With >400 recognized breeds worldwide and a recently completed draft genomic sequence, the dog has emerged as a model organism of choice for the genetic dissection of morphological traits and diseases (1–3). Of great interest is the fact that a number of disorders in purebred dogs are also of human medical relevance (1, 2). In this context, the study by Clark et al. (4) in this issue of PNAS is exciting because of its implications on morphological, medical, and evolutionary grounds. Indeed, the authors have discovered a mutation in the dog SILV gene that is responsible for merle, a color patterning in the coat of various canine breeds. Strikingly, merle dogs exhibit auditory and ophthalmologic abnormalities similar to those observed for the human auditory-pigmentation disorder Waardenburg syndrome, which accounts for 2–5% of all human cases of congenital deafness (5).
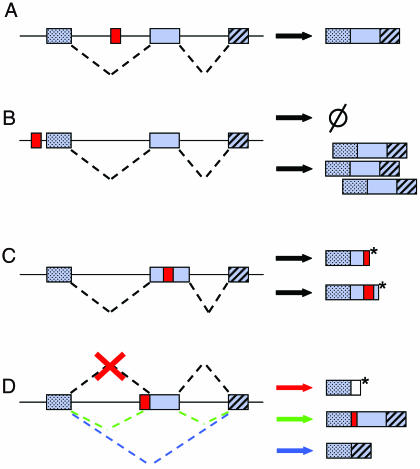
The different dog breeds were established within the past few centuries from small numbers of founding individuals. As a consequence, linkage disequilibrium within the genome extends over at least 50-fold greater distances in dogs as compared to humans (3). Taking advantage of this feature of the dog genome, Clark et al. (4) carried out a whole-genome scan using the Shetland Sheepdog and were able to map the merle locus to the dog homologue of human chromosome 12q13. Characterization of the pigment gene SILV (6) located in this region revealed the presence of a short interspersed element (SINE) inserted at an intron/exon boundary that segregates with the merle phenotype (4). SINEs are “jumping genes” belonging to the retrotransposon class of mobile elements that propagate in their host genomes via a “copy and paste” mechanism (7–11). Because only a small fraction of mammalian genomes is functional, most retrotransposon insertions occur in genomic regions where they essentially induce no damage to the host genome (12). However, retrotransposons occasionally insert in genomic regions where they can disrupt ORFs, alter splicing, or modulate gene expression (Fig. 1) (7–9, 11, 13, 14). In addition, retrotransposons may also create genomic deletions upon insertion into host genomes (15–18). Furthermore, because of their high copy number and sequence identity, SINEs can also have a postinsertional impact through unequal homologous recombination (7, 8, 19). In sum, the ongoing expansion of retrotransposons in humans and its consequences are responsible for a variety of human genetic disorders (19, 20). Considering that the mobility of SINEs in the canine lineage is even higher than in the human lineage (3, 21, 22), they may represent a very considerable source of genetic disorders in dogs (4, 23, 24). Interestingly, the results of Clark et al. (4) further suggest that SINE insertions may also play a significant role in the phenotypic diversity of dog breeds.
Fig. 1.
The impact of SINEs on gene structure and expression. A hypothetical gene constituted of three exons (gray boxes with and without patterns) along with its splicing pattern (dashed lines) is shown on the left; the resulting protein products are shown on the right. (A) A SINE element (red box) inserted within an intron essentially induces no damage. (B) A SINE inserts at or near a regulatory region of the gene and may decrease (dashed circle) or increase (as indicated by multiple protein products) gene expression levels. (C) A SINE inserts within an exon and may disrupt the open reading frame, thus introducing a premature stop codon (asterisks), resulting in a truncated and possibly nonfunctional protein. (D) A SINE inserts at an intron/exon boundary and may alter splicing patterns. This may result in intron sequence (white box) integrated to the protein product (red cross and arrow) with consequences analogous to C, exon skipping (blue splice variant and arrow) or the use of a cryptic splice site located within the SINE element (green splice variant and arrow), that may lead to SINE exonization if the open reading frame is preserved.
The question arises why SINE retrotransposition is so active in the dog genome. Repeated events of population size reduction, such as the bottleneck at the time of divergence between dog and wolf and the founder effects leading to the establishment of the different dog breeds (3), might have facilitated the significant expansion of dog SINEs. Similar to dogs, humans have also experienced bottlenecks during their recent history, and about three times as many Alu SINEs have inserted in the human genome as compared with the chimpanzee genome since their divergence ≈6 million years ago (25, 26). SINEs typically are comprised of subfamilies of closely related elements that are largely derived from small numbers of highly active source elements (7, 27, 28). Repeated demographic crashes in dog populations may have led to an increase of active SINEs as a result of enhanced genetic drift and reduced efficiency of selection against deleterious SINEs (25). The data presented by Clark et al. (4) provide some support for this hypothesis, because the authors show that the SINE insertion in the SILV gene responsible for the merle phenotype in the Shetland Sheepdog also is present and responsible for the merle phenotype in other dog breeds. This finding suggests that the SINE insertion occurred before the divergence of canine breeds. Because merle dogs exhibit a wide range of developmental defects, the SINE insertion in the SILV gene must be deleterious to some extent. Yet, the SINE insertion and the merle phenotype have not been eliminated in all dog breeds, possibly as a result, at least partly, of enhanced genetic drift and inefficient negative selection following the establishment of the breeds. In any case, the availability of the dog genome sequence (3), along with well defined breeds, makes the dog an excellent system in which to further test the contribution of host demographic factors in the triggering of the expansion of mobile elements.
Another intriguing aspect of the results of Clark et al. (4) is that, whereas the SINE insertion in the SILV gene was present in all merle dogs, some nonmerle dogs also had the retrotransposon. However, the SINE element in the nonmerle dogs had a much shorter poly(A) tail (≈60 nucleotides in nonmerle dogs vs. ≈100 nucleotides in merle dogs), a typical component of the 3′ end of SINEs (29). Therefore, the presence of the SINE element is necessary but not sufficient to generate the merle phenotype; a long poly(A) tail in the element is also required. This is an interesting observation for several reasons. First, given that the size of SINE poly(A) tails can vary over time (29), it offers a viable explanation as to why nonmerle-phenotype dogs sometimes produce merle offspring. Second, it raises exciting questions about the molecular mechanism by which the SINE insertion causes the merle phenotype. This SINE element is inserted at an intron/exon boundary. This finding suggests that the presence of the retrotransposon may affect splicing of the SILV gene by displacing the putative lariat branch point relative to the acceptor splice site that precedes the exon (Fig. 1D) (4). Interestingly, the acceptor splice site of the exon is duplicated as part of the SINE insertion [see figure 2 in Clark et al. (4)]. Therefore, it is also possible that splicing occurs in merle dogs using the acceptor splice site located upstream of the SINE insertion. This would result in the entire SINE insertion being integrated in the mature messenger RNA, presumably leading to a nonfunctional SILV protein as the result of a frameshift mutation from slight variation in the poly(A) tail length (Fig. 1C). How, then, is a functional SILV protein restored when the SINE insertion possesses a short poly(A) tail? It is noteworthy that the SINE insertion identified by Clark et al. (4) is in anti-sense orientation and contains cryptic acceptor splice sites (22). Therefore, it is possible that the shorter poly(A) tail of the SINE insertion brings the putative lariat branch point to a more favorable distance with a cryptic acceptor splice site located within the SINE element. If this splice site happens to preserve the reading frame of the SILV gene, then the SINE insert may be exonized (30), thereby generating a functional SILV protein (Fig. 1D). The findings of Clark et al. (4) thus offer an excellent substrate for further investigations related to splicing mechanisms.
Although mammalian genomes possess hundreds of thousands of SINEs (3, 12, 21), very few disease-causing insertions have been identified (20, 23, 24). Therefore, the discovery by Clark et al. (4) is quite exciting. It not only contributes insight into the potential impact of mobile elements on genome and phenotype variation but also demonstrates that the dog is an excellent model in which to determine the genetic basis of disorders of human biomedical importance, one more reason why dogs are mankind's best friends.
Acknowledgments
Research on mobile elements in the Batzer laboratory is supported by National Science Foundation Grants BCS-0218338 and EPS-0346411 and National Institutes of Health Grant GM59290.
Author contributions: R.C. and M.A.B. wrote the paper
Conflict of interest statement: No conflicts declared.
See companion article on page 1376.
References
- 1.Sutter, N. B. & Ostrander, E. A. (2004) Nat. Rev. Genet. 5, 900–910. [DOI] [PubMed] [Google Scholar]
- 2.Parker, H. G. & Ostrander, E. A. (2005) PLoS Genet. 1, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindblad-Toh, K., Wade, C. M., Mikkelsen, T. S., Karlsson, E. K., Jaffe, D. B., Kamal, M., Clamp, M., Chang, J. L., Kulbokas, E. J., Zody, M. C., et al. (2005) Nature 438, 803–819. [DOI] [PubMed] [Google Scholar]
- 4.Clark, L. A., Wahl, J. M., Rees, C. A. & Murphy, K. E. (2006) Proc. Natl. Acad. Sci. USA 103, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak, C. S. & Isaacson, G. (2003) Ann. Otol. Rhinol. Laryngol. 112, 817–820. [DOI] [PubMed] [Google Scholar]
- 6.Theos, A. C., Truschel, S. T., Raposo, G. & Marks, M. S. (2005) Pigment Cell Res. 18, 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batzer, M. A. & Deininger, P. L. (2002) Nat. Rev. Genet. 3, 370–379. [DOI] [PubMed] [Google Scholar]
- 8.Deininger, P. L., Moran, J. V., Batzer, M. A. & Kazazian, H. H., Jr. (2003) Curr. Opin. Genet. Dev. 13, 651–658. [DOI] [PubMed] [Google Scholar]
- 9.Kazazian, H. H., Jr., & Moran, J. V. (1998) Nat. Genet. 19, 19–24. [DOI] [PubMed] [Google Scholar]
- 10.Brookfield, J. F. (2005) Nat. Rev. Genet. 6, 128–136. [DOI] [PubMed] [Google Scholar]
- 11.Han, J. S. & Boeke, J. D. (2005) BioEssays 27, 775–784. [DOI] [PubMed] [Google Scholar]
- 12.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 13.Han, J. S., Szak, S. T. & Boeke, J. D. (2004) Nature 429, 268–274. [DOI] [PubMed] [Google Scholar]
- 14.Perepelitsa-Belancio, V. & Deininger, P. (2003) Nat. Genet. 35, 363–366. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, N., Lutz-Prigge, S. & Moran, J. V. (2002) Cell 110, 315–325. [DOI] [PubMed] [Google Scholar]
- 16.Symer, D. E., Connelly, C., Szak, S. T., Caputo, E. M., Cost, G. J., Parmigiani, G. & Boeke, J. D. (2002) Cell 110, 327–338. [DOI] [PubMed] [Google Scholar]
- 17.Callinan, P. A., Wang, J., Herke, S. W., Garber, R. K., Liang, P. & Batzer, M. A. (2005) J. Mol. Biol. 348, 791–800. [DOI] [PubMed] [Google Scholar]
- 18.Han, K., Sen, S. K., Wang, J., Callinan, P. A., Lee, J., Cordaux, R., Liang, P. & Batzer, M. A. (2005) Nucleic Acids Res. 33, 4040–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deininger, P. L. & Batzer, M. A. (1999) Mol. Genet. Metab. 67, 183–193. [DOI] [PubMed] [Google Scholar]
- 20.Chen, J. M., Stenson, P. D., Cooper, D. N. & Ferec, C. (2005) Hum. Genet. 117, 411–427. [DOI] [PubMed] [Google Scholar]
- 21.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A. L., Pop, M., Wang, W., Fraser, C. M., et al. (2003) Science 301, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 22.Wang, W. & Kirkness, E. F. (2005) Genome Res. 15, 1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, L., Faraco, J., Li, R., Kadotani, H., Rogers, W., Lin, X., Qiu, X., de Jong, P. J., Nishino, S. & Mignot, E. (1999) Cell 98, 365–376. [DOI] [PubMed] [Google Scholar]
- 24.Pele, M., Tiret, L., Kessler, J. L., Blot, S. & Panthier, J. J. (2005) Hum. Mol. Genet. 14, 1417–1427. [DOI] [PubMed] [Google Scholar]
- 25.Hedges, D. J., Callinan, P. A., Cordaux, R., Xing, J., Barnes, E. & Batzer, M. A. (2004) Genome Res. 14, 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chimpanzee Sequencing and Analysis Consortium (2005) Nature 437, 69–87. [DOI] [PubMed] [Google Scholar]
- 27.Cordaux, R., Hedges, D. J. & Batzer, M. A. (2004) Trends Genet. 20, 464–467. [DOI] [PubMed] [Google Scholar]
- 28.Deininger, P. L., Batzer, M. A., Hutchison, C. A., 3rd, & Edgell, M. H. (1992) Trends Genet. 8, 307–311. [DOI] [PubMed] [Google Scholar]
- 29.Roy-Engel, A. M., Salem, A. H., Oyeniran, O. O., Deininger, L., Hedges, D. J., Kilroy, G. E., Batzer, M. A. & Deininger, P. L. (2002) Genome Res. 12, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev-Maor, G., Sorek, R., Shomron, N. & Ast, G. (2003) Science 300, 1288–1291. [DOI] [PubMed] [Google Scholar]