Abstract
Deregulation of energy metabolism by external interventions or mutations in metabolic genes can extend lifespan in a wide range of species. We describe mutations in Drosophila melanogaster that confer resistance to oxidative stress and display a longevity phenotype. These phenotypes are associated with molecular lesions in a hitherto uncharacterized gene we named Enigma. We show that Enigma encodes a mitochondrial protein with homology to enzymes of the β-oxidation of fatty acids and that mutations in this locus affect lipid homeostasis. Our analysis provides further support to the notion that lipid metabolism may play a central role in metazoan lifespan regulation.
Keywords: β-oxidation, longevity, mitochondria
Aging is a complex phenomenon with a polygenic etiology. Several theories have been advanced over the years that attempt to explain how endogenous and exogenous factors impact on the progressive physical deterioration of organisms. Restricted dietary calorie uptake is known to impact lifespan in a wide range of species, including mammals, insects, nematodes, and yeast (1, 2). Consequently, mutations that mimic calorie restriction conditions show life extension phenotypes (3–5). Calorie restriction induces a series of metabolic responses, with loss of fat mass being the most prominent phenotypic change observed (6). Because reduction of fat tissue alone is sufficient to prolong lifespan in mice (7), lipid homeostasis seems to be a significant parameter of longevity regulation.
Lifespan extension of calorie restricted animals has been linked to decreased metabolism and a reduced generation of reactive oxygen species (ROS) (8). According to the “free-radical theory of aging,” ROS, which are predominantly by-products of mitochondrial metabolism, are thought to contribute to cellular aging through the oxidative damage they inflict on macromolecules (9). Supporting this theory, several studies have established a strong correlation between longevity and increased tolerance to oxidative stress, with long-lived organisms displaying augmented resistance to free-radical generators such as methyl viologen (paraquat) (9).
Here, we describe the isolation and characterization of mutations in Drosophila melanogaster that define a genetic locus we name Enigma (Egm) and that confer increased lifespan and resistance to oxidative stress. Egm loss-of-function mutations were first identified as modifiers of Notch signaling in a genetic screen for modulators of the rough eye phenotype elicited by the overexpression of a constitutively activated form of the Notch receptor (10). Whereas the molecular basis of the relationship between Egm and Notch remains enigmatic, our analysis reveals that modulation of Egm activity affects the lifespan of the fruit fly as well as its tolerance to oxidative stress. We establish that Egm encodes a mitochondrial protein, which affects lipid metabolism, and provide evidence suggesting that it is implicated in the β-oxidation pathway.
Breakdown of saturated fatty acids through the β-oxidation spiral takes place in the mitochondria and is a major energy-yielding cellular process (11). β-oxidation has been poorly characterized in Drosophila, but comparative sequence analysis clearly demonstrates that the fruit fly genome contains homologues for all of the known mammalian enzymes responsible for the activation, trafficking, and processing of fatty acids through the β-oxidation pathway. The identification in Drosophila of conserved metabolic processes that affect longevity is of special interest because it may uncover lifespan regulatory mechanisms across a wide range of organisms.
Results
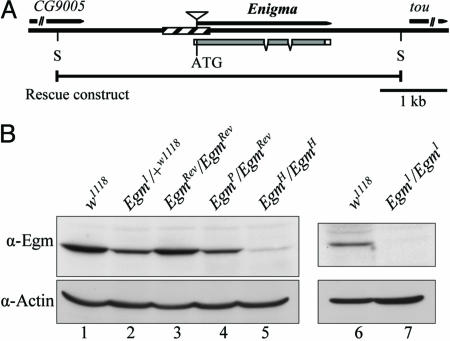
Egm Encodes a Mitochondrial Protein with Homology to Acyl-CoA Dehydrogenases (ACADs). The cloning of the Egm locus was based on the lethal phenotype of Egm1, an allele generated in ethyl-methyl sulfonate (EMS)-treated w1118 flies (10) (Fig. 1A). The lethality of Egm1 had been mapped to the base of the right arm of chromosome II. We screened a series of deletions that uncover this region and isolated three deficiency lines, which failed to complement the lethality of Egm1 [Df(2R)en-A, Df(2R)en-B, and Df(2R)en-SFX31]. Df(2R)stan1, which is also partially overlapping with the other three deficiencies, does complement Egm1. These deficiencies define a unique interval of 85 kb. Screening a series of lethal transposable-element insertions within this region identified a P-element mutant (EgmP) that disrupts the previously uncharacterized gene CG9006 (Fig. 1 A). On the basis of genetic criteria, both Egm1 and the P-element mutant EgmP are null alleles of the locus. To prove that the lethality of the Egm mutant flies is indeed caused by mutations in CG9006, we generated transgenic flies carrying a fragment of genomic DNA containing a WT copy of the gene (Fig. 1 A). Indeed, one copy of the construct was sufficient to rescue the lethality linked to either of the two null alleles (Egm1 and EgmP). The rescue experiments confirm that Egm encodes the CG9006 gene, which corresponds to a 639-aa protein with homology to ACADs, the enzymes that catalyze the first of four reactions that constitute one cycle of the β-oxidation pathway (12) (see Fig. 6, which is published as supporting information on the PNAS web site). To further characterize this gene, we raised an antibody against a recombinant Egm protein. The protein levels of Egm in whole lysates of Egm1 and EgmP heterozygous flies are halved compared with WT (Fig. 1B), indicating that these mutations are molecular nulls. By using the same antibody, we consistently did not detect immunoreactive material in extracts from Egm1/Egm1 homozygous null larvae, demonstrating the specificity of the antiserum (Fig. 1B). To rigorously characterize the phenotypes associated with mutants in the Egm locus, we generated additional alleles by mobilizing the P-element of EgmP. Partial excision of the transposable element created the hypomorphic allele EgmH, which expresses significantly reduced amounts of the protein (Fig. 1B), but in sufficient quantity, however, to produce viable adults. In addition, we isolated a precise excision of the P-element, which results in a viable WT revertant of EgmP (EgmRev) and restores protein expression to WT levels (Fig. 1B, compare lane 3 with lanes 1 and 4). Altogether, these data link unambiguously the Egm mutations to the ACAD homologous gene CG9006 (henceforth Egm).
Fig. 1.
The Enigma locus and molecular characterization of the Egm mutations. (A) Schematic diagram of the Egm gene region (chromosome II, 48A3) and the Egm mutations. Shaded and white boxes indicate coding and untranslated regions, respectively. Hatched rectangle indicates the deletion of the null allele Egm1 (–423 to +241 relative to the starting ATG). The triangle designates the P-element of EgmP at position –1. Allele EgmH contains a truncated form of the P-element (data not shown). The rescue construct pCaSpeR3-Enigma extends 2.2 kbp upstream of the starting ATG and 1.1 kbp downstream of the stop codon. S, restriction enzyme SacII. tou (toutatis) and CG9005 are the genes flanking Egm.(B) Western blot analysis of the different Egm alleles. Shown are Egm levels in WT flies w1118 and EgmRev/EgmRev, lanes 1 and 3, respectively; heterozygous flies for the null alleles Egm1 and EgmP, lanes 2 and 4, respectively, and homozygous for the hypomorphic allele EgmH(lane 5). Egm is undetected in late Egm1/Egm1 larvae (lane 7). Actin was used as a loading control.
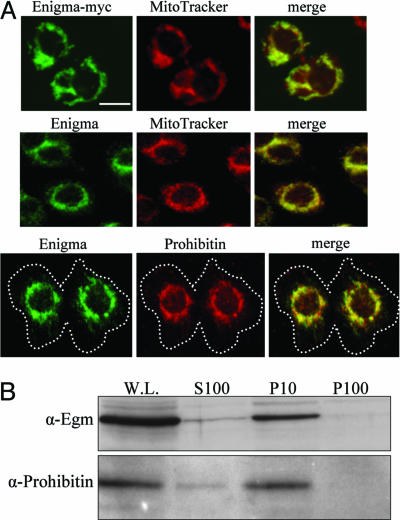
The intracellular sites of activity of all of the ACADs in metazoans are the peroxisomes and the mitochondria (11). Given the homology of Egm to this class of enzymes, we sought to determine its subcellular localization by immunocytochemical analysis. In Drosophila Kc-167 and SL2 cells, Egm is found almost exclusively in the mitochondrion. Transfected myc-tagged Egm in Kc-167 cells, stained with the monoclonal anti-myc antibody 9E10, localizes into the mitochondria, as highlighted by the mitochondrial marker MitoTracker (Fig. 2A). Using the antibody that we raised against the Enigma protein, the same localization pattern was observed for the endogenous protein, which colocalizes both with MitoTracker and the Drosophila homologue of mammalian prohibitin, a resident protein of the inner mitochondrial membrane (13) (Fig. 2 A). The mitochondrial localization of Egm was further demonstrated by cell fractionation experiments. Kc-167 cell extracts were subjected to differential centrifugation, and each fraction was analyzed by immunoblotting for the presence of Egm. As shown in Fig. 2B, the endogenous Egm protein is highly enriched in the fraction that contains the mitochondria (10,000 × g), as judged by the mitochondrial marker prohibitin, and is absent from the microsomal fraction (100,000 × g). The enrichment of Egm in the mitochondrial fraction, in combination with the immunostaining results, leads us to conclude that Egm resides in the mitochondria, where β-oxidation takes place.
Fig. 2.
Egm is a mitochondrial protein. (A) Transfected Egm-myc (Top, green) detected with anti-myc antibody overlaps with MitoTracker (Top, red). Endogenous Egm (Middle and Bottom, green) colocalizes with MitoTracker (Middle, red) and prohibitin (Bottom, red) in KC cells. White dashed line in the Bottom merged figure delineates the cell membrane. (Scale bar: 5 μm.) (B) Immunoblotting of cell fractions separated by centrifugation. W.L., whole lysate input; S100, cytosol; P10, pellet at 10,000 × g containing mitochondria; P100, pellet at 100,000 × g. Egm and the mitochondrial resident protein prohibitin are significantly enriched in the P10/mitochondrial fraction.
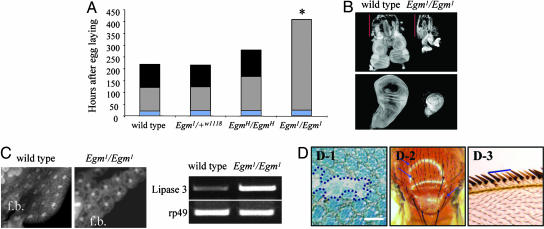
Egm Is an Essential Gene in Drosophila melanogaster. At the organismic level, Egm is an essential gene because homozygous null animals (Egm–/–) die during the late larval stage (third instar). Remarkably, although embryonic development seems to be normal, the mutant Egm–/– animals enter a prolonged larval lifespan that is extended, on average, by 3.9-fold compared with the WT (Fig. 3A). Invariably, late Egm–/– larvae develop subcuticular melanotic masses, have abnormally small eye and wing imaginal discs (Fig. 3B), and malformed fat bodies (Fig. 3C), conferring a transparent appearance to the animals. Consistent with this phenotype and in agreement with a role for Egm in lipid homeostasis, the stored fatty acids in the form of triglycerides are almost halved in larvae lacking zygotically expressed Egm (triglycerides: w1118 control = 70 ng/μg protein, SD ± 8.3, and Egm1/Egm1 = 37 ng/μg protein, SD ± 4.0). In parallel, the transcription levels of lipase-3, the enzyme responsible for triglyceride degradation and a molecular marker of starvation (14), are highly up-regulated (Fig. 3C).
Fig. 3.
Decreased amounts of Egm interfere with the normal development of the organism. (A) Reduced levels of Egm decelerate the rate of development in a dosage-dependent manner. Average larval-phase lengths are as follows: WT, 125 h; Egm1/Egm1, 408 h; EgmH/EgmH, 168 h. Maximal survival for Egm1/Egm1is 27 days. Average pupal-phase lengths are as follows: control, 98 h; EgmH/EgmH, 112 h. *, Egm1/Egm1 larvae invariably die before reaching the pupal stage.Each rectangle in a bar represents a different developmental stage: blue, embryonic; gray, larval; black, pupal. (B) Imaginal disk growth defects in the absence of Egm. The eye-antennal (Upper) and wing (Lower) imaginal discs remain underdeveloped in the homozygous Egm1 mutants. Red bars show the length of the larval mouth hooks for body-length comparison. (C) Normally fed Egm larvae show signs of lipid metabolism deregulation. The fat bodies (f.b.) in Egm1/Egm1third instar larvae are semitransparent and the cells contain large cytoplasmic vacuoles. RT-PCR shows up-regulation of lipase-3. Ribosomal protein rp49 was used as internal control. (D) Egm1/Egm1 mosaic clones in different tissues. (D-1) Adult eye retina. The boarders of the clone are indicated with blue dashed line. (Scale bar: 30 μm.) (D-2) Thorax. Blue arrows point mutant bristles. (D-3) Wing margin. Cells lacking Egm produce smaller wing margin bristles (yellow) than the WT (brown).
Low levels of Egm expressed in homozygous EgmH animals (Fig. 1B, compare lanes 3–5) are sufficient for survival but still result in a significant developmental delay and decreased amounts of stored fatty acids. EgmH/EgmH animals have a prolonged larval and pupal stage by 34% and 15%, respectively (Fig. 3A), and display, on average, a 39% decrease in triglyceride content compared with similarly aged control adult flies (triglycerides: EgmRev/EgmRev control = 56 ng/μg of protein, SD ± 4.9, and EgmH/EgmH = 34 ng/μg of protein, SD ± 8.2). The rate of development of the heterozygous Egm-null animals, however, is similar to WT (Fig. 3A). In conclusion, the developmental growth of the organism is sensitive to the dosage of the available Egm protein, although only under a certain threshold. In fact, overexpression of Egm, driven by the galactose-4 (GAL4) and upstream activation sequence (UAS) binary expression system (15), does not elicit any obvious phenotype (data not shown), an observation in agreement with other overexpression studies involving enzymes with saturation kinetics, such as most enzymes of intermediary metabolism (16).
Although overexpression of the protein does not interfere with the development of any organ in the fly, complete depletion of Egm impedes proper morphogenesis in all tissues analyzed. Clones of mutant Egm cells were generated by using the flipase-flipase recombination target (FLP/FRT) recombination system as described (17). In the eye, null Egm–/– clones show an aberrant morphology (Fig. 3D-1), indicating that Egm is required for eye development in a cell autonomous fashion. This is not an effect peculiar to this tissue, because further clonal analysis demonstrated that Egm is also necessary for the development of the bristles in the thorax (Fig. 3D-2) as well as in the wing margin (Fig. 3D-3). In addition, we found that mutant ovarioles do not produce viable eggs. These functional observations are compatible with Egm encoding for a ubiquitous mitochondrial protein that may be involved in a fundamental cellular process such as β-oxidation. This notion is strengthened by the phenotypic similarity of Egm–/– clones to mutant clones of scully (scu), an established component of the β-oxidation pathway (18).
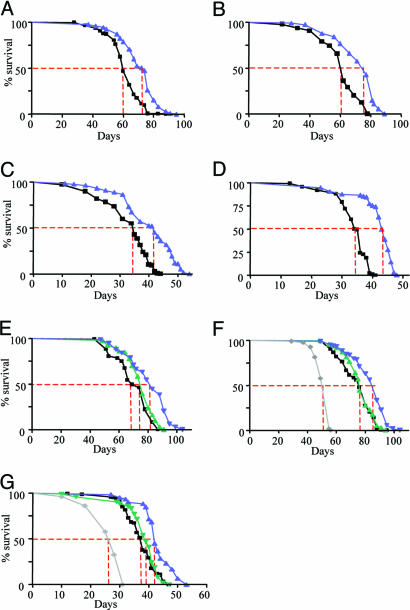
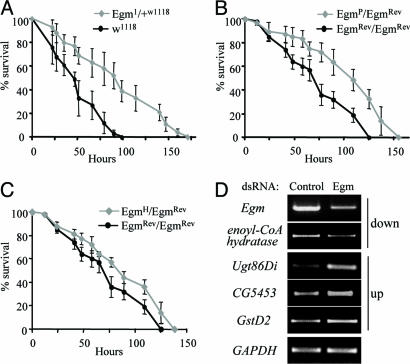
Lifespan Extension in Flies with Reduced Levels of Egm. The recessive phenotypes of the different alleles established Egm as an essential gene in Drosophila development that affects lipid homeostasis. Significantly for the present analysis, we have also uncovered a dominant longevity phenotype associated with the Egm mutations. At 25°C, the median survival of female flies heterozygous for the null allele Egm1 is on average 19.5% greater than their control flies, consisting of the parental w1118 strain in which the Egm1 mutation was originally generated (Fig. 4 A and B) (10).
Fig. 4.
Lifespan extension in Egm female flies. Survival curves for the two Egm allelic groups, group I (A–D) and group II (E–G), are shown. For each group, we present two independent trials at 25°C and two at 29°C, except for group II at 29°C where one trial is presented. (A–D) Longevity extension for Egm1/+w1118 mutant flies (group I) at 25°C (A and B) and 29°C (C and D). Median lifespan and sample sizes, respectively, are as follows: w1118, 60 days, n = 283 and Egm1/+w1118, 74 days, n = 398 (A); w1118, 62 days, n = 310 and Egm1/+w1118, 77 days, n = 362 (B); w1118, 35 days, n = 202 and Egm1/+w1118, 43 days, n = 103 (C). w1118, 36 days, n = 133 and Egm1/+w1118, 44 days, n = 87 (D). Black line, w1118; blue line, Egm1/+w1118. (E–G) Lifespan extension for group II at 25°C (E and F) and 29°C (G). Median lifespan and sample sizes, respectively, are as follows: EgmRev/EgmRev, 69 days, n = 149; EgmP/EgmRev, 83 days, n = 180; EgmH/EgmRev, 76 days, n = 177 (E); EgmRev/EgmRev, 76 days, n = 309; EgmP/EgmRev, 88 days, n = 327; EgmH/EgmRev, 76 days, n = 275; EgmH/EgmH, 55 days, n = 275 (F); EgmRev/EgmRev, 38 days, n = 141; EgmP/EgmRev, 42 days, n = 60; EgmH/EgmRev, 39 days, n = 95; EgmH/EgmH, 28 days, n = 128 (G). Black line, EgmRev/EgmRev; blue line, EgmP/EgmRev; green line, EgmH/EgmRev; gray line, EgmH/EgmH. All of the survivorship curves were analyzed with the GraphPad (San Diego) prism software. Log-rank tests were performed and a P < 0.001 was calculated for all of the curves. Dotted red line corresponds to the median lifespan for each strain.
To further explore the longevity phenotype and ensure that it is linked to Egm, we scored the effects on lifespan of other members of the allelic series, which have been generated in a distinct genetic background from that of the Egm1 and w1118 flies. We reasoned that monitoring the survival of Egm mutant flies for additional alleles in a diverse genetic profile would address the considerations of hybrid vigor effects on the observed longevity phenotype. Therefore, we scored the lifespan of flies carrying the null, P-element allele EgmP, the hypomorphic allele EgmH, or the WT allele EgmRev, which was isolated as a precise excision of the P-element and used as control. Analysis of these mutations further demonstrated that a significant lifespan extension is achieved when the levels of the Egm protein are halved; female flies carrying one copy of inactivated Egm by the P-element insertion (EgmP/EgmRev) live longer (18.4% on average) than the revertant, WT flies (EgmRev/EgmRev) (Fig. 4 E and F). A similar longevity phenotype is observed for Egm mutant flies kept at a higher temperature (29°C) (Fig. 4 C, D, and G). The sensitivity of the organism to the dosage of Egm is further demonstrated by the observation that a more drastic decrease in Egm protein, seen in EgmH homozygotes (Fig. 1B, lane 5), is deleterious for the organism, resulting in a 30% shorter lifespan at 25°C (Fig. 4 F and G).
The fact that the two different control strains (w1118 and EgmRev) have different survival curves (for example, compare Fig. 4 A and E) is indicative of the impact of genetic background on the lifespan of an organism. On the other hand, the extension in lifespan of Egm mutants compared with their respective controls in two different backgrounds emphasizes the importance of the activity of this locus in lifespan regulation. Furthermore, the fact that Egm male flies do not outlive their control, in either genetic background, provides additional evidence against the effect of hybrid vigor on the longevity phenotype. We conclude that Egm defines a critical factor for the control of lifespan in Drosophila.
The observation that Egm mutations do not have an obvious effect on the lifespan of male flies (Fig. 7, which is published as supporting information on the PNAS web site) could reflect sex-specific metabolic differences. Indeed, sex-related differences have been reported for mutations that affect lipid metabolism and specifically β-oxidation. Male flies, mutant for the acyl-CoA synthetase gene bubblegum (bgm) (Fig. 6B), show elevated levels of very long-chain fatty acids whereas the females with the same mutation do not (19). In addition, it is noteworthy that, in several cases, lifespan extension experiments by means of either environmental or genetic interventions have demonstrated a high degree of sex specificity (20–23). In light of our findings, it is also important to mention that female flies have been found to respond significantly stronger than males to dietary calorie restriction (21), which is the best characterized environmental intervention that prolongs lifespan. Differential, sex-dependent responses to modulations of the insulin pathway, as well as energy utilization and distribution, have been documented (21, 22, 24, 25). We should mention, however, that an explanation for the sex-related difference in lifespan observed in all organisms has yet to be provided.
Long-Lived Egm Flies Do Not Have Decreased Reproductive Potential. Lifespan extension has been suggested to be linked to a loss in reproductive output, because decreased fertility may reduce possible burdens associated with reproduction, thereby freeing resources for somatic maintenance (23). Given the female-specific lifespan extension observed in Egm mutants and the sterility of females with Egm-null germ-line clones, we examined the fertility of the Egm flies. The Egm long-lived heterozygous females do not show a loss in reproductive potential and have normal fertility and fecundity (Fig. 8, which is published as supporting information on the PNAS web site). Therefore, in agreement with studies carried out with other aging mutations, like the Krebs cycle-related gene Indy (4) or the ecdysone receptor gene EcR (26), the longevity phenotype in Egm mutant flies is not linked to an altered reproductive capability.
Egm Mutations Increase the Tolerance of the Organism to Oxidative Stress. Many long-lived organisms have increased resistance to free radical-generating chemicals such as paraquat (9). We first assayed the survival of WT flies at different concentrations of paraquat, and a working concentration of 7.5 mM was chosen, because it induces an intermediate stress response (Fig. 9A, which is published as supporting information on the PNAS web site). Under these experimental conditions, 50% of w1118 female flies die within 50 h after exposure. Egm mutant flies demonstrate a significantly increased resistance to the toxic effects of 7.5 mM paraquat compared with WT flies, as shown in Fig. 5 A–C. Consistent with the longevity phenotype, only female Egm flies show an increased tolerance to paraquat. Furthermore, the degree of stress tolerance correlates with the allelic strength of the different Egm mutants: heterozygous flies for null Egm mutations (Egm1/+w1118 and EgmP/EgmRev) show higher resistance than the flies heterozygous for the hypomorphic EgmH mutation, which, in turn, outperform WT flies (Fig. 5 A–C). These observations directly link Egm levels to the oxidative stress response of the organism.
Fig. 5.
Oxidative stress responses under decreased levels of Egm. (A) Survival curves of female flies heterozygous for Egm1 and the parental control (w1118) exposed to paraquat. (B) Survival curves of female flies heterozygous for EgmPand their control (EgmRev/EgmRev) exposed to paraquat. (C) Survival curves of female flies heterozygous for EgmH and their control (EgmRev/EgmRev) exposed to paraquat. All values in A, B, and C are presented as mean ± SEM. The results shown are for one experiment per genotype with four to six vials with 20 flies in each vial. (D) Transcriptional consequences of Egm reduction by RNA interference in KC cells. Shown is semiquantitative RT-PCR on transcripts displaying the largest alterations in expression in the microarray data analysis (for details, see Fig. 10, which is published as supporting information on the PNAS web site). GAPDH2 was used as internal control.
In an attempt to explore the mechanisms underlying the enhanced tolerance to oxidative stress conferred by low Egm levels, we examined the transcriptional consequences of decreased Egm using oligonucleotide arrays. Given that gene expression is dynamically regulated during the lifetime of an organism, comparative expression studies between WT and mutants animals with a delayed-aging phenotype are particularly complex. We reasoned, therefore, that tissue culture conditions would provide a more controlled system for studying the effects of Egm reduction at the transcriptional level, devoid of the physiological responses induced in the fly. To that end, we compared the transcriptional profile between Drosophila Kc-167 cells treated with Egm or control (β-lactamase) dsRNA. The reduction of Egm protein relative to the control was confirmed by Western blotting (Fig. 9B). Analysis of three data sets from microarrays shows that among ≈13,100 genes represented on the DrosGenome1 array (Affymetrix, Santa Clara, CA), changes were detected in a small subset of genes. Only 46 genes (0.35%) were down-regulated by 1.5-fold or more (P < 0.05), and 58 (0.44%) displayed an increase equal or >1.5-fold (P < 0.05). Egm mRNA, which was targeted by dsRNA, showed the largest decrease (6.6-fold) (Table 1, which is published as supporting information on the PNAS web site). We note that the second most down-regulated gene is a highly conserved homologue of the mammalian short chain enoyl-CoA hydratase (CG18645), an integral component of the β-oxidation pathway (Fig. 5D), further emphasizing the possible involvement of Egm in this process. A large percentage of up-regulated genes encodes for major detoxification enzymes, including several glutathione S-transferases (GstD2, GstD9, GstE6, and GstE2) and a cytochrome P450 (Cyp4p3) (27). Comparatively, the largest transcriptional induction corresponds to the expression of Ugt86Di (CG6658), one of the 33 fly homologues of UDG-glucosyltransferases (UGTs) (Fig. 5D) (28). These enzymes play a major role in the detoxification of a variety of endogenous and exogenous compounds. Up-regulation of UGT expression has also been observed in the long-lived insulin daf-2 mutants in Caenorhabditis elegans, and it has been proposed to promote longevity by augmenting the antioxidant and xenobiotic defense of the organism (27, 29). In spite of the obvious caveats associated with any extrapolation from a cell culture system to an in vivo situation, the transcriptional changes we detect in cell culture could possibly provide a physiological basis for the Egm-associated oxidative stress and by extension lifespan phenotypes (see Table 1 for the full list of results).
Discussion
This study indicates that Egm is a Drosophila longevity locus that encodes a mitochondrial protein. Moreover, indirect evidence suggests that this protein may be involved in β-oxidation. The genetic analysis of the allelic mutant series we generated has shown that the organism is particularly sensitive to the dosage of Egm. Complete loss of function affects the development of different tissues and is lethal at an organismic level. Low levels of Egm expressed by the hypomorphic allele EgmH are sufficient for life, but result in premature lethality. Intermediate amounts of the protein, found in heterozygous flies, increase lifespan and tolerance to oxidative stress. These phenotypes also demonstrate that precise regulation of Egm is an important factor for lifespan determination.
The analysis of the phenotypes associated with mutant Egm indicate an involvement of this locus in lipid metabolism. Therefore, it is reasonable to suggest that the impact of Egm on longevity is due to the modulation of factors that affect lipid homeostasis. The putative function of Egm was originally suggested by its sequence homology to ACADs. Consistent with an involvement in β-oxidation, Egm is localized in the primary site of β-oxidation, the mitochondrion, and Egm mutant animals display decreased levels of triglycerides, demonstrating a deregulation of the fatty acid degradation pathway. Indeed, reduced levels of Egm in tissue culture cells interfere with the normal expression of genes encoding for homologues of enzymes involved in lipid homeostasis, like a carnitine transporter (CG4630), a triglycerol lipase (CG11055), and the homologues of β-oxidation enzymes, enoyl-CoA hydratase (CG18645), and the short-branched chain ACAD (CG3902).
To date, there is no evidence to suggest a link between lifespan extension and β-oxidation regulation. However, two major factors affecting longevity, calorie restriction and modulation of the insulin-signaling pathway (6, 30), have a potent effect on lipid homeostasis, which is directly influenced by β-oxidation. Mutations in β-oxidation enzymes in humans are linked to a group of inherited, autosomal recessive diseases known as β-oxidation deficiencies. The clinical features of these diseases are very diverse, and patients are predominantly characterized by hypoglycemia, lipid accumulation, cardiomyopathy, and intolerance to prolonged fasting, whereas severe cases can cause acute morbidity and death (31). If our hypothesis is correct, it would be interesting to examine whether the heterozygous condition in humans positively affects lifespan.
In Drosophila, very little is known about the physiological effects of β-oxidation deficiencies. Mutant f lies for the β-oxidation-related gene bgm are characterized by progressive neurodegeneration and a shorter lifespan than WT flies (19). In addition, we have cloned and biochemically characterized the product of the Drosophila gene CG7461 and have shown that it is the fly orthologue of the human ACAD very-long chain dehydrogenase (VLCAD). We find that flies deficient in dVLCAD protein [i.e., homozygous for a null allele from the Exelixis Drosophila mutant collection (32–34)] die prematurely, similarly to the bgm mutants (data not shown). Together, these observations suggest that the modulation of lipid metabolism, specifically through mutations in genes involved in the β-oxidation pathway, can have a significant impact on the lifespan of the fruit fly. In the present context, it is noteworthy that treatment of Drosophila with 4-phenylbutyrate (PBA), a drug that has been reported to influence mitochondrial β-oxidation (35), can extend significantly the lifespan of the fly (36).
On the basis of the analysis presented here, we wish to raise the testable hypothesis that the β-oxidation pathway, a fundamental, evolutionary conserved mechanism of fatty acid degradation, might play a critical role in lifespan determination. It will therefore be important to investigate whether genes capable of modulating fatty acid breakdown may define a novel class of longevity mutations.
Materials and Methods
Drosophila Strains. The Egm1 allele was isolated in a genetic screen conducted in our laboratory (10) and found to be allelic to l(2)k14708 (henceforth EgmP) described in FlyBase. Viable lines were isolated by mobilization of the P-element in EgmP flies, and the precise excision of the P element in EgmRev was confirmed by PCR, sequencing, and Western blot analyses. Standard second chromosome FRT chromosomes described in FlyBase were used to generate mosaic clones. For the eye clones, FRT42D-Egm/CyO males were crossed to eyFLP; FRT42D/CyO females. For the wing and thorax clones, FRT42D-P(y+) Egm/CyO males were crossed to hsFLP; FRT42D/CyO females. To induce the expression of the FLP recombinase, larvae were heat-shocked twice for 1 h, 48 h, and 72 h after egg laying.
Immunofluorescence. An amount equal to 1 × 106 Kc-167 cells per ml was allowed to attach for 6 h on coverslips coated with 5 mg/ml Con A (catalog no. c2010, Sigma). For MitoTracker staining (catalog no. M-7513, Molecular Probes), live cells were incubated with 250 nM reagent in serum-free media for 25 min at room temperature. Cells were then fixed in 2% methanol-free formaldehyde for 20 min. Anti-Enigma antibody was diluted 1:500 in PBT (0.3% Triton X-100/PBS) with 5% goat serum for 2 h at room temperature. Anti-prohibitin antibody (catalog no. ab2996, Abcam, Cambridge, MA) was used at a concentration of 50 μg/ml in PBT plus 5% goat serum. Staining for prohibitin colocalizes with MitoTracker (data not shown).
Cell Fractionation. Cell fractionation was performed as described in ref. 37 with slight modifications. For a detailed protocol, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Triglyceride Assay. For the triglyceride assay, 10 15-day-old flies or 20 larvae were lysed with 200 μl of lysis solution (150 mM NaCl/10 mM Tris•HCl, pH 8.0/0.1% Triton X-100). Triglyceride concentration in the filtered homogenate was measured by using the Stanbio (Boerne, TX) LiquiColor assay kit, according to the manufacturer's instructions. The protein concentration of the same homogenate solution was measured by using a bicinchoninic acid (BCA) assay method.
Paraquat Assay. For measuring resistance to oxidative stress induced by paraquat, 10-day-old female flies were dry-starved for 5 h at 25°C and exposed to 7.5 mM paraquat (Sigma) in 5% sucrose solution. Dead flies were scored periodically.
RNA Interference in Tissue Culture Cells. dsRNA was produced by using the RiboMax RNA production system (Promega). The target sequence was 845 bp, corresponding to bases 94–939 of Egm cDNA (forward primer: T7-tccagcagcttggactcac; reverse primer: T7-gttgccatcgtgtggtagg). An amount equal to 0.5 ml of 1.6 × 106 cells was incubated for 1 h with 20 μg of dsRNA in serum-free Sang's M3 medium. Subsequently, an equal volume of 10% serum M3 medium was added, and the cells were collected after 5 days. The efficiency of the RNA interference (RNAi) was measured by immunoblotting.
Microarray Gene Expression Analysis. RNA samples from Kc-167 cells were subjected to analysis by Affymetrix high-density oligonucleotide arrays by using the DrosGenome1 array. Probe synthesis and microarray hybridization were performed according to standard Affymetrix protocols. External standards were included to control for hybridization efficiency and sensitivity. The chips were scanned with a Hewlett–Packard GeneArray laser scanner. We used the R-based analysis packages gene chip robust multiarray expression measure (gcrma) (38) and linear modes for microarray data (limma) (39) to extract and normalize signal levels and perform statistical analysis to identify differentially expressed genes. The microarray experiment was executed three times, by using three independent dsRNA preparations for β-lactamase/control and Egm.
Supplementary Material
Acknowledgments
We thank Stewart Frankel for critical comments. This work was supported by grants from the National Institutes of Health and the Ellison Medical Foundation.
Author contributions: P.M. and S.A.-T. designed research; P.M. performed research; P.M. and G.D.H. analyzed data; and P.M. and S.A.-T. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: Egm, Enigma; ACAD, acyl-CoA dehydrogenase.
Data deposition: The microarray data have been deposited in Gene Expression Omnibus (accession no. GSE3566).
References
- 1.Clancy, D. J., Gems, D., Hafen, E., Leevers, S. J. & Partridge, L. (2002) Science 296, 319. [DOI] [PubMed] [Google Scholar]
- 2.Tissenbaum, H. A. & Guarente, L. (2002) Dev. Cell 2, 9–19. [DOI] [PubMed] [Google Scholar]
- 3.Rogina, B., Helfand, S. L. & Frankel, S. (2002) Science 298, 1745. [DOI] [PubMed] [Google Scholar]
- 4.Rogina, B., Reenan, R. A., Nilsen, S. P. & Helfand, S. L. (2000) Science 290, 2137–2140. [DOI] [PubMed] [Google Scholar]
- 5.Lakowski, B. & Hekimi, S. (1998) Proc. Natl. Acad. Sci. USA 95, 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koubova, J. & Guarente, L. (2003) Genes Dev. 17, 313–321. [DOI] [PubMed] [Google Scholar]
- 7.Bluher, M., Kahn, B. B. & Kahn, C. R. (2003) Science 299, 572–574. [DOI] [PubMed] [Google Scholar]
- 8.Sohal, R. S. & Weindruch, R. (1996) Science 273, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel, T. & Holbrook, N. J. (2000) Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- 10.Verheyen, E. M., Purcell, K. J., Fortini, M. E. & Artavanis-Tsakonas, S. (1996) Genetics 144, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartlett, K. & Eaton, S. (2004) Eur. J. Biochem. 271, 462–469. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, S., Bartlett, K. & Pourfarzam, M. (1996) Biochem. J. 320, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nijtmans, L. G., de Jong, L., Artal Sanz, M., Coates, P. J., Berden, J. A., Back, J. W., Muijsers, A. O., van der Spek, H. & Grivell, L. A. (2000) EMBO J. 19, 2444–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinke, I., Kirchner, C., Chao, L. C., Tetzlaff, M. T. & Pankratz, M. J. (1999) Development (Cambridge, U.K.) 126, 5275–5284. [DOI] [PubMed] [Google Scholar]
- 15.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 16.Tatar, M. (1999) Am. Nat. 154, Suppl., S67–S68. [DOI] [PubMed] [Google Scholar]
- 17.Xu, T. & Rubin, G. M. (1993) Development (Cambridge, U.K.) 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 18.Torroja, L., Ortuno-Sahagun, D., Ferrus, A., Hammerle, B. & Barbas, J. A. (1998) J. Cell Biol. 141, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min, K. T. & Benzer, S. (1999) Science 284, 1985–1988. [DOI] [PubMed] [Google Scholar]
- 20.Burger, J. M. & Promislow, D. E. (July 14, 2004) SAGE KE, 10.1126/sageke.2004.28.pe30. [DOI] [PubMed]
- 21.Magwere, T., Chapman, T. & Partridge, L. (2004) J. Gerontol. A Biol. Sci. Med. Sci. 59, 3–9. [DOI] [PubMed] [Google Scholar]
- 22.Clancy, D. J., Gems, D., Harshman, L. G., Oldham, S., Stocker, H., Hafen, E., Leevers, S. J. & Partridge, L. (2001) Science 292, 104–106. [DOI] [PubMed] [Google Scholar]
- 23.Tatar, M., Kopelman, A., Epstein, D., Tu, M. P., Yin, C. M. & Garofalo, R. S. (2001) Science 292, 107–110. [DOI] [PubMed] [Google Scholar]
- 24.Gems, D. & Riddle, D. L. (2000) Genetics 154, 1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holzenberger, M., Dupont, J., Ducos, B., Leneuve, P., Geloen, A., Even, P. C., Cervera, P. & Le Bouc, Y. (2003) Nature 421, 182–187. [DOI] [PubMed] [Google Scholar]
- 26.Simon, A. F., Shih, C., Mack, A. & Benzer, S. (2003) Science 299, 1407–1410. [DOI] [PubMed] [Google Scholar]
- 27.McElwee, J. J., Schuster, E., Blanc, E., Thomas, J. H. & Gems, D. (2004) J. Biol. Chem. 279, 44533–44543. [DOI] [PubMed] [Google Scholar]
- 28.Luque, T. & O'Reilly, D. R. (2002) Insect Biochem. Mol. Biol. 32, 1597–1604. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., Li, H. & Kenyon, C. (2003) Nature 424, 277–283. [DOI] [PubMed] [Google Scholar]
- 30.Saltiel, A. R. & Kahn, C. R. (2001) Nature 414, 799–806. [DOI] [PubMed] [Google Scholar]
- 31.Rinaldo, P., Raymond, K., al-Odaib, A. & Bennett, M. J. (1998) Curr. Opin. Pediatr. 10, 615–621. [DOI] [PubMed] [Google Scholar]
- 32.Artavanis-Tsakonas, S. (2004) Nat. Genet. 36, 207. [DOI] [PubMed] [Google Scholar]
- 33.Thibault, S. T., Singer, M. A., Miyazaki, W. Y., Milash, B., Dompe, N. A., Singh, C. M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H. L., et al. (2004) Nat. Genet. 36, 283–287. [DOI] [PubMed] [Google Scholar]
- 34.Parks, A. L., Cook, K. R., Belvin, M., Dompe, N. A., Fawcett, R., Huppert, K., Tan, L. R., Winter, C. G., Bogart, K. P., Deal, J. E., et al. (2004) Nat. Genet. 36, 288–292. [DOI] [PubMed] [Google Scholar]
- 35.McGuinness, M. C., Zhang, H. P. & Smith, K. D. (2001) Mol. Genet. Metab. 74, 256–263. [DOI] [PubMed] [Google Scholar]
- 36.Kang, H. L., Benzer, S. & Min, K. T. (2002) Proc. Natl. Acad. Sci. USA 99, 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorstyn, L., Read, S., Cakouros, D., Huh, J. R., Hay, B. A. & Kumar, S. (2002) J. Cell Biol. 156, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, Z., LeBlanc, R. & Irizarry, R. A. (September 2003) Stochastic Models Based on Molecular Hybridization Theory for Short Oligonucleotide Microarrays (Collection of Biostatistics Research Archive, Berkeley Electronic Press, Berkeley, CA), The Johns Hopkins University Department of Biostatistics Working Paper No. 4.
- 39.Wettenhall, J. M. & Smyth, G. K. (2004) Bioinformatics 20, 3705–3706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.