitinated at low levels. Previous observations suggested that ubiquitination outside the p6 region may be functional, because the mutation of the two ubiquitinated lysine residues in p6 region did not affect virus release (37). However, in three independent experiments, the Gag ubiquitination was reduced in cells expressing ISG15 activating enzyme UBEL1 (40%) and in cells that overexpressed both ISG15 and UBEL1 (55%). The differences between the levels of Gag ubiquitination in the presence and absence of UBEL1 or ISG15 and UBEL1 were found to be statistically significant (P < 0.01). Overexpression ISG15 alone without UBEL1 was not inhibitory indicating that the cellular levels of UBEL1 and not ISG15 are limiting. These data indicate that activation of the ISG15 conjugation results in the inhibition of Gag ubiquitination. Although the ubiquitination of Gag was easily detected, we could not detect conjugation of ISG15 to Gag polyprotein or the stabilized GFP-p6 fusion protein (38).
Fig. 2.
ISG15 inhibits ubiquitination of Gag and Tsg101. (A) 293T cells were cotransfected with plasmids expressing optimized Gag or its p6 deletion mutant ΔGag. When indicated, cells were also transfected with Ub-HA-, ISG15-, and UBE1L-expressing plasmids. Cell lysates were analyzed at 48 h after transfection by Western blot with Gag polyclonal antibody (Upper), or immunoprecipitated with Gag and the precipitates analyzed by Western blot with anti-Ub monoclonal antibody (Lower). The integrated density value of bands is based on the comparison with Gag-Ub (100%). (B) 293T cells were transfected with GFP-Gag and GFP-ΔGag plasmids and the presence of Gag in lysates of transfected cells was determine by immunoblotting with Gag, GFP, or Ub antibodies. (C) Cells were cotransfected with Tsg101, Ub-HA, and, when indicated, ISG15 and UBE1L expression plasmids. Cell lysates were analyzed 24 h after transfection by Western blot with HA antibody (Top), or immunoprecipitated with Tsg101 antibody, and the precipitates were analyzed by immunoblotting with Tsg101 antibody (Middle) or HA antibodies (Bottom). All of the analyses were done in two independent experiments.
Tsg101 functions in sorting of vacuolar proteins (39). It contains, in its N-terminal region, a domain related to ubiquitin-conjugating enzyme E2, which is incapable of directly conjugating Ub but binds to p6 of Gag polyprotein. Tsg101 also binds Ub (40) and is itself ubiquitinated. Therefore, we examined whether ectopic ISG15 could also modulate ubiquitination of Tsg101 (Fig. 2C). Western blot analysis of cellular lysates revealed a profound increase in the ubiquitination of cellular proteins in Ub transfected cells. Expression of ectopic Ub resulted also in polyubiquitination of Tg101, which was inhibited in cells expressing ectopic ISG15 and UBEL1, but not ISG15 alone. Relative levels of Tsg101 were not significantly altered in cells expressing either ectopic Ub or ISG15, and immunodetection with ISG15 antibodies did not detect a presence of ISGylated Tsg101 (data not shown).
ISG15 Inhibits Gag-Tsg101 Interaction. HIV-1 Gag polyprotein specifically interacts through p6 PTAP motif with Tsg101, and this interaction is critical for HIV-1 budding (41). To understand the mechanism by which ISG15 affects HIV-1 release, we have examined whether ISG15 modulates the association of Gag with Tsg101. 293T cells were cotransfected with both HIV-1 provirus (NL43) and Tsg101-Flag or together with ISG15- and UBEL1-expressing plasmids. The transfected cells were treated with the proteasome inhibitor (MG132) to prevent possible degradation of Tsg101 and to stabilize Gag-Tsg101 complex. Immunoprecipitation with Tsg101 antibodies (Fig. 3A) and immunoblotting of the precipitates with Gag antibodies detected Gag in immunoprecipitates of cell lysates transfected with NL43 and Tsg101, but not in the cells that were also expressing ISG15 or ISG15 and UBE1L. When UBP43 was expressed together with ISG15, low levels of the association between Tsg101 and Gag could be detected. The association between Tsg101 and Gag was confirmed by the reciprocal immunoprecipitation. Tsg101 could be detected in Gag precipitates of NL43 and Tsg101-transfected cell lysates, but not in lysates of cells that expressed ISG15 or ISG15 and UBP43. The levels of Gag were comparable in all cells transfected with NL43 proviral DNA, and ectopic Tsg101 was expressed at high levels in transfected cells. These data indicate that ectopic ISG15 inhibits association of Tsg101 with Gag and that this inhibition depends on ISG15 conjugation to the targeted proteins.
Fig. 3.
ISG15 inhibits Gag-Tsg101 interaction. (A) 293T cells were cotransfected with NL43 proviral DNA and Tsg101 expressing plasmid and, when indicated, also with ISG15 and UBE1L or UBP43 plasmids. Cell lysates (1 mg), prepared 48 h after transfection, were immunoprecipitated with Tsg101 or Gag antibody. The precipitates were analyzed by Western blot with Gag- or Tsg101-specific antibodies. (B) 293T cells were cotransfected with NL43 proviral DNA and, and when indicated, also with Tsg101-, UBE1L-, and UbcH8-expressing plasmids. Twenty-four hours after transfection, cells were treated with IFN-α (500 units/ml) for 48 h and then cell lysates (1 mg) were immunoprecipitated with Tsg101 antibody. The precipitates were analyzed by Western blot with Gag antibody (Top) or Tsg101 antibody (Middle). Presence of Gag in the cell lysates of transfected cells was determined by Western blot (Bottom). All of the analysis was done in two independent experiments.
Inhibition of the Tg101-Gag association could be also seen in IFN-treated cells that were cotransfected with NL43 proviral DNA. Cells were transfected with NL43 DNA and, 24 h later, treated with human IFN-α for additional 48 h (when the ISGylation to cellular proteins is substantially increased). Lysates from IFN-treated and untreated cells were then immunoprecipitated with Tsg101 antibody and the presence of Gag in the precipitates was detected by immunoblotting with the Gag-specific antibodies (Fig. 3B). Although the association of Tsg101 with Gag was detected in untreated cells, IFN treatment disrupted this association. Expression of ectopic Tsg101 increased the levels of Gag associated with Tsg101, but this association was again disrupted in IFN-treated cells. These data show that the levels of endogenous ISG15 induced by IFN are sufficient enough to disrupt the Tsg101-Gag binding.
The ESCRT-I complex that interacts with ubiquinated Gag protein comprises Tsg101, Vps28, and Vps 37 (22), where both Vps37 and Vps28 bind to the C-terminal and central region of Tsg101 (42). Ectopic ISG15 did not inhibit Tsg101-Vp28 interaction (Fig. 5, which is published as supporting information on the PNAS web site).
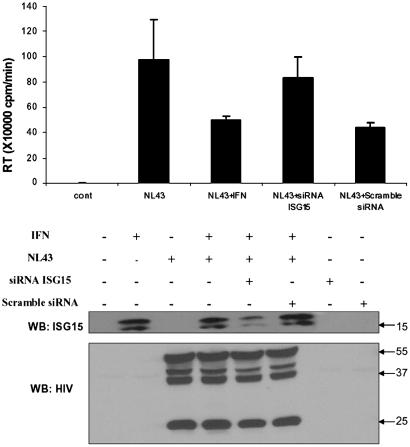
ISG15 siRNA Reverses IFN-Mediated Inhibition. IFN induces a large number of ISG proteins with antiviral functions that could also contribute to the IFN-mediated inhibition of HIV-1 replication (43-45). To determine the critical role of ISG15 in IFN-mediated inhibition of HIV-1 assembly and release, we have depleted the intracellular pool of ISG15 in IFN-treated, HIV-1-infected cells. 293T cells were cotransfected with ISG15-specific siRNA and HIV-1 provirus NL43 and, 48 h after transfection, treated with IFN-α for 24 h. As a control, cells were also cotransfected with scrambled siRNA and HIV-1 provirus NL43 (Fig. 4). The relative levels of ISG15 protein in the IFN-treated cells transfected with ISG15-specific siRNA were substantially lower in the presence of ISG15-specific siRNA than in its absence or in cells that were transfected with the scrambled siRNA. Transfection of IRF-5 siRNA or the scrambled siRNA did not stimulate induction of ISG15. Neither IFN treatment nor ISG15 or scrambled siRNA down-regulated the levels of Gag proteins in the cells.
Fig. 4.
Rescue of the IFN-mediated inhibition of HIV-1 release by ISG15 siRNA. 293T cells were cotransfected with NL43, ISG15 siRNA, or scrambled siRNA, and 48 h after transfection, cells were treated/untreated with IFN-α (500 units/ml) for 24 h. HIV-1 virions in the supernatants were analyzed by the RT assay. Cell lysates (20 μg of proteins) were analyzed by Western blot with ISG15 monoclonal antibody and the presence of Gag proteins was detected by immunoblotting with human HIV-1 antiserum (Tulpin). As a control, cells were transfected either with 10 nmol of siRNA or scrambled siRNA, and 48 h after transfection, cell lysates were analyzed by Western blot with ISG15 monoclonal antibodies.
Although the release of HIV-1 virions was inhibited in IFN-treated control cells, transfection of the ISG15 specific siRNA reversed this inhibition. The IFN effect was not reversed in cells transfected with the scrambled siRNA. These data indicate that ISG15 is the key mediator of IFN-induced inhibition of HIV-1 release.
Discussion
We have shown in this study that the antiviral immune response targets the ubiquitin-dependent pathway of HIV-1 budding and release and that IFN-mediated inhibition is mediated by the IFN-induced protein, ISG15. The data have also revealed that expression of ectopic ISG15 and UBEL1 mimics the antiviral effect of IFN and that the inhibition of ISG15 expression in IFN treated cells by ISG15 specific siRNA, reverses IFN inhibition, and rescues HIV-1 release.
Addressing the molecular mechanism of this inhibition, we have shown that this inhibition occurs by altering protein interaction and ubiquitination steps that are required for virus release. Ectopic UBEL1 or UBEL1 and ISG15 inhibits the ubiquitination of HIV-1-encoded Gag polypeptide and Tsg101 and the interaction of Tsg101, a central component of ESCRT-1 with the p6 domain of the Gag polyprotein. Tsg101 N-terminal UEV domain homologous to E2 enzymes (46) binds to Ub, and this interaction is essential for the targeting of Ub-conjugated proteins to multivesicular bodies (47). However, Tsg101 UEV motif binds also to the P(T/S)AP motif in the L domain of HIV-1 Gag polypeptide, and the crystal structure analysis revealed that the UEV domain can bind both Ub and PTAP peptide simultaneously (48). The observation that ISG15 interferes with ubiquitination of Gag and consequent release of virus particles correlates with the previous observation, which showed the inhibition of HIV-1 release in the presence of proteasome inhibitors (49).
The precise mechanism by which ISG15 and UBEL1 interfere with the HIV-1 Vps pathway is unclear. At this stage, we have shown that expression of ectopic ISG15 and IFN-induced ISG15 prevents the Gag-Tsg101 interaction. Although we have not detected ISGylation of Gag or Tsg101, ISG15 was shown to conjugate to Ubc13 (7), Ub carrier protein E2. Ubc13 forms heterodimers with Mms2, and these catalyze assembly of K63-linked polyubiquitination (50). While this study was underway, it was shown that ISGylation of Ubc13 suppresses its ability to form thioester intermediates with Ub (51), which leads to inhibition of K63-linked ubiquitination. Thus, the inhibition of Gag or Tsg101 ubiquitination may be a consequence of ISGylation of Ubc13. The decrease in ubiquitination of Gag and Tsg101 may then alter the binding affinity of Tsg101 to Gag and destabilize the Tsg101-Gag complex. In many retroviruses, there is a correlation between the L domain and ubiquitination, where conjugated lysines are in a close proximity to the L domain. Further investigations of the ISG15-mediated effects are clearly warranted.
The functional consequences of ISG15 conjugation remain unknown. ISG15 targets large numbers of cellular proteins, and many of the ISGylated proteins are components of the antiviral innate signaling pathway (7). Although it has been suggested that ISGylation may increase the antiviral effect of IFN (52), mice with homozygous deletion of ISG15 show neither defect in the antiviral response against VSV and LCM nor enhanced STAT1 signaling (53). However, although a large number of cellular proteins are ISGylated, the ISGylation of a virus-encoded protein has not yet been demonstrated. Modification of Gag protein by SUMO was described (54), but the conjugation of ISG15 to Gag or GFP-p6 was not detected.
Several ISGs, such as PKR, ISG20, and 2-5AOS were implicated in the inhibition of the early steps of HIV-1 replication (43-45). Recently, expression of three cellular proteins, APOBEC3G (A3G), APOBEC3F (A3F), and TRIM 5α, was associated with restriction of HIV-1 replication (55). TRIM 5α belongs to a family of proteins with tripartite motif (56), some of which are stimulated by IFN (57). TRIM blocks the early step of HIV-1 replication, possibly by ubiquitination of the HIV-1 capsid protein (58). A3G and A3F are APOBEC family proteins containing the cytidine deaminase domain. A3G and A3F convert cytidine to uridine in the single-stranded proviral DNA, which results in the hypermutation of the HIV-1 genome (59). Although expression of A3G is not stimulated by IFN in T cell line H9 (60), one of the ISGs, adenine deaminase, catalyzes deamination of adenosine to inosine in viral RNAs (61). Interestingly, the HIV-1-encoded Vif protein degrades A3G by the Ub-mediated proteasome pathway by activated E3 ligase, Cullin 5 (17). Whether ISG15 affects also ubiquitination and degradation of A3G is not known.
In conclusion, our data uncovered a mechanism in which the innate antiviral response targets ubiquitination steps in HIV-1 replication cycle and identified an IFN-induced cellular protein, ISG15, as the inhibitor of HIV-1 assembly pathway. The effect of ISG15 may be at least partially related to the inhibition of Gag and Tsg101 ubiquitination and to disruption of the interaction of Gag L domain with Tsg101, although additional mechanisms cannot be excluded. Number of retroviruses and negative strand enveloped RNA viruses contain the L domains that have a similar role in the endosomal trafficking pathway (62). Thus, ISG15 may affect replication not only of HIV-1, but also of a broad group of RNA viruses. Inhibition of murine leukemia virus assembly in IFN treated cells has been demonstrated (63). These results advance the understanding of previous findings and uncover a target for anti-HIV-1 intervention. Possible implications of these findings are the development of more effective clinical therapies that will not have the side effects associated with IFN treatment.
Materials and Methods
Cell Culture, Plasmids, and Virus. 293T cells were cultured in DMEM with 10% FBS. PM-1, U937, and U1.1 cells were cultured in RPMI medium 1640 with 10% FBS. The histidine-tagged Vps 28 (His-Vps28) plasmid was generated by insertion of Vps28 cDNA into the HindII and XhoI restriction sites of pcDNA 3.1 vector (Invitrogen). The hemagglutinin (HA)-tagged Ub (Ub-HA) plasmid was obtained from Heinrich Gottlinger (Harvard Medical School, Boston), and pISG15, UBE1L and the histidine-tagged ISG15 (ISG15-His) have been described (64). The Flag-tagged Tsg101 plasmid (Tsg101 Flag) was from Seth Welles (Harvard University, Boston), the optimized GFP-Gag plasmid was from George Pavlakis (National Cancer Institute, Frederick, MD), and the GFP-p6 fusion plasmid was from Jeremy Luban (Columbia University, New York). The lentiviral vector expression cassette 301 was obtained from Y. N. Chang (Lentigen, Catonsville, MD). The HIV-1 provirus NL43 and the macrophage-tropic HIV-1 AD8 were obtained from Malcolm Martin (National Institutes of Health, Bethesda). Infectious virus was produced and purified as described in Supporting Text, which is published as supporting information on the PNAS web site. HIV-1-NL43 provirus was derived from two North American strains of HIV-1; its sequence and characterization are described in ref. 65.
Lentivirus Vector Containing ISG15cDNA. The ISG15 cDNA was cloned to NotI and BstEII restriction sites of lentiviral vector 301, containing a CMV-driven enhanced GFP (EGFP). Vector 301-ISG15 was cotransfected to 293T cells with the helper plasmids pCMVR8.2 carrying Gag, pol, tat, and rev, the accessory genes vif, vpr, vpu, and nef, and the pMDG plasmid encoding VSV-G protein. Preparation of virus stock was done according to Davis et al. (66) and is described in Supporting Text.
Antibodies, Immunoprecipitation, Western Blot, and Statistical Analysis. The Tsg101, and ubiquitin antibodies were purchased from Santa Cruz Biotechnology, HA antibodies were purchased from Roche, Flag antibodies were purchased from Sigma, and the Gag-specific antibodies were purchased from Advanced Biotechnologies (Columbia, MD). ISG15 monoclonal antibody was kindly provided by E. Borden (Cleveland Clinic, Cleveland). Human serum, Tulpin, which detects HIV-1 proteins, was generous gift from Malcolm Martin. The Western blot analysis of cell lysates and immunoprecipitations (67) are described in Supporting Text. Statistical analysis by t test and Student's t test were performed in excel (Microsoft).
RT Assay. Supernatants collected from HIV-1 infected cells or from cells transfected with HIV-1 proviral DNA at times indicated were clarified by low-speed centrifugation and passed through 0.45-μm-pore-size filters. Virus was concentrated by centrifugation on 20% sucrose cushions at 25,000 rpm for 1 h at 4°C. Pelleted virus was analyzed by RT assay as described (68).
ISG15 siRNA. ISG15-synthetic siRNA (G1P2) was purchased from Ambion (Austin, TX) as annealed oligonucleotide and was resuspended in RNase-free H2O. 293T cells were cotransfected with ISG15 siRNA (10 nmol), scramble siRNA (5 or 10 nmol as indicated, purchased from Dharmacon RNA Technologies), and NL43 by using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were treated with IFN-β for 24 h. The levels of ISG15 in cell lysates were determined by Western blot analyses.
Supplementary Material
Acknowledgments
We thank Dr. E. Borden for the generous gift of ISG15 antibodies and Drs. Y.-N. Chang, H. Gottlinger, J. Luban, M. Martin, G. Pavlakis, and S. Welles for the respective plasmids. These studies were supported by National Institute of Allergy and Infectious Diseases Grants AI054537 and AI054276 (to P.M.P.).
Author contributions: A.O. and P.M.P. designed research; A.O. and G.L. performed research; A.O. and I.P.-R. contributed new reagents/analytic tools; G.L., I.P.-R., and P.M.P. analyzed data; I.P.-R. and P.M.P. wrote the paper; and G.L. made a graphical presentation of the data.
Conflict of interest statement: No conflicts declared.
Abbreviations: ISG, IFN-induced gene; Ub, ubiquitin; siRNA, small interfering RNA; RT, reverse transcriptase; VSV, vesicular stomatitis virus; TPA, phorbol 12-tetradecanoate 13-acetate; HA, hemagglutinin.
References
- 1.de Veer, M. J., Holko, M., Frevel, M., Walker, E., Der, S., Paranjape, J. M., Silverman, R. H. & Williams, B. R. (2001) J. Leukoc. Biol. 69, 912-920. [PubMed] [Google Scholar]
- 2.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227-264. [DOI] [PubMed] [Google Scholar]
- 3.Nyman, T. A., Matikainen, S., Sareneva, T., Julkunen, I. & Kalkkinen, N. (2000) Eur. J. Biochem. 267, 4011-4019. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, P. J., Broeze, R. J. & Lengyel, P. (1979) Nature 279, 523-525. [DOI] [PubMed] [Google Scholar]
- 5.Zhao, C., Beaudenon, S. L., Kelley, M. L., Waddell, M. B., Yuan, W., Schulman, B. A., Huibregtse, J. M. & Krug, R. M. (2004) Proc. Natl. Acad. Sci. USA 101, 7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochrainer, K., Mayer, H., Baranyi, U., Binder, B., Lipp, J. & Kroismayr, R. (2005) Genomics 85, 153-164. [DOI] [PubMed] [Google Scholar]
- 7.Zhao, C., Denison, C., Huibregtse, J. M., Gygi, S. & Krug, R. M. (2005) Proc. Natl. Acad. Sci. USA 102, 10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson, K. D. (1997) FASEB J. 11, 1245-1256. [DOI] [PubMed] [Google Scholar]
- 9.Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D. E. (2002) J. Biol. Chem. 277, 9976-9981. [DOI] [PubMed] [Google Scholar]
- 10.Yuan, W. & Krug, R. M. (2001) EMBO J. 20, 362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, M., Li, X. L. & Hassel, B. A. (2003) J. Biol. Chem. 278, 1594-1602. [DOI] [PubMed] [Google Scholar]
- 12.Finley, D., Ciechanover, A. & Varshavsky, A. (2004) Cell 116, S29-S32, 2 pages following S32. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Neriah, Y. (2002) Nat. Immunol. 3, 20-26.11753406 [Google Scholar]
- 14.Boutell, C., Sadis, S. & Everett, R. D. (2002) J. Virol. 76, 841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu, Y., Wang, S. E. & Hayward, G. S. (2005) Immunity 22, 59-70. [DOI] [PubMed] [Google Scholar]
- 16.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646-650. [DOI] [PubMed] [Google Scholar]
- 17.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056-1060. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dussart, S., Courcoul, M., Bessou, G., Douaisi, M., Duverger, Y., Vigne, R. & Decroly, E. (2004) Biochem. Biophys. Res. Commun. 315, 66-72. [DOI] [PubMed] [Google Scholar]
- 20.Vogt, V. M. (2000) Proc. Natl. Acad. Sci. USA 97, 12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed, E. O. (1998) Virology 251, 1-15. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., Babst, M. & Emr, S. D. (2001) Cell 106, 145-155. [DOI] [PubMed] [Google Scholar]
- 23.Levy, J. A., Scott, I. & Mackewicz, C. (2003) Clin. Immunol. 108, 167-174. [DOI] [PubMed] [Google Scholar]
- 24.Shirazi, Y. & Pitha, P. M. (1992) J. Virol. 66, 1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gendelman, H. E., Baca, L. M., Turpin, J., Kalter, D. C., Hansen, B., Orenstein, J. M., Dieffenbach, C. W., Friedman, R. M. & Meltzer, M. S. (1990) J. Immunol. 145, 2669-2676. [PubMed] [Google Scholar]
- 26.Poli, G., Orenstein, J. M., Kinter, A., Folks, T. M. & Fauci, A. S. (1989) Science 244, 575-577. [DOI] [PubMed] [Google Scholar]
- 27.Pitha, P. M. (1994) Antiviral Res. 24, 205-219. [DOI] [PubMed] [Google Scholar]
- 28.Bednarik, D. P., Mosca, J. D., Raj, N. B. & Pitha, P. M. (1989) Proc. Natl. Acad. Sci. USA 86, 4958-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlinger, H. G., Dorfman, T., Sodroski, J. G. & Haseltine, W. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda, Y., Miyake, S., Kato, S., Kita, M., Kishida, T., Kimura, T. & Ikuta, K. (1990) J. Acquired Immune Defic. Syndr. 3, 1046-1051. [PubMed] [Google Scholar]
- 31.Smith, M. S., Thresher, R. J. & Pagano, J. S. (1991) Antimicrob. Agents Chemother. 35, 62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitha, P. M., Wivel, N. A., Fernie, B. F. & Harper, H. P. (1979) J. Gen. Virol. 42, 467-480. [DOI] [PubMed] [Google Scholar]
- 33.Kunzi, M. S. & Pitha, P. M. (1996) J. Interferon Cytokine Res. 16, 919-927. [DOI] [PubMed] [Google Scholar]
- 34.Popik, W. & Pitha, P. M. (2000) Virology 276, 1-6. [DOI] [PubMed] [Google Scholar]
- 35.Kunzi, M. & Pitha, P. M. (1998) in Topley and Wilson's Microbiology and Microbial Infections, eds. Collier, L., Balows, A. & Sussman, M. (Edward Arnold Ltd., London), Vol. 1, pp. 193-210. [Google Scholar]
- 36.Strack, B., Calistri, A. & Gottlinger, H. G. (2002) J. Virol. 76, 5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E., Coren, L. V., Chertova, E. N., Gagliardi, T. D. & Schubert, U. (2000) Virology 278, 111-121. [DOI] [PubMed] [Google Scholar]
- 38.Gurer, C., Berthoux, L. & Luban, J. (2005) J. Virol. 79, 910-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop, N. & Woodman, P. (2001) J. Biol. Chem. 276, 11735-11742. [DOI] [PubMed] [Google Scholar]
- 40.Bishop, N., Horman, A. & Woodman, P. (2002) J. Cell Biol. 157, 91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55-65. [DOI] [PubMed] [Google Scholar]
- 42.Babst, M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, 248-258. [DOI] [PubMed] [Google Scholar]
- 43.Carpick, B. W., Graziano, V., Schneider, D., Maitra, R. K., Lee, X. & Williams, B. R. (1997) J. Biol. Chem. 272, 9510-9516. [DOI] [PubMed] [Google Scholar]
- 44.Montefiori, D. C., Sobol, R. W., Jr., Li, S. W., Reichenbach, N. L., Suhadolnik, R. J., Charubala, R., Pfleiderer, W., Modliszewski, A., Robinson, W. E., Jr., & Mitchell, W. M. (1989) Proc. Natl. Acad. Sci. USA 86, 7191-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Espert, L., Degols, G., Lin, Y. L., Vincent, T., Benkirane, M. & Mechti, N. (2005) J. Gen. Virol. 86, 2221-2229. [DOI] [PubMed] [Google Scholar]
- 46.Ponting, C. P., Cai, Y. D. & Bork, P. (1997) J. Mol. Med. 75, 467-469. [PubMed] [Google Scholar]
- 47.Bilodeau, P. S., Winistorfer, S. C., Kearney, W. R., Robertson, A. D. & Piper, R. C. (2003) J. Cell Biol. 163, 237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundquist, W. I., Schubert, H. L., Kelly, B. N., Hill, G. C., Holton, J. M. & Hill, C. P. (2004) Mol. Cell 13, 783-789. [DOI] [PubMed] [Google Scholar]
- 49.Schubert, U., Ott, D. E., Chertova, E. N., Welker, R., Tessmer, U., Princiotta, M. F., Bennink, J. R., Krausslich, H. G. & Yewdell, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hofmann, R. M. & Pickart, C. M. (2001) J. Biol. Chem. 276, 27936-27943. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi, T. & Yokosawa, H. (2005) Biochem. Biophys. Res. Commun. 336, 9-13. [DOI] [PubMed] [Google Scholar]
- 52.Malakhova, O. A., Yan, M., Malakhov, M. P., Yuan, Y., Ritchie, K. J., Kim, K. I., Peterson, L. F., Shuai, K. & Zhang, D. E. (2003) Genes Dev. 17, 455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osiak, A., Utermohlen, O., Niendorf, S., Horak, I. & Knobeloch, K. P. (2005) Mol. Cell. Biol. 25, 6338-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nevels, M., Brune, W. & Shenk, T. (2004) J. Virol. 78, 7803-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goff, S. P. (2004) Nature 427, 791-793. [DOI] [PubMed] [Google Scholar]
- 56.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Der, S. D., Zhou, A., Williams, B. R. & Silverman, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848-853. [DOI] [PubMed] [Google Scholar]
- 59.Yu, Q., Chen, D., Konig, R., Mariani, R., Unutmaz, D. & Landau, N. R. (2004) J. Biol. Chem. 279, 53379-53386. [DOI] [PubMed] [Google Scholar]
- 60.Rose, K. M., Marin, M., Kozak, S. L. & Kabat, D. (2004) J. Biol. Chem. 279, 41744-41749. [DOI] [PubMed] [Google Scholar]
- 61.George, C. X. & Samuel, C. E. (1999) Proc. Natl. Acad. Sci. USA 96, 4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morita, E. & Sundquist, W. I. (2004) Annu. Rev. Cell Dev. Biol. 20, 395-425. [DOI] [PubMed] [Google Scholar]
- 63.Pitha, P. M., Rowe, W. P. & Oxman, M. N. (1976) Virology 70, 324-338. [DOI] [PubMed] [Google Scholar]
- 64.Pitha-Rowe, I., Hassel, B. A. & Dmitrovsky, E. (2004) J. Biol. Chem. 279, 18178-18187. [DOI] [PubMed] [Google Scholar]
- 65.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis, B. M., Humeau, L. & Dropulic, B. (2004) Mol. Ther. 9, 160-172. [DOI] [PubMed] [Google Scholar]
- 67.Lubyova, B., Kellum, M. J., Frisancho, A. J. & Pitha, P. M. (2004) J. Biol. Chem. 279, 7643-7654. [DOI] [PubMed] [Google Scholar]
- 68.Vicenzi, E. & Poli, G. (1994) Chem. Biol. Interact. 91, 101-109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.