Abstract
The use of chemokine receptors as cell recognition signals is a property common to several lentiviruses, including feline, human, and simian immunodeficiency viruses. Previously, two feline immunodeficiency virus (FIV) isolates, V1CSF and Petaluma, were shown to use chemokine receptors in a strain-dependent manner to infect human peripheral blood mononuclear cells (PBMC) (J. Johnston and C. Power, J. Virol. 73:2491-2498, 1999). Since the sequences of these viruses differed primarily in regions of the FIV envelope gene implicated in receptor use and cell tropism, envelope chimeras of V1CSF and Petaluma were constructed to investigate the role of envelope diversity in the profiles of chemokine receptors used by FIV to infect primate cells. By use of a receptor-blocking assay, all viruses were found to infect human and macaque PBMC through a mechanism involving the CXCR4 receptor. However, infection by viruses encoding the V3-to-V5 region of the V1CSF surface unit was also inhibited by blockade of the CCR3 or CCR5 receptor. Similar results were obtained with GHOST cells, human osteosarcoma cells expressing specific combinations of chemokine receptors. CXCR4 was required for infection by all FIV strains, but viruses expressing the V3-to-V5 region of V1CSF required the concurrent presence of either CCR3 or CCR5. In contrast, CXCR4 alone was sufficient to allow infection of GHOST cells by FIV strains possessing the V3-to-V5 region of Petaluma. To assess the role of primate chemokine receptors in productive infection, Crandell feline kidney (CrFK) cells that expressed human CXCR4, CCR3, or CCR5 in addition to feline CXCR4 were generated. Sustained infection by viruses encoding the V3-to-V5 region of V1CSF was detected in CrFK cells expressing human CCR3 or CCR5 but not in cells expressing CXCR4 alone, while all CrFK cell lines were permissive to viruses encoding the V3-to-V5 region of Petaluma. These results indicate that FIV uses chemokine receptors to infect both human and nonhuman primate cells and that the profiles of these receptors are dependent on envelope sequence, and they provide insights into the mechanism by which xenoinfections may occur.
A large body of evidence supports the ability of retroviruses to cross species. For example, it is accepted widely that human immunodeficiency virus type 1 (HIV-1) is the result of the propagation and adaptation of a simian immunodeficiency virus (SIV) strain (SIVcpz) from chimpanzees to humans, following an earlier cross-species transmission from monkeys to chimpanzees (15). However, little is known about the mechanisms by which viruses adapt to and spread in a new host. Feline immunodeficiency virus (FIV) is a retrovirus that causes persistent infection and an immunodeficiency syndrome in domestic cats similar to those caused by HIV and simian SIV in their respective hosts (1). Although FIV is antigenically distinct from the primate lentiviruses, it shares many biological properties that manifest in its ability to infect productively both primary and immortalized primate cell lines in vitro (10, 20, 22, 28, 44). Thus, it is likely that the determinants of feline cell tropism, such as envelope-mediated entry of target cells and long terminal repeat-mediated transcription of the viral genome following entry, may also influence infection of primate cells by FIV.
The requirement for specific cell surface molecules to act as receptors for viral entry represents a major obstacle to lentiviral xenoinfection (17). Within its natural host, FIV possesses a broad cell tropism, infecting many blood- and brain-derived cell types with various degrees of efficiency (5, 7, 9, 13, 35). In contrast, a pattern of in vitro cell tropism exists in which most primary isolates of FIV infect primary lymphocytes, but fewer viral strains exhibit the ability to adapt to and replicate in fibroblast-derived Crandell feline kidney (CrFK) cells (8). The diverse nature of the cells permissive to FIV suggests that a widely expressed host molecule serves as a cell recognition signal for infection; however, the primary receptor used by FIV has not been identified. Although FIV preferentially targets CD4+ lymphocytes during infection (49), the feline CD4 molecule is not the primary FIV receptor. As with HIV and SIV, however, recent studies have implicated the chemokine receptors, a family of G-protein-coupled integral membrane proteins that mediate the biological activities of chemokines, in infection by both primary isolates and molecular clones of FIV (38). For example, CXCR4 antibodies (50), agonists (18), and antagonists (11) have been shown to block FIV infection and FIV-mediated fusion, while expression of CXCR4 on nonpermissive cell lines has been shown to increase their susceptibility to FIV infection (36, 50). Although the potential for other host molecules to serve as receptors for FIV infection remains unclear, several lines of evidence suggest that chemokine receptors other than CXCR4 may be involved. For example, the inhibition of FIV infection by CXCR4 inhibitors and agonists is incomplete and varies between primary and culture-adapted FIV strains (38). In addition, the presence of CXCR4 alone on target cells is not sufficient to allow infection by all FIV strains (18, 26), and previous reports have demonstrated that FIV can interact with both CXC and CC chemokine receptors in a viral-strain-dependent manner during the infection of feline (26) and nonfeline (22) cells. However, it is not known whether chemokine receptors are the primary FIV receptors or whether they have accessory functions as coreceptors.
The capacity of both primate and nonprimate lentiviruses to interact with chemokine receptors suggests common determinants in the pathogenesis of these viruses that may facilitate cross-species transmission. For example, the CD4-independent use of human chemokine receptors has been shown to mediate infection of primary human cells by both SIV (21) and FIV (22) through a mechanism that is viral-strain dependent. Similarly, FIV infection of human and rodent cell lines expressing primate CXCR4 has been demonstrated (36). Since the primary function of the envelope glycoproteins is host cell recognition and entry, the mutability of these proteins provides a molecular basis for differences in target cell tropism and receptor usage (1). For example, single amino acid changes in the V3 region of the FIV surface unit (SU) envelope glycoprotein (47) or transmembrane (TM) glycoprotein (46) determine whether FIV strains are preferentially tropic for interleukin-2-independent cell lines, such as CrFK cells, or primary lymphocytes. Similarly, residues in the V3 and V4 domains of the SU determine FIV tropism for macrophages (45). Despite wide variations in sequence, the major functions of the envelope are maintained across genetically distinct viruses (32), suggesting that lentiviral envelope proteins possess a conserved structural framework that may promote similarities in biological properties conducive to xenoinfection. For example, specific envelope sequences have been shown to determine the ability of SIV to infect human cells (21) and to influence the development of disease in primate zoonoses involving chimeras of SIV and HIV (SHIVs) (6, 40).
Previously, it was demonstrated that infection of primate cells by FIV is viral-strain dependent and involves human chemokine receptors (22). Moreover, xenoinfection of nonhuman primates by FIV has been reported recently (24). In the present study, these results were extended by showing that chemokine receptors also mediate FIV infection of primary cells from a nonhuman primate species. Furthermore, by use of envelope chimeras of blood-derived Petaluma and cerebrospinal fluid (CSF)-derived V1CSF, the V3-to-V5 region of the FIV SU was found to be the principal determinant of this property. Although CXCR4 was essential to infection by all FIV strains, the requirement for the concurrent presence of additional chemokine receptors, such as CCR3 and CCR5, to allow infection by chimeras expressing the V3-to-V5 region of V1CSF suggested the cooperative use of multiple chemokine receptors by some FIV strains. These results indicate that common factors mediate the ability of FIV to infect primary cells from different primate species, and they may provide insight into the mechanism of cross-species infection and the role of diversity in the lentiviral envelope gene in this process.
MATERIALS AND METHODS
Cell culture.
Peripheral blood mononuclear cells (PBMC) used for infection studies and preparation of viral stocks were isolated from blood obtained from specific-pathogen-free adult felines by density-gradient centrifugation, as described previously (22). PBMC were initially stimulated for 3 days with 5 μg of concanavalin A/ml and then maintained in RPMI 1640 medium with 15% fetal calf serum (FCS) and 100 IU of interleukin-2/ml. CrFK cells were obtained from the American Type Culture Collection (Manassas, Va.) and cultured in minimal essential medium (MEM) with 10% horse serum, 2 mM l-glutamine, and 1 mM sodium pyruvate. CrFK cells transfected with human chemokine receptors (trCrFK) were cultured in the same media with the addition of 700 μg of G418/ml (Gibco, Burlington, Ontario, Canada). Human osteosarcoma cells expressing specific combinations of chemokine receptors (GHOST cells; National Institutes of Health [NIH] AIDS Reagent Program) were cultured in MEM supplemented with 10% FCS, 100 μg of hygromycin/ml, 500 μg of G418/ml, and 1 μg of puromycin/ml. Parental GHOST cells expressing only CD4 were cultured in the absence of puromycin. All cells were grown in the presence of penicillin and streptomycin.
Construction of trCrFK cells.
CrFK cells (4 × 105) were seeded in 6-well plates and cultured for 24 h to achieve 50% confluence. Plasmid DNAs (2 μg) encoding the human CCR3, CCR5, or CXCR4 chemokine receptor (NIH AIDS Reagent Program) and Lipofectin reagent (10 μl; Gibco) were each diluted in 100 μl of Opti-MEM serum-free medium (Gibco) and incubated at room temperature for 30 min. The two solutions were combined, incubated for 10 min at room temperature, and added to CrFK cells in 2 ml of Opti-MEM. Cells were incubated for 8 h at 37°C, washed twice with phosphate-buffered saline (PBS), and cultured in MEM containing 10% horse serum for 3 days. After 3 days, stable transfectants were obtained by selection for 4 weeks in medium containing 700 μg of G418/ml. Following selection, chemokine receptor expression in each cell line was confirmed by Western blot analysis (23) and flow cytometry. Mock-transfected cells were generated by using an empty vector (pcDNA3.1) lacking an expression cassette for any of the chemokine receptors and were analyzed in the same manner as transfected cells.
Viruses.
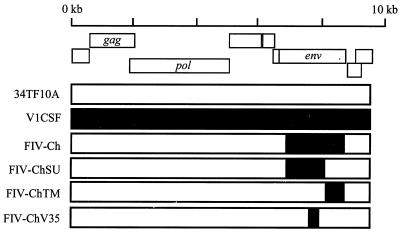
The FIV strains used in this study included the primary isolate V1CSF, which was derived from the CSF of a cat with encephalopathy (37) and underwent fewer than 10 passages in feline PBMC prior to the present experiments to minimize the effects of culture adaptation. Virus was also obtained from infectious molecular clones of FIV by transfection into CrFK cells and selection in feline PBMC, as described previously (23). These viruses included a hybrid of two molecular clones of the blood-derived Petaluma strain of FIV (34), pFIV-34TF10 and pFIV-14 (NIH AIDS Reagent Program), which was constructed by replacing sequences encoding the ORFA gene (nucleotides [nt] 5328 to 6393) of pFIV-34TF10 with equivalent sequences from pFIV-14. The resulting hybrid lacked both the 8 kb of extraneous cellular DNA incorporated into pFIV-14 and the stop codon present in the ORFA gene of pFIV-34TF10. Similarly, an envelope chimera of V1CSF and Petaluma was generated by replacing specific regions of the envelope gene of the Petaluma hybrid with equivalent regions from V1CSF (23). These chimeras included viruses expressing the entire envelope (FIV-Ch, nt 6393 to 8906), the SU (FIV-ChSU, nt 6363 to 8287) or TM (FIV-ChTM, nt 8287 to 8906) glycoprotein, or the V3-to-V5 region (FIV-ChV35, nt 7325 to 8200) of V1CSF (Fig. 1). Culture supernatants from FIV-infected feline PBMC, which served as sources of infectious virus for these experiments, were cleared of cellular debris by centrifugation and titered by limiting dilution as described previously (22). For infection, cells were inoculated with 200 μl of viral stock (102.5 to 104.5 50% tissue culture infective doses [TCID50]/ml), incubated for 2 h at 37°C, washed twice, and cultured as described above until supernatants and cells were harvested. Uninfected cells or cells treated with heat-inactivated virus served as controls.
FIG. 1.
The V1CSF-Petaluma envelope chimeras. Fragments representing specific regions of the FIV envelope gene (env) were removed from the Petaluma molecular clone (34TF10A) and replaced with equivalent sequences from V1CSF that were amplified and cloned from genomic DNA from V1CSF-infected feline PBMC. The resulting chimeras included viruses encoding the entire envelope (FIV-Ch), the SU (FIV-ChSU) or TM (FIV-ChTM) glycoprotein alone, or only the V3-to-V5 region of the SU (FIV-ChV35) from V1CSF.
PCR analyses.
Genomic DNA was isolated from cultured cells by using DNAzol reagent (Gibco) according to the manufacturer's protocols. PCRs were performed in a mixture containing 0.2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, 0.2 μM both sense and antisense primers, 5 mM KCl, 1 mM Tris-HCl, 0.25 U of Taq DNA polymerase, and 2 μl of template (300 to 400 ng) in a total volume of 25 μl. For semiquantitative PCR, template DNA or cDNA was amplified by 1 cycle of 95°C for 5 min (denaturing), 30 cycles of 95°C for 1 min, 50 to 65°C for 1 min, and 72°C for 2 min (amplification), and 1 cycle of 72°C for 10 min (extension). Nested PCR was performed by using 2 μl of product amplified in the first PCR with the same reagents and reaction conditions. FIV infection was detected by amplification of a conserved region of the FIV pol gene using primers 5′-GAAGGATCCAGAAAAGATACTATGG-3′ and 5′-GGCAACATTAGCTTTACCCCTGTTGG-3′, generating a 770-bp fragment corresponding to region 3361 to 4131 following 30 cycles of PCR. Nested PCR with primers 5′-CCAGATATGATGGAGGGAATCT-3′ and 5′-CATATCCTGCATCTTCTGAACT-3′ generated a 192-bp fragment corresponding to region 3860 to 4052. To minimize the possibility of contamination, PCRs were prepared in one room, templates were added in a second, and the actual PCR amplification was conducted in a third area. As a contamination control for each reaction, 2 μl of water was added to the reaction mixture instead of the template. Loading of equal amounts of template from each sample was ensured by amplification of the host glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene using primers 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ and 5′-CGGAGTCAACGGATTTGGTCG-3′ at an annealing temperature of 50°C. To optimize the number of cycles and amount of template necessary for amplification within the linear range of detection of the PCR protocol, standard curves were generated for all primer pairs by using serial dilutions of template from control samples. The limit of detection for primers used to amplify the FIV pol gene was determined previously to be 10 to 25 proviral copies (22). Genomic DNA or cDNA derived from FIV-infected feline PBMC served as a positive control for all studies involving detection of FIV infection. Templates derived from uninfected cell cultures served as negative controls.
Southern blot and densitometric analyses.
PCR products were separated by agarose gel electrophoresis, transferred to a nylon membrane via capillary action, and probed with an [α-32P]dCTP-labeled oligonucleotide probe specific for the FIV pol gene (5′-TGTCAAACAATGATGATAATAGAAGG-3′) or the GAPDH gene (5′-ATCAATGGAAATCACCATCTTCCAG-3′), as described previously (22). DNA and cDNA levels were quantitated by densitometric analysis of a minimum of three Southern blots from separate experiments. Briefly, blots were scanned in grayscale at high resolution (>600 dpi; Hewlett-Packard ScanJet 6300C), and optical density was measured by using Scion Image image processing and analysis software. Levels of each host or viral gene were normalized to the corresponding GAPDH level for statistical analysis and compared to standards with known concentrations. Standards were included on all membranes to calibrate for relative transfer and exposure efficiency and to allow comparison between blots.
Chemokine receptor-blocking assay.
Cells were incubated at 37°C for 1.5 h with monoclonal antibodies recognizing specific chemokine receptors, washed twice, and then resuspended in 0.3 ml of medium containing both virus and antibody (half the original blocking concentration). Following incubation at 37°C for 2 h, cells were washed twice and cultured for 24 h. Infected cells were harvested, and FIV proviral levels in genomic DNA were determined by semiquantitative PCR and densitometric analysis of the FIV pol gene, as described above. Uninfected cells and cells infected in the absence of antibodies to any of the chemokine receptors served as negative and positive controls, respectively. FIV DNA levels were normalized to the corresponding GAPDH level for statistical analysis and were compared to the positive controls for each strain. A minimum of two blots from separate experiments were analyzed. To investigate the effect of a nonspecific protein on FIV infection, human PBMC were incubated with 1 μg of either bovine serum albumin or an anti-mouse immunoglobulin G (IgG) antibody/ml and viral DNA levels were assessed in the same manner as with the chemokine receptor antibodies. Antibodies to human CCR3, CCR5, and CXCR4 were obtained from the NIH AIDS Reagent Program (catalogue numbers 3932, 3933, and 4084) and used at concentrations of 3, 3, and 5 μg/ml, respectively. To determine optimal concentrations, defined as the concentration at which maximal inhibition of FIV infection was observed, dose-response studies were conducted using human PBMC infected with V1CSF in the presence of varying concentrations (0.5 to 5.0 μg/ml) of antibody.
Flow cytometry.
CrFK and trCrFK cells were harvested and pelleted by centrifugation, and approximately 106 cells were resuspended in PBS containing 0.1% sodium azide. Cells were incubated for 20 min at room temperature with the monoclonal antibody to human CXCR4 (10 μg/ml), CCR5 (5 μg/ml), or CCR3 (5 μg/ml) described above, washed twice, and then incubated for 20 min with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (0.25 μg/μl; Becton Dickinson, San Jose, Calif.). Labeled cells were washed twice and resuspended in 0.5 ml of 1% formalin in PBS for analysis. By using a FacScan flow cytometer (Becton Dickinson) with the argon laser excitation set at 488 nm, data were collected from approximately 15,000 events for each experimental condition, and results were expressed as a single-parameter log fluorescence histogram. Aliquots of each cell line incubated in the absence of antibodies or with 1 μg of FITC-labeled, isotype-matched murine IgG1 (Becton Dickinson)/ml served as controls.
RT assay.
Reverse transcriptase (RT) activity in culture supernatants was measured by using a protocol described previously (23). Briefly, 10 μl of culture supernatant was cleared of cellular debris by centrifugation and incubated with 40 μl of a reaction cocktail containing [α-32P]TTP for 2 h at 37°C. Samples were blotted on DE81 Ion Exchange Chromatography Paper (Whatman International Ltd., Maidstone, United Kingdom) and washed three times for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and twice for 5 min in 95% ethanol. RT levels were measured by liquid scintillation counting. All assays were performed in duplicate and repeated a minimum of two times.
Statistical analyses.
Statistical analyses were performed by using Graphpad Prism, version 3.0 for Windows (Graphpad Software, San Diego, Calif.). For comparisons of three or more unmatched groups, one-way analysis of variance (ANOVA) was performed followed by a Tukey-Kramer multiple-comparison posttest to determine differences between specific groups. For data derived from two groups, Student's t test was used. P values below 0.05 were considered significant for all tests.
RESULTS
Feline cell tropism of the parent and chimeric FIV strains.
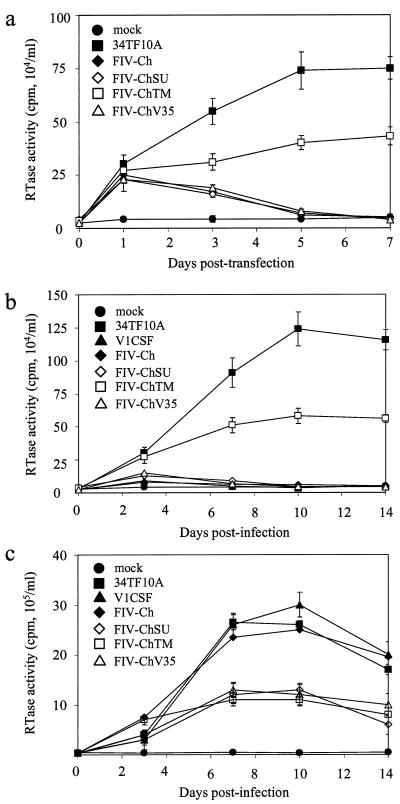
To investigate the infectivity and cell tropism of the parent and chimeric viruses (Fig. 1), CrFK cells were transfected with plasmids encoding each virus, and RT activity was measured in culture supernatants at successive days posttransfection. Compared to mock-transfected cells, RT activity that increased over a 1- week time course was detected in CrFK cultures transfected with either the hybrid molecular clone of Petaluma or the FIV-ChTM chimera (Fig. 2a). In contrast, RT activity was increased to similar levels immediately following transfection with the other chimeras but declined to background levels by day 5 posttransfection. To investigate the relative abilities of the viruses to replicate in feline cells following cell-free infection, CrFK cells and primary feline PBMC were infected with parent or chimeric virus and replication was assessed over a 2-week period. Analysis of RT activity in culture supernatants confirmed infection of feline PBMC by all viruses, with peak RT activity detected at day 10 postinfection (p.i.) (Fig. 2c). However, the chimeras other than FIV-Ch replicated to a lesser extent than either of the parent viruses (P < 0.005), whereas FIV-Ch infected PBMC as effectively as V1CSF and Petaluma. Compared to mock-infected controls, elevated RT activity that increased over a 2-week time course was also detected in CrFK cultures infected with Petaluma or FIV-ChTM (Fig. 2b), but as with PBMC, RT activity in CrFK cells infected with FIV-ChTM was lower than that in Petaluma-infected cultures (P < 0.005). In contrast, increased RT activity was not detectable after 3 days p.i. in CrFK cells infected with the other viruses (Fig. 2b). These results, which supported the findings of the transfection studies, indicated that FIV-ChTM possessed a feline cell tropism that resembled that of Petaluma, while the other chimeras more closely resembled the V1CSF isolate.
FIG. 2.
Replication of parent and chimeric FIV in feline cells following transfection and cell-free infection. Replication was investigated by RT assay of culture supernatants from transfected CrFK cells (a) or from CrFK cells (b) and primary feline PBMC (c) infected in a cell-free manner by each virus. RT activity for each clone is expressed as the mean ± standard error of the mean (counts per minute per milliliter) of triplicate samples from three independent wells. Significant differences in RT activity relative to control cultures were determined by ANOVA and the Tukey-Kramer posthoc test. (a) Increased RT activity was detected beyond 3 days only in CrFK cells transfected with Petaluma or FIV-ChTM. Significant differences between mock- and FIV-transfected cultures were observed at day 1 for all clones (P < 0.01) and at days 3, 5, and 7 for 34TF10A and FIV-ChTM (P < 0.001). (b) Elevated RT activity relative to controls was detected at all days p.i. (P < 0.001) in CrFK cells infected with Petaluma or FIV-ChTM. Activity in CrFK cells infected with FIV-ChSU or FIV-ChV35 was increased at day 1 p.i. only (P < 0.001). RT levels in Petaluma-infected cells exceeded thoaw in FIV-ChTM-infected cells at days 7, 10, and 14 (P < 0.005). (c) Compared to mock-infected controls, increased RT activity that peaked at day 10 p.i. was detected in all FIV-infected PBMC cultures. (P < 0.001 at all days). RT activity was significantly greater in PBMC infected with V1CSF, Petaluma, or FIV-Ch than in cells infected with the other viruses at days 7, 10, and 14 p.i. (P < 0.005).
Infection of primate PBMC by FIV involves chemokine receptors.
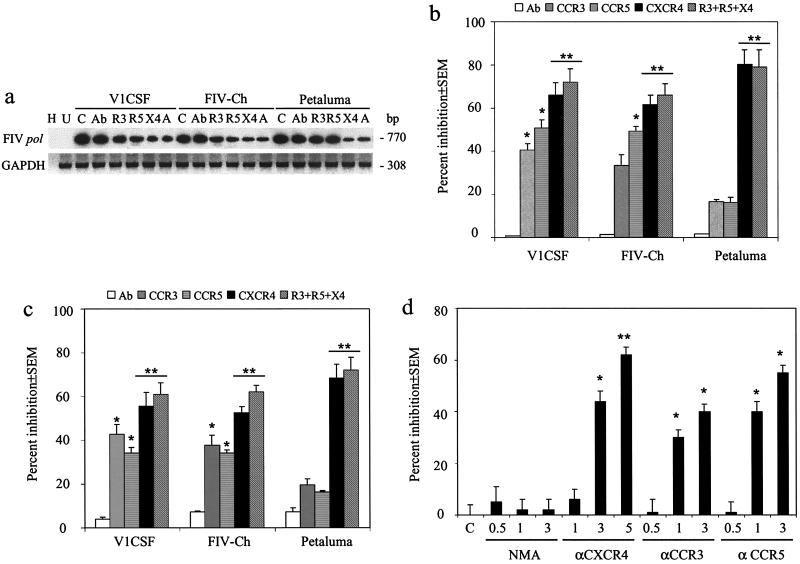
Previously, we demonstrated that blockade of human chemokine receptors inhibited infection of human PBMC by primary isolates of V1CSF and Petaluma in a viral-strain-dependent manner (22). To assess the role of envelope diversity in this process, a receptor-blocking assay (22) was employed in which human or cynomolgus macaque PBMC were infected with FIV in the presence or absence of blocking antibodies specific to the human CCR3, CCR5, or CXCR4 chemokine receptor. Viral entry was assessed by semiquantitative PCR and densitometric analyses of the levels of the FIV pol gene in antibody-treated and untreated cultures following a single round of infection, as shown for macaque PBMC (Fig. 3a). For all viruses, treatment with anti-CXCR4 antibodies resulted in the greatest decrease in viral DNA levels, but differences were observed in the extent of inhibition depending on the FIV strain. FIV genome levels in Petaluma-infected macaque (Fig. 3b) and human (Fig. 3c) PBMC were decreased by 68.4% ± 6.3% and 80.3% ± 6.8%, respectively, while infection by V1CSF (66.0% ± 5.7% and 55.6% ± 4.5%) and FIV-Ch (61.5% ± 4.5% and 52.6% ± 4.5%) was reduced to a lesser extent. With V1CSF, antibodies to the CCR5 and CCR3 chemokine receptors inhibited infection of PBMC from either species by approximately 50 and 40%, respectively (Fig. 3b and c). Although similar decreases in FIV DNA were detected in both FIV-Ch-infected human PBMC and FIV-Ch-infected macaque PBMC following treatment with anti-CCR5 antibodies (approximately 50%), anti-CCR3 antibodies significantly inhibited infection of human PBMC (P < 0.05) but not macaque cells (33.3% ± 5.1%; P = 0.06). Of interest, simultaneously pretreating primate PBMC with antibodies to all three chemokine receptors inhibited infection by both V1CSF and FIV-Ch to a greater extent than treatment with any single antibody, but no significant differences were observed between these cells and cultures treated with anti-CXCR4 antibodies alone (P = 0.7) (Fig. 3b and c). In contrast, infection of primate PBMC by Petaluma was not inhibited significantly by antibodies against either CCR3 or CCR5, nor did the combination of these antibodies and anti-CXCR4 antibodies increase the degree of inhibition detected with antibodies to CXCR4 alone. To ensure that antibodies shown to block HIV infection would also specifically inhibit infection by FIV, human PBMC were infected with V1CSF in the presence of varying concentrations of anti-chemokine receptor antibodies. A dose-dependent decrease in V1CSF proviral DNA levels was observed with each of the antibodies, confirming a role for primate chemokine receptors in FIV infection of primate cells (Fig. 3d). These findings demonstrated that FIV infection of both human and nonhuman primate cells was mediated by several primate chemokine receptors. Furthermore, diversity within the FIV envelope gene was found to determine the profile of chemokine receptors used to infect primate cells.
FIG. 3.
Infection of primate PBMC by FIV involves chemokine receptors. Primary macaque and human PBMC were infected with parent or chimeric FIV strains (104.5 TCID50/ml) in the presence or absence of antibodies recognizing the human CCR3 (R3, 3 μg/ml), CXCR4 (X4, 5 μg/ml), or CCR5 (R5, 3 μg/ml) chemokine receptor. Additional wells were infected in the presence of all three antibodies (A). Genomic DNA was isolated after 24 h, and infection was assessed by semiquantitative PCR detection of the FIV pol gene and densitometric analysis. Uninfected PBMC (U), PBMC infected in the presence of an anti-mouse IgG antibody (Ab), and PBMC infected without pretreatment with anti-chemokine receptor antibodies (C) served as controls. Lane H, water blank. FIV DNA levels were normalized to the corresponding GAPDH levels in each sample, and inhibition of FIV infection was represented as percent change in proviral DNA levels relative to levels in untreated controls. Values are mean percentages ± standard errors of the means from three experiments, for which representative blots are shown. Statistical significance was determined by ANOVA and the Tukey-Kramer posthoc test, with differences between treated and untreated groups indicated (∗, P < 0.05; ∗∗, P < 0.01). (a) Representative blot demonstrating PCR detection of the FIV pol gene in treated and untreated PBMC from cynomolgus macaques. Representative blots for human cells are not shown. (b) Densitometric analysis of the relative FIV proviral DNA levels in macaque PBMC treated with anti-chemokine receptor antibodies. (c) Inhibition of FIV infection of human PBMC by anti-chemokine receptor antibodies. (d) Human PBMC were infected with V1CSF in the presence or absence of varying concentrations of antibodies against the human CCR3 (0.5 to 3.0 μg/ml), CCR5 (0.5 to 3.0 μg/ml), or CXCR4 (1.0 to 5.0 μg/ml) chemokine receptor. NMA, non-chemokine receptor isotype-matched antibody.
Infection of human GHOST cells by FIV.
Several mechanisms can be envisaged to explain the ability to attenuate FIV infection of PBMC by inhibiting both CXC and CC chemokine receptors. Specific envelope sequences may confer the ability to use multiple chemokine receptors independently for cell entry. Alternatively, infection by V1CSF and FIV-Ch may require the presence of more than one chemokine receptor that interact to allow infection or to increase the efficiency of infection. For example, heterodimerization of chemokine receptors, which has been demonstrated previously for polymorphic CCR5 receptors (2), could result in the formation of a functional receptor complex for FIV entry. Similarly, different regions of the FIV envelope may interact with different chemokine receptors concurrently, as proposed for HIV (39). To address this question, we investigated the abilities of parent and chimeric viruses to infect GHOST cells, a series of cell lines derived from human osteosarcoma cells (29). In addition to being transfected stably with human CCR3, CCR5, and/or CXCR4, all cells in this series express endogenous levels of CXCR4. The nontransfected parent GHOST cell line served as a control. As with primate PBMC, FIV infection of GHOST cells was assessed by PCR detection of FIV-specific sequences in genomic DNA isolated 24 h after infection.
Nested PCR analyses revealed infection of all GHOST cell lines by Petaluma that could be eliminated by pretreatment with anti-CXCR4 antibodies (Fig. 4). In contrast, antibodies to either CCR3 or CCR5 did not affect the ability of Petaluma to infect any of the GHOST cell lines. These findings likely reflected the presence of endogenous CXCR4 on GHOST cells and agreed with the results observed for primate PBMC indicating that CXCR4 is the chemokine receptor used exclusively by Petaluma to infect primate cells. Following infection with V1CSF or FIV-Ch, however, FIV-specific sequences were detected in GHOST cells expressing CCR3 or CCR5, but parent GHOST cells or cells transfected with CXCR4 alone were refractory to infection by either virus (Fig. 4). As with primary PBMC, antibodies against either CCR3 or CCR5 inhibited infection by V1CSF and FIV-Ch of cells transfected with the corresponding chemokine receptor (Fig. 4). In GHOST cells expressing all three chemokine receptors, pretreatment with antibodies to CCR3 and CCR5 together, but not individually, prevented infection by V1CSF and FIV-Ch, indicating that these viruses were able to use either receptor for entry. Surprisingly, infection of all three cell lines expressing CC chemokine receptors was also inhibited by anti-CXCR4 antibodies. Taken together, these results suggested that expression of CXCR4 was necessary, but not sufficient, to permit infection of GHOST cells by V1CSF and FIV-Ch. Rather, the concurrent presence of a second receptor, such as CCR3 or CCR5, was also required.
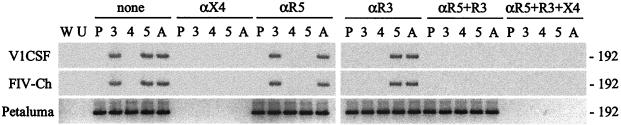
FIG. 4.
FIV infection of human GHOST cells. Human GHOST cells were infected with 104.5 TCID50 of parent or chimeric FIV strains/ml, and infection was assessed after 24 h by nested PCR amplification of the FIV pol gene from genomic DNA. GHOST cell lines used included nontransfected parent cells (P), cells transfected with the human CCR3 (3), CXCR4 (4), or CCR5 (5) receptor, and cells transfected with all three chemokine receptors (A). Mock-infected cells (U) served as negative controls, and amplification of a water blank (W) was performed to ensure the absence of contamination. The role of specific chemokine receptors in FIV infection was determined by infecting cells in the absence (none) or presence of antibodies to CCR3 (αR3), CCR5 (αR5), or CXCR4 (αX4), alone or in combination. Although Petaluma infected all cell lines, infection by either V1CSF or FIV-Ch was restricted to cells expressing CCR3 or CCR5. For all viruses, anti-CXCR4 antibodies inhibited infection, but antibodies to CCR3 or CCR5 affected infection by V1CSF and FIV-Ch.
Infection of trCrFK cells by FIV.
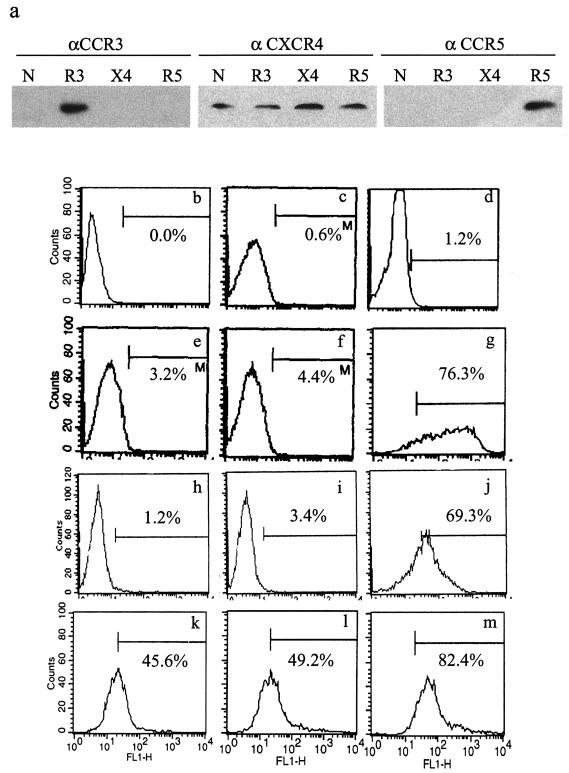
The previous experiments allowed an assessment of FIV entry under conditions in which replication was limited to a single round but did not provide insight into the role of chemokine receptors in productive infection. To answer this question, feline cells, termed trCrFK cells, were generated by stably transfecting CrFK cells with plasmids encoding human CCR3, CCR5, or CXCR4. Following neomycin selection, trCrFK cells were analyzed by Western blotting (Fig. 5a) and flow cytometry (Fig. 5b through m), and a minimum of 45% of cells in each line were found to express their respective receptor. In addition, like human GHOST cells and in agreement with previously published reports, fluorescence-activated cell sorter (FACS) analysis confirmed the presence of endogenous feline CXCR4 on each of the trCrFK cell lines. In contrast, neither mock-transfected cells nor CrFK cells not transfected with human CCR3 or CCR5 were labeled with antibodies against receptors other than CXCR4.
FIG. 5.
Chemokine receptor expression on trCrFK cells. Expression of chemokine receptors on nontransfected CrFK cells (N) and on CrFK cells transfected stably with the human CCR3 (R3), CXCR4 (X4), or CCR5 (R5) receptor was determined by Western blot analysis (a) and FACS analysis (b through m). (a) Expression of CXCR4 was evident in all cells when anti-CXCR4 (αCXCR4) antibodies were used, but CCR3 and CCR5 proteins were detectable only in cell lines transfected with these receptors. (b through m) Surface expression of chemokine receptors was assessed by FACS analysis using nontransfected cells incubated without antibodies (b), with the secondary antibody alone (c), or with an isotype-matched antibody (d) as a control. Mock-transfected and nontransfected CrFK cells expressed CXCR4 abundantly (g and j), but minimal expression of CCR3 (e and h) or CCR5 (f and i) was detected. In contrast, a minimum of 45% of cells transfected with CCR3 (k), CCR5 (l), or CXCR4 (m) expressed their respective receptor. Representative histograms from triplicate experiments are shown.
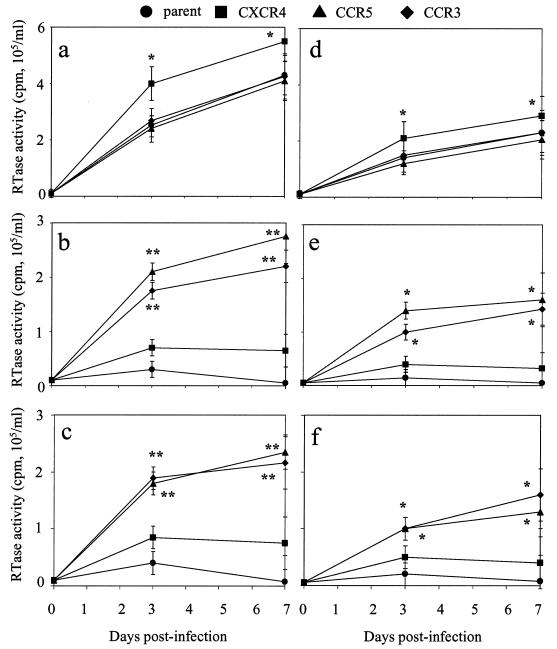
The ability of FIV to replicate in trCrFK cells was assessed by RT assay of culture supernatants (Fig. 6) and PCR analysis of genomic DNA (Table 1) following infection with the parent and chimeric viruses. Compared to mock-infected controls, RT activity that increased from day 3 to day 7 p.i. was observed in all CrFK cell lines infected with Petaluma (Fig. 6a). However, the levels of RT activity detected in CrFK cells transfected with human CXCR4 were increased significantly at both time points (P < 0.01) compared to the other cell lines. RT activity was also increased in nontransfected CrFK cells and in CrFK cells transfected with human CXCR4 following infection with V1CSF (Fig. 6b) or FIV-Ch (Fig. 6c) relative to that in controls, but no significant differences were detected between the two cell lines. In contrast, RT activity in trCrFK cells expressing CCR3 or CCR5 was elevated significantly following infection with V1CSF or FIV-Ch (P < 0.005) and, unlike the activity detected in the other cell lines, increased from day 3 to day 7 p.i. However, RT activity was approximately 50% lower than that detected in the same cell lines following infection with Petaluma.
FIG. 6.
RT activity in FIV-infected trCrFK cells. trCrFK cells were infected with Petaluma (a), V1CSF (b), or FIV-Ch (c) at 104.5 TCID50/ml or with FIV-ChTM (d), FIV-ChSU (e), or FIV-ChV35 (f) at 102.5 TCID50/ml, and RT activity was measured in culture supernatants at days 0, 3, and 7 p.i. trCrFK cell lines included nontransfected parent cells and cells transfected with human CCR3, CXCR4, or CCR5. RT activity is expressed as counts per minute per milliliter of medium, with the mean ± standard error of the mean from triplicate samples shown. Significant differences relative to parent cells were determined by ANOVA and the Tukey-Kramer posthoc test (∗, P < 0.01; ∗∗, P < 0.005). Increased RT activity was detected over the time course in all Petaluma- and FIV-ChTM-infected cell lines, but following infection with the other viruses, increased RT activity was detected only in cells transfected with CCR3 or CCR5.
TABLE 1.
Infection of trCrFK cells by FIV
| Cella | Abb | Infectionc by the following virus:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Mock | VICSF | Ch | Petaluma | ChSU | ChTM | ChV35 | ||
| Parent | None | − | + | + | +++ | + | ++ | + |
| X4 | − | − | − | − | − | − | − | |
| R3 | − | + | + | +++ | + | ++ | + | |
| R5 | − | + | + | +++ | + | ++ | + | |
| CXCR4 | None | − | + | + | ++++ | + | ++ | + |
| X4 | − | − | − | + | − | − | − | |
| R3 | − | + | + | ++++ | + | ++ | + | |
| R5 | − | + | + | ++++ | + | ++ | + | |
| CCR3 | None | − | +++ | +++ | ++++ | +++ | ++ | +++ |
| X4 | − | − | − | − | − | − | − | |
| R3 | − | ++ (53) | ++ (49) | ++++ | ++ (47) | ++ | ++ (44) | |
| R5 | − | +++ | +++ | ++++ | +++ | ++ | +++ | |
| CCR5 | None | − | +++ | +++ | ++++ | +++ | ++ | +++ |
| X4 | − | − | − | − | − | − | − | |
| R3 | − | +++ | +++ | ++++ | +++ | ++ | +++ | |
| R5 | − | ++ (62) | ++ (61) | ++++ | ++ (59) | ++ | ++ (54) | |
trCrFK cells used in this study included nontransfected cells (parent) or cells transfected with the human CCR3, CCR5, or CXCR4 chemokine receptor. All trCrFK cell lines expressed endogenous feline CXCR4.
Ab, antibody trCrFK cells were infected in the absence of anti-chemokine receptor antibodies (none) or in the presence of antibodies to human CCR3 (R3), CXCR4 (X4), or CCR5 (R5).
trCrFK cells were infected with either 104.5 TCID50 of V1CSF, FIV-Ch (Ch), or Petaluma/ml or 102.5 TCID50 of FIV-ChSU (ChSU), FIV-ChTM (ChTM), or FIV-ChV35 (ChV35)/ml. Infection was assessed after 72 h by PCR amplification of the FIV pol gene from genomic DNA (−, not detectable; +, nested PCR required; ++++, +++, and ++, detectable by single-round PCR with decreasing abundance. Graded responses are measured relative to cells infected in the absence of antibodies, with percent decreases in viral DNA given in parentheses.
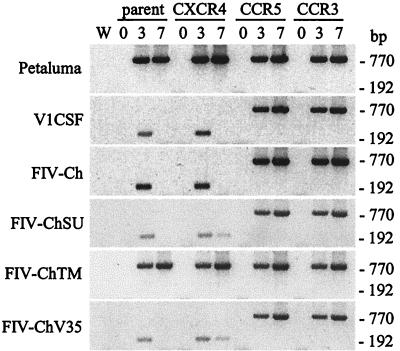
PCR analysis of genomic DNA from these cells revealed that FIV DNA was detectable in all cultures in which increased RT activity was observed (Fig. 7). Furthermore, a single round of PCR was sufficient to amplify FIV sequences at days 3 and 7 p.i. from all Petaluma-infected cell lines and from trCrFK cells expressing CCR3 or CCR5 that were infected with V1CSF or FIV-Ch (Fig. 7). However, nested PCR was required to detect virus in parent CrFK cells or cells expressing human CXCR4 following infection with V1CSF and FIV-Ch. Even with nested PCR, viral DNA was not detected in these cultures beyond 3 days p.i., suggesting that V1CSF and FIV-Ch were unable to replicate and spread efficiently in cells lacking chemokine receptors other than CXCR4 (Fig. 7). Confirmation that FIV infection of trCrFK cells was mediated by chemokine receptors was obtained by infecting these cells in the presence of anti-chemokine receptor antibodies. As with the human GHOST cells, antibodies to CXCR4 inhibited infection by V1CSF and FIV-Ch of CrFK cells expressing CCR3 or CCR5 (Table 1). Similarly, anti-CCR3 and -CCR5 antibodies also inhibited infection of the cell lines expressing the corresponding chemokine receptors (50% for CCR3 and 60% for CCR5 with either V1CSF or FIV-Ch). In contrast, only antibodies to CXCR4 significantly influenced the ability of Petaluma to infect any of the cell lines (Table 1).
FIG. 7.
Detection of FIV sequences in trCrFK cells. Cells were infected with 104.5 (V1CSF, Petaluma, and FIV-Ch) or 102.5 (FIV-ChSU, -ChTM, and -ChV35) TCID50 of FIV/ml, and the FIV pol gene was detected in genomic DNA by single-round (770 bp) or nested (192 bp) PCR at 3 and 7 days p.i. trCrFK cell lines used included nontransfected parent cells and cells transfected with human CCR3, CXCR4, or CCR5. Amplification of a water blank (W) ensured the lack of contamination. FIV sequences were detected by a single round of PCR amplification in all Petaluma- and FIV-ChTM-infected cell lines and in cells expressing either CCR3 or CCR5 that were infected with the other viruses. In contrast, nested PCR was required to detect FIV DNA in parent cells and in cells expressing human CXCR4 following infection with V1CSF, FIV-Ch, FIV-ChSU, or FIV-ChV35.
Envelope domains mediating chemokine receptor use by FIV.
Based on the experiments investigating feline cell tropism, FIV-ChTM was posited to possess a Petaluma phenotype, whereas the other chimeras resembled V1CSF more closely. In addition, the sequence of the V3-to-V5 region of the SU glycoprotein was implicated as a determinant of these properties. Thus, the trCrFK cells were used as a feline cell model to determine the profile of primate chemokine receptors used by these chimeras. As was observed with cytotropism, FIV-ChTM exhibited the same ability to replicate in all trCrFK cell lines as the parent Petaluma strain, with the greatest levels of RT activity detected in cells transfected with human CXCR4 (Fig. 6d). Like V1CSF and FIV-Ch, both FIV-ChSU (Fig. 6e) and FIV-ChV35 (Fig. 6f) exhibited limited ability to replicate in the parent and CXCR4-transfected CrFK cells, but significantly increased RT activity relative to that in the parent cell line was detected in CrFK cells expressing human CCR3 or CCR5 at both day 3 and day 7 p.i. (P < 0.01). A similar pattern was observed following PCR amplification of the FIV pol gene from genomic DNA. Nested PCR was required to detect FIV DNA in cells lacking human CC chemokine receptors following infection with FIV-ChSU or FIV-ChV35, whereas a single round of PCR was sufficient to amplify FIV sequences from all other infected cultures (Fig. 7). Furthermore, antibodies to CXCR4 inhibited infection of permissive cells by any of the chimeras, but antibodies to CCR3 or CCR5 only affected the ability of FIV-ChSU and FIV-ChV35 to infect trCrFK cells transfected with those receptors (44 to 59%) (Table 1). These results suggested that, like the ability to replicate in CrFK cells, the profile of primate chemokine receptors used by FIV for infection was determined by the sequence of the V3-to-V5 region.
DISCUSSION
The present study demonstrated that the use of chemokine receptors by FIV to infect primate cells is a property conserved between different primates. In agreement with earlier reports demonstrating that all FIV strains can use the feline CXCR4 receptor to infect feline cells (38), infection of primate cells by FIV also involved the preferential use of CXCR4. The ability of the primate CXCR4 receptor to mediate infection by FIV is not surprising given that the sequence of this receptor shares 95% similarity with the feline homologue (25), and that feline CXCR4 binds with high affinity to human stromal cell-derived factor-1α and -1β, the principal ligands for this receptor (18). Although CXCR4 played a pivotal role in FIV infection of primate cells, viruses that exhibited the ability to interact with human CC chemokine receptors, such as V1CSF, failed to infect human GHOST cells that expressed CXCR4 only. It is unlikely that this observation simply reflected the density of CXCR4 on target cells, since the same profile was observed in GHOST cells transfected with, and expressing high levels of, CXCR4. Rather, these results suggest that CXCR4 is necessary, but not sufficient, to allow infection of human cells by FIV. In contrast, Petaluma could gain entry to parent CrFK cells that expressed feline CXCR4 only, possibly indicating that this receptor alone is sufficient to mediate infection of some feline cell lines by FIV. The independent use of a single chemokine receptor for cell entry has been demonstrated previously for other lentiviruses, most notably the use of CCR5 by SIV (27) and of CXCR4 by HIV-1 and -2 (12, 19). Therefore, it is possible that CXCR4 alone can mediate FIV infection, as suggested for the Petaluma strain (26, 48). However, these results might also reflect the presence of an unidentified FIV receptor on CrFK cells or nonspecific cell entry that occurred through an envelope-independent mechanism, as has been reported for HIV (14, 33).
In addition to CXCR4, some FIV strains were also found to interact with primate CCR3 and CCR5. Although the sequences of CCR3 and CCR5 are highly conserved between primates, the similarity with their feline counterparts is not as pronounced as that observed with CXCR4 (approximately 75%) (25). Therefore, a rationale for the ability of FIV to use primate CC receptors to infect primate cells is not as obvious as that for CXCR4, and it is possible that the use of CC receptors by FIV is a phenomenon restricted to primate cells. However, recent studies have demonstrated that blocking CC chemokine receptors inhibits infection of feline cells by some FIV isolates (26) and that macrophage- or T-lymphocyte-tropic FIV strains that use feline CC or CXC receptors, respectively, may also exist (25). Because the primary receptor for FIV has not been defined, it is conceivable that primate CCR3 and CCR5 closely resemble another feline cell surface molecule with which FIV interacts; therefore, the results observed with feline cells may not be specific to the CC receptors. However, CCR3 and CCR5 are sufficiently diverse (25) that it is unlikely that they would mimic the same receptor. Since complete inhibition of infection of primate PBMC was not observed with anti-CCR3, anti-CCR5, or anti-CXCR4 alone or in combination, it is also plausible that other receptors for FIV infection are present on primate cells. Although blockade of chemokine receptors was able to completely eliminate the detection of FIV sequences in specific human GHOST cell lines and, in some cases, in trCrFK cells, these results likely reflected the lack of alternate receptors on these cell lines.
The results presented here are also consistent with cooperativity between two or more chemokine receptors during FIV infection of primate cells. In this model, CXCR4 functions as the principal receptor by which FIV infects primate cells, analogous to CD4 for HIV. However, an additional chemokine receptor, such as CCR3 or CCR5, is required to allow cell entry to occur, possible by acting as a coreceptor to promote the conformational changes required to initiate the fusion process. This model explains the failure of V1CSF to infect human GHOST cells lacking CCR3 or CCR5, despite the ability of anti-CXCR4 antibodies to block infection of all cell lines. Similarly, it is consistent with the results obtained from primate PBMC in which antibodies to CCR3, CCR5, or CXCR4 could each inhibit infection by VICSF, but all three antibodies together did not exceed the inhibition obtained with CXCR4 alone. Moreover, this model allows for FIV strains capable of interacting with human CXCR4 in the absence of supporting receptors, such as Petaluma, to also infect primate cells. The potential cooperative use of chemokine receptors by some FIV strains is also consistent with the finding that diversity in the V3-to-V5 region of the FIV envelope gene influenced chemokine receptor use, since the determinants required to bind to CXCR4 and induce a conformational change necessary to expose the TM fusion domain are contained within the V3-to-V5 region of the FIV SU (48). In contrast to the SU encoded by Petaluma, which can assume an appropriate conformation in the absence of binding to an additional receptor, exposure of the V1CSF fusion domain may require a second conformational change induced by binding to an additional receptor. These findings complement previous reports demonstrating that the V3-to-V5 region of FIV determines tropism for feline cells (3, 31, 41, 45, 47, 50) and extend those results to show that the same regions also determine the ability of FIV to infect primate cells and interact with primate chemokine receptors.
Because both primate and nonprimate lentiviruses have been shown to interact with chemokine receptors during cell entry, these receptors have been proposed to represent a common mechanism of infection and an evolutionary link in the pathogenesis of divergent lentiviruses (50). However, as the present study illustrates, a receptor system that is shared by multiple species also has the potential to mediate the initial cross-species transmission of pathogens, possibly providing insight into the evolution of lentiviral infections. For example, the capacity for SIV strains to use both CXCR4 and CCR5 independently of CD4 has been proposed to facilitate infection of human cells by these viruses by obviating the need to adapt to the human CD4 molecule (4, 21). The feline CD4 homologue shares less than 60% sequence similarity with the primate molecules (30); therefore, the advantage of a mechanism permitting infection of human CD4+ cells without using CD4 becomes evident. Moreover, the ability of HIV to infect cells via CD4-independent mechanisms, such as the use of chemokine receptors (12, 19) or galactosylceramide (16), may represent a conserved property originating from the SIV strains from which HIV evolved. Analogous to the use of chemokine receptors by FIV, the capacity for feline and primate retroviruses to use other common, widely expressed surface molecules as cell recognition signals has been shown previously. For example, the feline endogenous retrovirus RD114 and simian type D retroviruses employ a common neutral amino acid transporter for infection (42), while some feline and primate leukemia viruses share the gibbon ape leukemia virus (GALV) receptor (43). In each case, the presence of these receptors on human cells facilitates cross-species infections by these feline pathogens. Recently, we demonstrated xenoinfection of cynomologus macaques by the FIV strain Petaluma, which was accompanied by viral replication and the development of an anti-FIV immune response (24). The results of the present study implicate chemokine receptors as a potential mechanism by which FIV establishes an infection in cells from nonfeline species, possibly allowing the virus to adapt to its new host. However, it should be noted that the ability of chemokine receptors to mediate the spread and replication of FIV in vivo has not been demonstrated in either felines or primates.
The present findings may also be relevant to the mechanism by which FIV infects feline cells. In agreement with previous reports that specific sequences in the FIV envelope gene determine the ability to replicate in CrFK cells (26, 46, 47), V1CSF and all chimeras encoding the V3-to-V5 domains of the V1CSF SU were found to infect primary feline PBMC but not CrFK cells. In contrast, only the Petaluma parent and FIV-ChTM, which encodes the SU of Petaluma, replicated efficiently in both cell-types, indicating that this property mapped primarily to the SU of V1CSF. However, the finding that FIV-ChSU, -ChTM, and -ChV35 replicated to a lesser extent than FIV-Ch or the parent viruses suggests that sequences in both the SU and TM are essential to the tropisms of V1CSF and Petaluma. For example, it is conceivable that structural differences in the envelope glycoproteins of V1CSF and Petaluma precluded optimal association within the viral envelope and reduced infectivity. In addition, the facts that all chimeras possessed the same long terminal repeat and that infectious virus could be obtained from CrFK cultures transfected with any of the chimeras, but only FIV-ChTM replicated efficiently in CrFK cells, support a receptor-based mechanism for these differences. However, the presence of increased RT activity in CrFK cultures immediately following cell-free infection with V1CSF or the chimeras suggests that receptors capable of interacting with the V1CSF SU are present on CrFK cells, but that replication occurs inefficiently compared to that of Petaluma. Like the requirement for multiple chemokine receptors on human cells, these observations may indicate that V1CSF requires the concurrent expression of additional receptors for cell entry that are not all present on CrFK cells. V1CSF could infect CrFK cells that expressed CXCR4 as their sole chemokine receptor, but replication and spread were reduced markedly compared to those in CrFK cells expressing other chemokine receptors. Thus, CrFK cells may lack a coreceptor to complement CXCR4, a function that was performed by human CCR3 and CCR5 in the trCrFK cells. However, similar functions for feline CCR3 and CCR5 may not apply to infection of feline cells, since another feline receptor may play an equivalent role. Therefore, the decreased ability of V1CSF to replicate in trCrFK cells compared to that of Petaluma despite the presence of CCR3 and CCR5 could indicate that these receptors cooperate with feline CXCR4 less efficiently than the feline receptor.
Zoonoses continue to be a major concern as a potential source of new human pathogens, particularly in light of the use of viruses for human gene therapy and the use of nonhuman cells and organs for xenotransplantation. Recent studies demonstrating the capacity of viruses to cross species in vitro and in vivo raise questions about ongoing transmissions and render the study of the adaptations required for a virus to be transmitted from one host species to another increasingly relevant. Although feline tissues are unlikely candidates for use in human transplantation, the results of the present study may provide insight into the mechanisms by which xenoinfections by other pathogens may occur. For example, recent studies have demonstrated that infection of primate cells by porcine endogenous retroviruses (PERV), viruses implicated in cross-species transmissions following xenotransplantation, exhibits species- and envelope-dependent properties similar to those of FIV (51). Similarly, infections of human cells by FIV and SIV incorporate many common properties, such as CD4-independent chemokine receptor use and dependence on envelope sequence (21). Consequently, FIV infection of primates may provide a model in which to study the process by which HIV was acquired in the human population. Thus, FIV infection of primate cells provides a system to assess both the mechanism of cross-species infection and the role of envelope diversity in the process of lentivirus adaptation to new hosts.
REFERENCES
- 1.Bendinelli, M., M. Pistello, S. Lombardi, A. Poli, C. Garzelli, D. Matteucci, L. Ceccherini-Nelli, G. Malvaldi, and F. Tozzini. 1995. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin. Microbiol. Rev. 8:87-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benkirane, M., D. Y. Jin, R. F. Chun, R. A. Koup, and K. T. Jeang. 1997. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J. Biol. Chem. 272:30603-30606. [DOI] [PubMed] [Google Scholar]
- 3.Billaud, J. N., D. Selway, N. Yu, and T. R. Phillips. 2000. Replication rate of feline immunodeficiency virus in astrocytes is envelope dependent: implications for glutamate uptake. Virology 266:180-188. [DOI] [PubMed] [Google Scholar]
- 4.Borsetti, A., C. Parolin, B. Ridolfi, L. Sernicola, A. Geraci, B. Ensoli, and F. Titti. 2000. CD4-independent infection of two CD4− CCR5− CXCR4+ pre-T-cell lines by human and simian immunodeficiency viruses. J. Virol. 74:6689-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner, D., and N. C. Pedersen. 1989. Infection of peritoneal macrophages in vitro and in vivo with feline immunodeficiency virus. J. Virol. 63:5483-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cayabyab, M., G. B. Karlsson, B. A. Etemad-Moghadam, W. Hofmann, T. Steenbeke, M. Halloran, J. W. Fanton, M. K. Axthelm, N. L. Letvin, and J. G. Sodroski. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J. Virol. 73:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danave, I. R., E. Tiffany-Castiglioni, E. Zenger, R. Barhoumi, R. C. Burghardt, and E. W. Collisson. 1994. Feline immunodeficiency virus decreases cell-cell communication and mitochondrial membrane potential. J. Virol. 68:6745-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, G. A., S. Himathongkham, and E. E. Sparger. 1999. Differential cell tropism of feline immunodeficiency virus molecular clones in vivo. J. Virol. 73:2596-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dow, S. W., M. J. Dreitz, and E. A. Hoover. 1992. Feline immunodeficiency virus neurotropism: evidence that astrocytes and microglia are the primary target cells. Vet. Immunol. Immunopathol. 35:23-35. [DOI] [PubMed] [Google Scholar]
- 10.Dow, S. W., M. L. Poss, and E. A. Hoover. 1990. Feline immunodeficiency virus: a neurotropic lentivirus. J. Acquir. Immune Defic. Syndr. 3:658-668. [PubMed] [Google Scholar]
- 11.Egberink, H. F., E. De Clercq, A. L. Van Vliet, J. Balzarini, G. J. Bridger, G. Henson, M. C. Horzinek, and D. Schols. 1999. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J. Virol. 73:6346-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 13.English, R. V., C. M. Johnson, D. H. Gebhard, and M. B. Tompkins. 1993. In vivo lymphocyte tropism of feline immunodeficiency virus. J. Virol. 67:5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fackler, O. T., and B. M. Peterlin. 2000. Endocytic entry of HIV-1. Curr. Biol. 10:1005-1008. [DOI] [PubMed] [Google Scholar]
- 15.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 16.Harouse, J. M., S. Bhat, S. L. Spitalnik, M. Laughlin, K. Stefano, D. H. Silberberg, and F. Gonzalez-Scarano. 1991. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science 253:320-323. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosie, M. J., N. Broere, J. Hesselgesser, J. D. Turner, J. A. Hoxie, J. C. Neil, and B. J. Willett. 1998. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor. J. Virol. 72:2097-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoxie, J. A., C. C. LaBranche, M. J. Endres, J. D. Turner, J. F. Berson, R. W. Doms, and T. J. Matthews. 1998. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J. Reprod. Immunol. 41:197-211. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, Y., K. Tomonaga, Y. Kawaguchi, M. Kohmoto, Y. Inoshima, Y. Tohya, T. Miyazawa, C. Kai, and T. Mikami. 1996. Feline immunodeficiency virus can infect a human cell line (MOLT-4) but establishes a state of latency in the cells. J. Gen. Virol. 77:1623-1630. [DOI] [PubMed] [Google Scholar]
- 21.Iyengar, S., D. H. Schwartz, J. E. Clements, and J. E. Hildreth. 2000. CD4-independent, CCR5-dependent simian immunodeficiency virus infection and chemotaxis of human cells. J. Virol. 74:6720-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, J., and C. Power. 1999. Productive infection of human peripheral blood mononuclear cells by feline immunodeficiency virus: implications for vector development. J. Virol. 73:2491-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston, J. B., Y. Jiang, G. van Marle, M. B. Mayne, W. Ni, J. Holden, J. C. McArthur, and C. Power. 2000. Lentivirus infection in the brain induces matrix metalloproteinase expression: role of envelope diversity. J. Virol. 74:7211-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, J. B., M. E. Olson, E. W. Rud, and C. Power. 2001. Xenoinfection of nonhuman primates by feline immunodeficiency virus. Curr. Biol. 11:1109-1113. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs, E. M., G. D. Baxter, and W. F. Robinson. 1999. Feline peripheral blood mononuclear cells express message for both CXC and CC type chemokine receptors. Arch. Virol. 144:273-285. [DOI] [PubMed] [Google Scholar]
- 26.Lerner, D. L., and J. H. Elder. 2000. Expanded host cell tropism and cytopathic properties of feline immunodeficiency virus strain PPR subsequent to passage through interleukin-2-independent T cells. J. Virol. 74:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, K. A., R. Wyatt, M. Farzan, H. Choe, L. Marcon, E. Desjardins, J. Robinson, J. Sodroski, C. Gerard, and N. P. Gerard. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278:1470-1473. [DOI] [PubMed] [Google Scholar]
- 28.Miyazawa, T., Y. Kawaguchi, M. Kohmoto, J. Sakuragi, A. Adachi, M. Fukasawa, and T. Mikami. 1992. Production of feline immunodeficiency virus in feline and non-feline non-lymphoid cell lines by transfection of an infectious molecular clone. J. Gen. Virol. 73:1543-1546. [DOI] [PubMed] [Google Scholar]
- 29.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norimine, J., T. Miyazawa, Y. Kawaguchi, Y. Tohya, C. Kai, and T. Mikami. 1992. A cDNA encoding feline CD4 has a unique repeat sequence downstream of the V-like region. Immunology 75:74-79. [PMC free article] [PubMed] [Google Scholar]
- 31.Pancino, G., S. Castelot, and P. Sonigo. 1995. Differences in feline immunodeficiency virus host cell range correlate with envelope fusogenic properties. Virology 206:796-806. [DOI] [PubMed] [Google Scholar]
- 32.Pancino, G., H. Ellerbrok, M. Sitbon, and P. Sonigo. 1994. Conserved framework of envelope glycoproteins among lentiviruses. Curr. Top. Microbiol. Immunol. 188:77-105. [DOI] [PubMed] [Google Scholar]
- 33.Pang, S., D. Yu, D. S. An, G. C. Baldwin, Y. Xie, B. Poon, Y. H. Chow, N. H. Park, and I. S. Chen. 2000. Human immunodeficiency virus Env-independent infection of human CD4− cells. J. Virol. 74:10994-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen, N. C., J. K. Yamamoto, T. Ishida, and H. Hansen. 1989. Feline immunodeficiency virus infection. Vet. Immunol. Immunopathol. 21:111-129. [DOI] [PubMed] [Google Scholar]
- 36.Poeschla, E. M., and D. J. Looney. 1998. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J. Virol. 72:6858-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Power, C., R. Buist, J. B. Johnston, M. R. Del Bigio, W. Ni, M. R. Dawood, and J. Peeling. 1998. Neurovirulence in feline immunodeficiency virus-infected neonatal cats is viral strain specific and dependent on systemic immune suppression. J. Virol. 72:9109-9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson, J., G. Pancino, R. Merat, T. Leste-Lasserre, A. Moraillon, J. Schneider-Mergener, M. Alizon, P. Sonigo, and N. Heveker. 1999. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J. Virol. 73:3661-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross, T. M., and B. R. Cullen. 1998. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc. Natl. Acad. Sci. USA 95:7682-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinohara, K., K. Sakai, S. Ando, Y. Ami, N. Yoshino, E. Takahashi, K. Someya, Y. Suzaki, T. Nakasone, Y. Sasaki, M. Kaizu, Y. Lu, and M. Honda. 1999. A highly pathogenic simian/human immunodeficiency virus with genetic changes in cynomolgus monkey. J. Gen. Virol. 80:1231-1240. [DOI] [PubMed] [Google Scholar]
- 41.Siebelink, K. H., J. A. Karlas, G. F. Rimmelzwaan, A. D. Osterhaus, and M. L. Bosch. 1995. A determinant of feline immunodeficiency virus involved in Crandell feline kidney cell tropism. Vet. Immunol. Immunopathol. 46:61-69. [DOI] [PubMed] [Google Scholar]
- 42.Tailor, C. S., A. Nouri, Y. Zhao, Y. Takeuchi, and D. Kabat. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tochikura, T. S., A. Tanabe Tochikura, K. A. Hayes, A. Lazo, R. T. Bailer, J. R. Blakeslee, Jr., L. J. Lafrado, P. Roy Burman, R. Pandey, R. G. Olsen, et al. 1993. Fusion activity dissociated from replication ability in feline immunodeficiency virus (FIV) in human cells. J. Acquir. Immune Defic. Syndr. 6:1301-1310. [PubMed] [Google Scholar]
- 45.Vahlenkamp, T. W., A. de Ronde, N. N. Schuurman, A. L. van Vliet, J. van Drunen, M. C. Horzinek, and H. F. Egberink. 1999. Envelope gene sequences encoding variable regions 3 and 4 are involved in macrophage tropism of feline immunodeficiency virus. J. Gen. Virol. 80:2639-2646. [DOI] [PubMed] [Google Scholar]
- 46.Vahlenkamp, T. W., E. J. Verschoor, N. N. Schuurman, A. L. van Vliet, M. C. Horzinek, H. F. Egberink, and A. de Ronde. 1997. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J. Virol. 71:7132-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verschoor, E. J., L. A. Boven, H. Blaak, A. L. van Vliet, M. C. Horzinek, and A. de Ronde. 1995. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J. Virol. 69:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willett, B. J., and M. J. Hosie. 1999. The role of the chemokine receptor CXCR4 in infection with feline immunodeficiency virus. Mol. Membr. Biol. 16:67-72. [DOI] [PubMed] [Google Scholar]
- 49.Willett, B. J., M. J. Hosie, T. H. Dunsford, J. C. Neil, and O. Jarrett. 1991. Productive infection of T-helper lymphocytes with feline immunodeficiency virus is accompanied by reduced expression of CD4. AIDS 5:1469-1475. [DOI] [PubMed] [Google Scholar]
- 50.Willett, B. J., L. Picard, M. J. Hosie, J. D. Turner, K. Adema, and P. R. Clapham. 1997. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J. Virol. 71:6407-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]