Abstract
Generation time is an important determinant of a neutral molecular clock. There are several human-specific life history traits that led to a substantially longer generation time in humans than in other hominoids. Indeed, a long generation time is considered an important trait that distinguishes humans from their closest relatives. Therefore, humans may exhibit a significantly slower molecular clock as compared to other hominoids. To investigate this hypothesis, we performed a large-scale analysis of lineage-specific rates of single-nucleotide substitutions among hominoids. We found that humans indeed exhibit a significant slowdown of molecular evolution compared to chimpanzees and other hominoids. However, the amount of fixed differences between humans and chimpanzees appears extremely small, suggesting a very recent evolution of human-specific life history traits. Notably, chimpanzees also exhibit a slower rate of molecular evolution compared to gorillas and orangutans in the regions analyzed.
Keywords: comparative genomics, generation time, hominoid evolution, primate genomics
Humans (Homo sapiens) have a longer generation time than any other extant hominoid because of differences in several life history traits. Humans take almost twice as long to reach sexual maturity as chimpanzees (Pan troglodytes) and gorillas (Gorilla gorillas) (1), have a longer lifespan, and have a longer gestation period as compared to any nonhuman hominoid (2). These traits are believed to have played important roles in human evolution. In particular, life span and the length of gestation are highly correlated with the size of the brain, which is approximately three times larger in humans than in other hominoids (2–4).
Difference in generation times may leave a molecular signature by affecting evolutionary rates. Specifically, species with longer generation times are expected to exhibit slower rates of molecular evolution than those with shorter generation times. This hypothesis, called the “generation-time effect hypothesis” (5–7), is based on the idea that most germ-line mutations originate from errors in DNA replication. Because species with longer generation times go through fewer numbers of replications in germ cells per unit time, fewer substitutions will accumulate. This hypothesis has been strongly supported by molecular data (5–9).
Therefore, the human lineage may exhibit a slower rate compared to other hominoids. However, genomic sequences of humans and other hominoids are extremely similar. For example, the alignable portions of human and chimpanzee genomes are 98.8% identical (9–13). Therefore, to detect subtle changes in the neutral substitution rates between these species, we need to perform high-quality, large-scale sequence comparisons. The recent accumulation of genomic sequence data from the chimpanzee genome (12, 13) and from several other catarrhines provide the opportunity to perform such comparisons.
In this study, we analyzed an extensive amount of sequence data from humans and other hominoids to determine whether there was a further slowdown in rate along the lineage leading to modern humans, as expected under the generation-time effect hypothesis. First, we constructed and analyzed large-scale human–chimpanzee–baboon and human–chimpanzee–rhesus alignments from various genomic data sources (see Materials and Methods). Together, these encompass ≈63 million base pairs (Mbps) of the human genome. In addition, we analyzed ≈2 Mbps of available BAC-based genome sequences from gorilla and orangutan to investigate variation in molecular clock among different hominoid genomes.
These analyses have revealed several interesting observations. We found a slight but significant substitution rate difference between human and chimpanzee genomes, suggesting a very recent evolution of human-specific life history traits. We also discovered significant rate variation among the other hominoids. Intriguingly, both humans and chimpanzees appear to have evolved slower than gorillas and orangutans. These findings suggest that life history traits may vary substantially among nonhuman hominoids.
Results
Slower Molecular Clock in Humans than in Chimpanzees. For the human–chimpanzee-baboon comparisons, we used two data sets obtained from different genomic data sources (data sets 1 and 2, see Materials and Methods). The average genetic distance between human and chimpanzee based on the BAC-based data from chromosomes 7 and 21 (data set 1) is 1.06 (±0.0037%) in introns, 1.17 (±0.0049%) in intergenic regions, and 1.10% (±0.11%) when introns and intergenic regions are concatenated. When we use data set 2, this estimate is 1.28 (±0.01%), 1.36 (±0.01%), and 1.34 (±0.01%) for introns, intergenic regions, and concatenated data, respectively.
To test the rate difference between the genomes of human and chimpanzee, we first performed a relative rate test (7). From data set 1, we found that the chimpanzee lineage has accumulated significantly more substitutions than the human lineage (Table 1). Both intergenic regions and introns show slower rates in the human lineage, although the extent of rate slowdown in the intergenic regions (4.2%) is slightly higher than that of the introns (3.1%). These differences were significant by the relative rate test (P < 0.05 for each comparison, Table 1). The rate slowdown pattern is common in both chromosome 7 and chromosome 21, although it is not significant in chromosome 21, presumably because of the 10-fold smaller size of the alignment (a total of 760,942 and 560,555 aligned sites in introns and intergenic regions from chromosome 21 compared to 7,078,758 and 4,394,569 aligned sites in introns and intergenic regions from chromosome 7; Table 1) or chance effects. The results remained unchanged when repetitive elements were removed from the alignments (Table 1).
Table 1. Number of sites compared, rate difference between human and chimpanzee (estimated by d), and the ratio of chimpanzee-specific vs. human-specific nucleotide substitutions from data set 1.
| Introns
|
Intergenic regions
|
|||||
|---|---|---|---|---|---|---|
| Fragments | Number of sites compared | d = Kbh – Kbc (per 10 kb) | KOC/KOH | Number of sites compared | d = Kbh – Kbc (per 10 kb) | KOC/KOH |
| Chromosome 7 | ||||||
| 1 | 648,094 (414,899) | –1.133 ± 1.356 (–0.181 ± 1.618) | 1.021 (1.003) | 443,956 (239,010) | –1.635 ± 1.689 (–0.585 ± 2.229) | 1.030 (1.011) |
| 2 | 1,307,590 (837,209) | –2.940 ± 0.926** (–1.451 ± 1.104) | 1.061 (1.032) | 686,506 (329,767) | –0.253 ± 1.394 (2.294 ± 1.926) | 1.004 (0.959) |
| 3 | 1,973,488 (1,262,416) | 0.662 ± 0.817 (1.021 ± 0.992) | 0.989 (0.981) | 1,602,991 (838,644) | –2.359 ± 0.951* (–1.990 ± 1.281) | 1.037 (1.033) |
| 4 | 2,350,686 (1,388,607) | –3.489 ± 0.678** (–3.507 ± 0.840)** | 1.075 (1.082) | 1,469,895 (714,791) | –4.521 ± 0.894** (–3.742 ± 1.225)** | 1.090 (1.080) |
| 5 | 534,707 (222,425) | –1.691 ± 1.586 (0.000 ± 2.307) | 1.030 (1.000) | 191,221 (58,686) | 0.895 ± 2.788 (2.444 ± 4.611) | 0.985 (0.956) |
| 6 | 264,193 (138,064) | 0.418 ± 2.167 (–0.473 ± 2.848) | 0.992 (1.010) | – | – | – |
| Total | 7,078,758 (4,263,620) | –1.734 ± 0.410** (–1.160 ± 0.506)* | 1.033 (1.025) | 4,394,569 (2,180,898) | –2.549 ± 0.548** (–1.649 ± 0.750)* | 1.044 (1.030) |
| Chromosome 21 | ||||||
| ENm005 | 760,942 (442,909) | –0.816 ± 1.310 (0.850 ± 1.588) | 1.014 (0.983) | 560,555 (303,620) | –1.289 ± 1.567 (–0.822 ± 1.996) | 1.021 (1.015) |
| Total | 7,839,700 (4,706,529) | –1.645 ± 0.391** (–0.971 ± 0.482)* | 1.031 (1.020) | 4,955,124 (2,484,518) | –2.408 ± 0.517** (–1.549 ± 0.703) | 1.042 (1.029) |
Intergenic regions from fragment 6 of chromosome 7 are not considered in this analysis because of gaps in the mapping. Results excluding repetitive elements are shown in parentheses. Kij is the average genetic distance between i and j. Human, chimpanzee, common ancestor of human-chimpanzee, and baboon are represented as h, c, o, and b, respectively. *, P < 0.05; **, P < 0.01 by the relative rate test.
We obtained similar results from the human–chimpanzee–rhesus comparison (data set 3). Humans had 3.02% fewer substitutions than chimpanzees in introns (among 2,479,142 intron sites, 15,815 substitutions were human-specific and 16,275 substitutions were chimpanzee-specific). In intergenic regions, humans had 5.37% fewer substitutions than chimpanzees (among 5,522,390 intergenic sites, 36,153 substitutions were human-specific and 38,017 substitutions were chimpanzee-specific). The average genetic distance in introns for human–chimpanzee, human–rhesus, and chimpanzee–rhesus pairs are 1.36 (±0.007%), 6.68 (±0.01%) and 6.71 (±0.01%), respectively. The average genetic distance in intergenic regions for human–chimpanzee, human–rhesus, and chimpanzee–rhesus pairs are 1.41 (±0.005%), 6.91 (±0.011%), and 6.94 (±0.011%), respectively.
We also performed maximum likelihood analyses with data set 1 to test how the data fits two different models. In the first model, which we refer to as the “two-rate model,” rates of the human and chimpanzee lineages are assumed to be the same, whereas it is allowed to differ in the lineage leading to baboon, following the well established “hominoid rate slowdown” (5, 6, 8, 9, 14). An alternative model, which we refer to as the “three-rate model,” allows the human, chimpanzee, and baboon lineages to have different rates. The two-rate model was rejected both in introns and intergenic regions (P < 0.05 and P < 0.01, respectively) when tested against the three-rate model, again supporting a slower substitution rate in the human lineage.
Rate slowdown in the human lineage is also found in other chromosomes (data set 2, Table 2). In all of the nine chromosomes studied, introns of the human lineage have evolved at slower rates than the orthologous chimpanzee introns, five of which are significant by the relative rate test. In intergenic regions, six of the eight chromosomes studied have evolved at slower rates in humans than chimpanzees, four of which are significant by the relative rate test. Among the chromosomes that show significant rate slowdown in humans, chromosome 12 exhibits the greatest slowdown both in introns and intergenic regions (32.3% and 31.4%, respectively), chromosome 10 exhibits the lowest rate slowdown in introns (10.8%), and chromosome 22 exhibits the lowest rate slowdown in intergenic regions (9.5%). The introns and intergenic regions of humans have evolved slower than the orthologous regions in chimpanzees by 18% and 10.6%, respectively (P < 0.01 for relative rate test in both introns and intergenic regions). This rate slowdown is also supported by maximum likelihood analysis (two-rate model was rejected with P < 0.01 when tested against three-rate model for both introns and intergenic regions) and Tajima's nonparametric relative rate tests (Supporting Text and Table 5, which are published as supporting information on the PNAS web site).
Table 2. Number of sites compared, rate difference between human and chimpanzee, and the ratio of chimpanzee-specific versus human-specific substitutions from data set 2.
| Introns
|
Intergenic regions
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | Number of sites compared | d = Kbh – Kbc (per 10 kb) | KOC/KOH | Number of sites compared | d = Kbh – Kbc (per 10 kb) | KOC/KOH |
| 2 | 41,571 (25,457) | –8.639 ± 5.864 (–9.751 ± 7.238) | 1.146 (1.177) | 143,774 (112,336) | 2.911 ± 2.918 (1.900 ± 3.157) | 0.949 (0.963) |
| 4 | 36,539 (18,257) | –3.040 ± 6.383 (–1.209 ± 8.457) | 1.048 (1.021) | 55,184 (19,556) | –7.973 ± 6.202 (–6.889 ± 10.292) | 1.093 (1.081) |
| 8 | 25,135 (17,869) | –14.124 ± 7.646 (–24.117 ± 8.961)** | 1.251 (1.482) | 55,151 (32,783) | 1.207 ± 6.206 (–0.671 ± 7.897) | 0.987 (1.007) |
| 10 | 33,533 (26,022) | –1.6288 ± 6.633 (–1.680 ± 7.542) | 1.025 (1.026) | 91,889 (45,965) | –6.388 ± 4.082 (–2.123 ± 5.719) | 1.099 (1.032) |
| 12 | 49,266 (46,909) | –12.497 ± 4.452** (–12.434 ± 4.581)** | 1.323 (1.318) | 113,283 (91,873) | –13.55 ± 3.127** (–16.419 ± 3.387)** | 1.314 (1.414) |
| 16 | 136,363 (68,749) | –7.487 ± 3.495* (–10.113 ± 4.438)* | 1.108 (1.183) | 48,813 (16,310) | –16.701 ± 6.070** (–3.345 ± 9.107) | 1.244 (1.058) |
| 18 | 145,283 (121,412) | –2.141 ± 2.819 (–5.097 ± 2.964) | 1.041 (1.110) | – | – | – |
| 21 | 132,603 (74,660) | –9.888 ± 3.826** (0.597 ± 5.201) | 1.125 (0.993) | 41,290 (25,445) | –25.768 ± 7.379** (–36.296 ± 9.799)** | 1.306 (1.418) |
| 22 | 443,452 (264,488) | –15.681 ± 1.780** (–15.797 ± 2.190)** | 1.292 (1.327) | 214,581 (116,725) | –6.156 ± 2.711* (–1.983 ± 3.583) | 1.095 (1.031) |
| Total | 1,043,745 (663,823) | –10.607 ± 1.181** (–10.222 ± 1.408)** | 1.180 (1.191) | 763,965 (460,993) | –6.873 ± 1.427** (5.961 ± 1.755)** | 1.106 (1.099) |
Intergenic regions from the chromosome 18 are shorter than 250 bp and not included in this analysis. Notations are the same as in Table 1. *, P < 0.05; **, P < 0.01 by the relative rate test.
Rate Heterogeneity Among Genomic Regions. The noncoding segments in data set 1 exhibit a wide range of variation in genetic distance between human and chimpanzee (0% to 1.92 ± 0.03% in intergenic regions and 0% to 5.2 ± 0.11% in introns). What causes such heterogeneity in rate is an important question (15–19). In accord with previous results, we observed that the numbers of nucleotide substitutions between human and chimpanzee are significantly positively correlated with the GC contents of the segments analyzed (R2 = 10.04%, P < 10-10), as well as with the average rate of recombination (R2 = 9.61%, P < 10-10). We found similar results in human–baboon and chimpanzee–baboon comparisons (results not shown).
The average human-chimpanzee genetic distance also varied among the chromosomes (data set 2). Among the nine chromosomes compared, chromosome 21 had the highest divergence (1.73 ± 0.03%) and chromosome 12 had the lowest divergence (0.99 ± 0.02%). In addition, the ratios of human- vs. chimpanzee-specific nucleotide substitutions (inferred by using parsimony) vary significantly among chromosomes (Table 5; P < 0.001 for both intergenic regions and introns by G test of heterogeneity, ref. 20). The disparity is not caused by the fact that the data from the chromosome 7 has the smallest difference (due to the better quality of data; data for chromosome 7 was obtained from data set 1, whereas the data for other chromosomes were obtained from data set 2) in the numbers of nucleotide substitutions between the human and chimpanzee lineages (P < 0.001 when the chromosome 7 is removed from the analysis). We found no evidence of the effect of chromosomal lengths on the rate variation, unlike the recent finding in birds by Axelsson et al. (21). This may be due to the limited statistical power in our study.
Variable Molecular Clocks in Hominoids. To better understand the dynamics of hominoid molecular clocks, we expanded our analysis to include ≈2 Mbps of high-quality sequence data from gorilla (Gorilla gorilla) and orangutan (Pongo pygmaeus) for the region orthologous to human chromosome 7: 115404472–117281897 (Encode Region ENm001, data set 4). A five-species sequence alignment of human, chimpanzee, gorilla, orangutan, and baboon (see Materials and Methods) was then used to calculate the pairwise genetic distances between different hominoids (Table 3) within the noncoding regions. To our knowledge, this study analyzed the largest amount of noncoding sequences to provide comprehensive estimates of neutral evolutionary rates among hominoids.
Table 3. Average pairwise distance (per 1,000 nucleotides) between the five species studied, using data from ENm001.
| Species | Human | Chimpanzee | Gorilla | Orangutan | Baboon |
|---|---|---|---|---|---|
| Human | - | - | - | - | - |
| Chimpanzee | 10.960 (±0.011) | - | - | - | - |
| Gorilla | 13.668 (±0.014) | 13.832 (±0.014) | - | - | - |
| Orangutan | 29.135 (±0.030) | 29.308 (±0.031) | 29.815 (±0.031) | - | - |
| Baboon | 58.704 (±0.063) | 58.851 (±0.063) | 59.460 (±0.064) | 59.644 (±0.0643) | - |
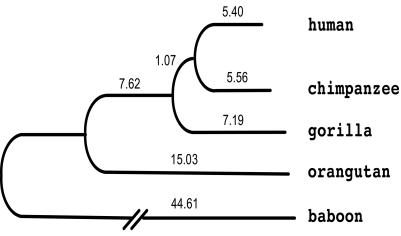
Fig. 1 shows the neighbor-joining tree (22) generated from this analysis. The human–chimpanzee clade is supported by 100% bootstrap value, providing further evidence for the sister grouping of these two species (23, 24). The ((human, chimpanzee), gorilla) clade is also supported by 100% bootstrap value.
Fig. 1.
Neighbor-joining tree of the five species compared in this study, based on data from the noncoding regions of EnM001. Branch lengths are the number of substitutions per 1,000 bp.
Using the distances in Table 3, we performed relative rate tests with all of the 10 combinations of three species (Table 4). The rate comparison between humans and chimpanzees was not significant in this analysis, presumably because of the smaller size of data set 4 (≈2 Mbps compared to ≈24 Mbps in data set 1) for human–chimpanzee rate comparison. Nevertheless, the pattern of higher rate in chimpanzees than humans (≈3% higher in data set 4) is consistent with the results from the larger data set 1.
Table 4. Ratios of species-specific branch lengths from all three species comparisons, using the data from ENm001.
| 5-species | HCG | HCO | HCB | HGO | HGB | HOB | CGO | CGB | COB | GOB |
|---|---|---|---|---|---|---|---|---|---|---|
| Introns | 1.014 (1.009) | 1.013 (1.005) | 1.019 (0.998) | 1.104** (1.089**) | 1.107** (1.084**) | 1.045** (1.038**) | 1.092** (1.084**) | 1.089** (1.086**) | 1.0372* (1.039*) | 0.996 (0.999) |
| Intergenic | 1.057 (1.065) | 1.063 (1.079) | 1.039 (1.051) | 1.106** (1.107**) | 1.134** (1.123**) | 1.101** (1.091**) | 1.051 (1.038) | 1.097** (1.076**) | 1.084** (1.069**) | 1.038 (1.033) |
| Total | 1.030 (1.029) | 1.032 (1.031) | 1.027 (1.017) | 1.105** (1.096**) | 1.117** (1.098**) | 1.067** (1.057**) | 1.076** (1.068**) | 1.092** (1.082**) | 1.057** (1.050**) | 1.012 (1.011) |
Results with repetitive sequences excluded are shown in parentheses. H, human; C, chimpanzee; G, gorilla; O, orangutan; B, baboon. The first two species are compared using the third species as an outgroup. *, P < 0.05; **, P < 0.01 by relative rate test.
Interestingly, both humans and chimpanzees show slower evolutionary rates compared to either gorillas or orangutans. In particular, the human lineage shows ≈11% rate slowdown than the gorilla lineage, when either orangutan or baboon is chosen as an outgroup. Also, the chimpanzee lineage shows ≈8% rate slowdown compared to the gorilla lineage, when either orangutan or baboon is chosen as an outgroup. In other words, our analysis clearly shows significant rate differences between the Homo–Pan and the gorillas and orangutans. There is no significant rate difference between the gorilla and orangutan lineages, at least in the current sequence comparison (data set 4).
We further tested the validity of this observation by using a maximum likelihood method. We first calculated the likelihood of a model in which the four hominoid species have evolved at the same rate while the branch leading to baboon has evolved at a different rate, taking into account the well supported hominoid-rate slowdown (5, 6, 9). We then calculated the likelihood of a model in which all seven branches are allowed to evolve at a different rate. The latter model fitted the data significantly better than the former (P < 0.05, likelihood ratio test). Two alternative models in which the human and chimpanzee branches are given the same rate and allowed to vary from gorillas and orangutans also performed better than the first model (P < 0.05 for either comparison). In conclusion, models that assume slower rates in the human and chimpanzee lineages than the gorilla and orangutan lineages generally perform significantly better than a model that assumes a uniform rate among hominoids, supporting the idea that molecular clocks in hominoids vary significantly.
Recent Evolution of Human-Specific Life History Traits. Given that molecular clocks in human and chimpanzee are only slightly different (see above), the life history traits that led to the current difference in generation time between humans and chimpanzees may have been established very recently during the evolution of humans. Analyses of fossil hominins have indeed suggested that the origins of human-specific life history traits are recent (25–28). Our results indicating a slight difference between the molecular clocks of human and chimpanzee agrees with the findings of these studies.
If we assume that the observed rate difference between humans and chimpanzees is caused solely by the difference in generation times and that the difference in generation times evolved instantaneously, we can estimate when the human-specific life history traits evolved (Supporting Text). Using 15 years as the generation time for chimpanzees and ancient humans and 20 years for that of modern humans, the estimated time of the evolution of long generation time in the modern humans is approximately one million years.
Discussion
The genetic distance between the genomes of humans and chimpanzees has been intensely investigated (e.g., refs. 9–12, 29, and 30). Our results confirm that there is very little difference in the alignable regions of the human and chimpanzee genomes. Despite such a small difference, we detect a significant rate slowdown in the human lineage when compared to the chimpanzee lineage, both in intergenic regions and introns (including and excluding repetitive portions). Interestingly, two main data sets (data sets 1 and 2) exhibit noticeably different levels of rate slowdown in the human genome (data set 2 shows a greater slowdown: Tables 1 and 2). This is likely to be a consequence of different qualities of the data used. The chimpanzee sequences in data set 2 are from the current chimpanzee genome shotgun assembly, which is only ≈3.6× coverage (12, 31) and may contain errors caused by either assembly or sequencing. For this reason, we may take the estimate from data set 1 as a more accurate estimate.
For closely related species such as human and chimpanzee, the inferred level of divergence is greatly affected by the existing levels of polymorphism in both species and the polymorphism that existed in their common ancestor (ancestral polymorphism). Therefore, the observed number of differences must be corrected for these polymorphisms, to infer the number of fixed differences between the populations. For this purpose, we used polymorphism levels from African humans (Πh) and Central African chimpanzees Πc), which reflect deeper coalescent times. Specifically, we used the values from Yu et al. (32) estimated from targeted high-quality sequencing of 49 noncoding intergenic regions (Πh = 0.115 and Πc = 0.130). Using the average rate difference between human–chimpanzee (from data sets 1 and 2) and 1.23% divergence between human and chimpanzee, the corrected rate difference (Supporting Text) between the human and chimpanzee is 11% if we assume the level of ancestral polymorphism to be the same as that in the current chimpanzee population. When we use the conservative estimate of 3% rate difference between human and chimpanzee (see above), the corrected rate is ≈2%.
Some studies suggest a greater level of polymorphism in Central African chimpanzees than the one used in the above calculation (e.g., Πc = 0.174; ref. 12), which will reduce the rate difference to 6% (using 10% observed difference) and suggest a rate increase in the human genome if we use 3% difference. This emphasizes the importance of accurate knowledge on the levels of polymorphisms (in different genomic regions) in inferring the exact rate difference between human and chimpanzee. In this regard, we point out that many studies on the molecular evolution of Y-linked regions resulted in a shorter human branch compared to the chimpanzee branch (e.g., refs. 10, 11, and 33). Because Y-linked regions are relatively free from the effect of ancestral and current polymorphisms, these observations suggest that the human lineage has indeed evolved more slowly than the chimpanzee lineage.
Several small-scale studies have previously reported rate slowdown in the human lineage as compared to other hominoids. For example, the η-globin pseudogene region (34–36), Xq13.3 region (37), the last intron of ZFX region (38), introns 7 and 44 of the dmd gene (39), and the ZFY region (11) exhibit rate slowdown in humans. Shi et al. (30) also reported a smaller number of substitutions in humans than in chimpanzees in functional regions of chromosome 21. In our study, we used a large amount of data from various regions to confirm that the rate slowdown in the human lineage compared to the chimpanzee lineage is a genome-wide phenomenon. However, humans and chimpanzees have accumulated only a slightly different number of single-nucleotide substitutions since their divergence from a common ancestor. Therefore, human-specific life history traits that led to longer generation time could have evolved only recently during human evolution, potentially around one million years ago (see Results). We emphasize again that this approximation is based on the simplifying assumptions that the generation time difference arose in the population instantaneously at some point of time after the human chimpanzee split, and the observed rate difference is caused solely by the difference in generation time.
When two other hominoid species (gorilla and orangutan) were included in the analysis, we found that both the human and the chimpanzee lineages exhibit slower molecular clocks compared to either gorilla or orangutan. This finding contradicts the view that humans differ greatly from all other hominoids in generation-time related life history traits. Rather, it suggests that life history traits that affect generation time may have evolved more than once during the evolution of hominoids, including the recent evolution of human-specific life history traits. The most parsimonious explanation for the slower molecular clocks in both humans and chimpanzees as compared to gorilla and orangutan is a slowdown in the ancestral lineage leading to the common ancestor of humans and chimpanzees. Yet another possibility is that both humans and chimpanzees went through independent rate slowdowns. To distinguish these two hypotheses, we will need an independent calibration point before the divergence of human and chimpanzee, which currently is unavailable.
On the other hand, the current study is based on a single genomic region (ENm001 region from chromosome 7; data set 4). As more genomic sequences accumulate from nonhuman hominoids in the near future, we can determine whether the findings from this study truly reflect a genome-wide pattern. If the analyzed region had indeed evolved at a different rate than other genomic regions, the underlying basis for such difference will be of great interest.
To further investigate differences in generation-time related life history traits, we examined currently known differences between the four hominoid species analyzed in this study (Table 6, which is published as supporting information on the PNAS web site, compiled from ref. 40). Some life history traits show similar patterns as the results of molecular clock analysis in this study, e.g., the age females reach sexual maturity and the age at first birth. Such differences theoretically could give rise to the fastest molecular clock of gorillas among the hominoid species compared. However, such an inference should be taken with caution, because it is unknown when such traits were established during the evolution of each lineage. Analyses and dating of fossil nonhuman hominoids may shed light on the time scale of such events. In this light, the recent discovery of the first fossil chimpanzee is extremely encouraging (41). With more such discoveries, we may uncover changes that led to rate variation within hominoids, in particular the slowdown in the human and chimpanzee lineages.
Materials and Methods
Human–Chimpanzee–Baboon Comparison. We have two sources of data for human-chimpanzee-baboon comparison, named data set 1 and data set 2 (shown in Tables 1 and 2, and Table 7, which is published as supporting information on the PNAS web site).
Data Set 1. This data set consists of sequences from orthologous BAC clones from chimpanzee (Pan troglodytes) and baboon (Papio anubis), corresponding to ≈22 Mbps of human chromosome 7 (Table 7) and ≈2 Mbps of chromosome 21 (Encode region ENm005; ref. 42). Chimpanzee and baboon BAC clones orthologous to regions in human chromosome 7 were isolated and sequenced as described by Thomas et al. (43, 44). For ENm005, we obtained the orthologous chimpanzee sequence from the high-quality BAC-based sequences of the chimpanzee chromosome 22 (13). The corresponding sequence from baboon was obtained by the BAC-based procedure described by Thomas et al. (43, 44).
Data Set 2. We obtained ≈4 Mbps of data from nine other chromosomes, by mapping 22 complete baboon BAC clone sequences available in the GenBank database [ref. 45; all available complete baboon BAC clones as of April 2005, excluding those orthologous to human chromosome 7, because most of them were incorporated in the data set 1] to the human and chimpanzee genome (Table 8, which is published as supporting information on the PNAS web site). We first used the megablast program (www.ncbi.nlm.nih.gov) to find the region in the human genome (hg17, corresponds to NCBI build 35) orthologous to the baboon sequence. Only the colinear nonoverlapping high scoring segment pairs (HSPs) >250 bp in length were used for further analysis. The orthologous chimpanzee sequence for each HSP was obtained from the “Chimp Chain” track in the University of California, Santa Cruz (UCSC) genome browser (46).
Human–Chimpanzee–Rhesus Comparison. In addition to the analysis of human–chimpanzee–baboon sequences, we compared the rate of molecular evolution of the whole human chromosome 21 (hs21, ≈35 Mbps of data) with chimpanzee chromosome 22 (ptr22; ref. 13) using the draft sequence of rhesus macaque (Macaca mulatta) (rheMac1, UCSC Genome Browser) as an outgroup. Because the sequences of hs21 and ptr22 are of high quality, random sequencing errors in the draft sequence of rhesus macaque (the outgroup) are not expected to affect the conclusions. Henceforth, we call this data set “data set 3.”
Data Set 3. We first aligned hs21 and ptr22. ptr22 was cut into five segments, each ≈7 Mb in length. Each segment was then aligned to orthologous regions in hs21 by using blastz (47) with the same parameters and substitution matrix used in the UCSC genome browser for the “Chimp Chain” track. The alignments were converted into chains by using the axtchain program (48). The longest chain for each segment was extracted manually and concatenated to form one single chain for the whole chromosome.
To obtain rhesus sequences orthologous to hs21, all of the chains in the “Rhesus Chain” track of UCSC genome browser that mapped to hs21 were filtered such that the length of the chain is (i) at least 100 kbp and (ii) >95% of the total length of the original rhesus scaffold that produced the chain. Overlapping chains, chains in ENm005 and chains near the telomeres were removed manually. This resulted in a set of 58 chains (Table 9, which is published as supporting information on the PNAS web site), encompassing a total of ≈10 Mbps in hs21. The orthologous chimpanzee region for each chain was extracted from the human–chimpanzee whole chromosome alignment based on the mapping coordinates of the chain on hs21.
Data from Other Hominoids. We analyzed ≈2 Mbps of BAC-based sequences from human, chimpanzee, gorilla, orangutan, and baboon corresponding to the region orthologous to human chromosome 7: 115404472–117281897 in hg17 (Encode Region ENm001; refs. 42 and 49). Henceforth, we call this data set “data set 4.”
Alignment and Annotation. Orthologous regions were aligned by using the Threaded Blockset Aligner (50). From the alignment, we extracted only introns and intergenic regions, using gene annotations included in the Known Genes and Ensembl Genes tables of the UCSC Genome Browser (hg17 assembly). Intergenic and intronic sequences likely to be selectively constrained [the 5′ and 3′ untranslated regions, and small (<250 bp) introns or intergenic intervals] were also excluded. Repetitive sequences were detected by using the repeatmasker program (www.repeatmasker.org). Recombination rates were obtained from the annotations in UCSC genome browser (“Recomb Rates” track created from ref. 51).
Distance Calculation and Statistical Tests. The Jukes–Cantor (52) method was used to correct for multiple hits. A relative rate test (7) was used to test for rate difference between any two species using a third outgroup species. We also performed maximum likelihood analyses to compare rates, using the baseml program in PAML package (53). The neighbor-joining tree was constructed by using mega software (54).
Supplementary Material
Acknowledgments
We thank Morris Goodman, Wen-Hsiung Li, Derek Wildman, and three anonymous reviewers for comments on the manuscript, Seong-Ho Kim for discussions, and Gregory Cooper for sharing his data. We thank the Baylor College of Medicine Human Genome Sequencing Center (www.hgsc.bcm.tmc.edu) for providing rhesus macaque genome sequence assembly. S.V.Y. is supported by the Georgia Institute of Technology.
Author contributions: J.W.T. and S.V.Y. designed research; N.E. and S.V.Y. performed research; J.W.T. and N.C.S.P. contributed new reagents/analytic tools; N.E. and S.V.Y. analyzed data; and N.E., J.W.T., and S.V.Y. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: UCSC, University of California, Santa Cruz.
References
- 1.Wood, B. & Collard, M. (1999) Science 284, 65-71. [DOI] [PubMed] [Google Scholar]
- 2.Harvey, P. H. & Clutton-Brock, T. H. (1985) Evolution (Lawrence, Kans.) 39, 559-581. [DOI] [PubMed] [Google Scholar]
- 3.Smith, B. H. & Tompkins, R. L. (1995) Ann. Rev. Anthropol. 24, 257-259. [Google Scholar]
- 4.Preuss, T. M., Caceres, M., Oldham, M. C. & Geschwind, D. H. (2004) Nat. Rev. Genet. 5, 850-860. [DOI] [PubMed] [Google Scholar]
- 5.Goodman, M. (1961) Hum. Biol. 33, 131-162. [PubMed] [Google Scholar]
- 6.Goodman, M. (1962) Hum. Biol. 34, 104-150. [PubMed] [Google Scholar]
- 7.Wu, C. I. & Li, W. H. (1985) Proc. Natl. Acad. Sci. USA 82, 1741-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, W. H., Ellsworth, D. L., Krushkal, J., Chang, B. H. & Hewett-Emmett, D. (1996) Mol. Phylogenet. Evol. 5, 182-187. [DOI] [PubMed] [Google Scholar]
- 9.Yi, S., Ellsworth, D. L. & Li, W. H. (2002) Mol. Biol. Evol. 19, 2191-2198. [DOI] [PubMed] [Google Scholar]
- 10.Chen, F. C. & Li, W. H. (2001) Am. J. Hum. Genet. 68, 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebersberger, I. & Meyer, M. (2005) Mol. Biol. Evol. 22, 1240-1245. [DOI] [PubMed] [Google Scholar]
- 12.The Chimpanzee Genome Sequencing Consortium (2005) Nature 437, 69-87. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T. D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R., et al. (2004) Nature 429, 382-388. [DOI] [PubMed] [Google Scholar]
- 14.Steiper, M. E., Young, N. M. & Sukarna, T. Y. (2004) Proc. Natl. Acad. Sci. USA 101, 17021-17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellmann, I., Ebersberger, I., Ptak, S. E., Pääbo, S. & Przeworski, M. (2003) Am. J. Hum. Genet. 72, 1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellmann, I., Prufer, K., Ji, H., Zody, M. C., Pääbo, S. & Ptak, S. E. (2005) Genome Res. 15, 1222-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meunier, J. & Duret, L. (2004) Mol. Biol. Evol. 21, 984-990. [DOI] [PubMed] [Google Scholar]
- 18.Webster, M. T., Smith, N. G. & Ellegren, H. (2003) Mol. Biol. Evol. 20, 278-286. [DOI] [PubMed] [Google Scholar]
- 19.Webster, M. T., Smith, N. G., Lercher, M. J. & Ellegren, H. (2004) Mol. Biol. Evol. 21, 1820-1830. [DOI] [PubMed] [Google Scholar]
- 20.Sokal, A. K. & Rolf, F. J. (1995) Biometry: The Principles and Practice of Statistics in Biology Research (Freeman, New York), pp. 738.
- 21.Axelsson, E., Webster, M. T., Smith, N. G., Burt, D. W. & Ellegren, H. (2005) Genome Res. 15, 120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, M., Grossman, L. I. & Wildman, D. E. (2005) Trends Genet. 21, 511-517. [DOI] [PubMed] [Google Scholar]
- 24.Wildman, D. E., Uddin, M., Liu, G., Grossman, L. I. & Goodman, M. (2003) Proc. Natl. Acad. Sci. USA 100, 7181-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bermudez de Castro, J. M., Rosas, A., Carbonell, E., Nicolas, M. E., Rodriguez, J. & Arsuaga, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4210-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogin, B. & Smith, B. H. (1996) Am. J. Hum. Biol. 8, 703-716. [DOI] [PubMed] [Google Scholar]
- 27.Dean, C., Leakey, M. G., Reid, D., Schrenk, F., Schwartz, G. T., Stringer, C. & Walker, A. (2001) Nature 414, 628-631. [DOI] [PubMed] [Google Scholar]
- 28.Tardieu, C. (1998) Am. J. Phys. Anthropol. 107, 163-178. [DOI] [PubMed] [Google Scholar]
- 29.Chen, F. C., Vallender, E. J., Wang, H., Tzeng, C. S. & Li, W. H. (2001) J. Hered. 92, 481-489. [DOI] [PubMed] [Google Scholar]
- 30.Shi, J., Xi, H., Wang, Y., Zhang, C., Jiang, Z., Zhang, K., Shen, Y., Jin, L., Zhang, K., Yuan, W., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8331-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, M. V. & Varki, A. (2003) Nat. Rev. Genet. 4, 20-28. [DOI] [PubMed] [Google Scholar]
- 32.Yu, N., Jensen-Seaman, M. I., Chemnick, L., Kidd, J. R., Deinard, A. S., Ryder, O., Kidd, K. K. & Li, W. H. (2003) Genetics 164, 1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makova, K. D. & Li, W. H. (2002) Nature 416, 624-626. [DOI] [PubMed] [Google Scholar]
- 34.Bailey, W. J., Fitch, D. H., Tagle, D. A., Czelusniak, J., Slightom, J. L. & Goodman, M. (1991) Mol. Biol. Evol. 8, 155-184. [DOI] [PubMed] [Google Scholar]
- 35.Graur, D. & Li, W. H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA), pp. 147-148.
- 36.Li, W. H., Tanimura, M. & Sharp, P. M. (1987) J. Mol. Evol. 25, 330-342. [DOI] [PubMed] [Google Scholar]
- 37.Kaessmann, H., Heissig, F., von Haeseler, A. & Pääbo, S. (1999) Nat. Genet. 22, 78-81. [DOI] [PubMed] [Google Scholar]
- 38.Jaruzelska, J., Zietkiewicz, E. & Labuda, D. (1999) Mol. Biol. Evol. 16, 1633-1640. [DOI] [PubMed] [Google Scholar]
- 39.Nachman, M. W. & Crowell, S. L. (2000) Genetics 155, 1855-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowe, N. (1996) in The Pictorial Guide to the Living Primates (Pogonias, Charlestown, RI), pp. 219-233.
- 41.McBrearty, S. & Jablonski, N. G. (2005) Nature 437, 105-108. [DOI] [PubMed] [Google Scholar]
- 42.The ENCODE (Encyclopedia of DNA Elements) Project (2004) Science 306, 636-640. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788-793. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, J. W., Prasad, A. B., Summers, T. J., Lee-Lin, S. Q., Maduro, V. V., Idol, J. R., Ryan, J. F., Thomas, P. J., McDowell, J. C. & Green, E. D. (2002) Genome Res. 12, 1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Wheeler, D. L. (2005) Nucleic Acids Res. 33, D34-D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M. & Haussler, D. (2002) Genome Res. 12, 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C., Haussler, D. & Miller, W. (2003) Genome Res. 13, 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kent, W. J., Baertsch, R., Hinrichs, A., Miller, W. & Haussler, D. (2003) Proc. Natl. Acad. Sci. USA 100, 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper, G. M., Stone, E. A., Asimenos, G., Green, E. D., Batzoglou, S. & Sidow, A. (2005) Genome Res. 15, 901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanchette, M., Kent, W. J., Riemer, C., Elnitski, L., Smit, A. F., Roskin, K. M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E. D., et al. (2004) Genome Res. 14, 708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong, A., Gudbjartsson, D. F., Sainz, J., Jonsdottir, G. M., Gudjonsson, S. A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. (2002) Nat. Genet. 31, 241-247. [DOI] [PubMed] [Google Scholar]
- 52.Jukes, T. H. & Cantor, C. R. (1969) in Mammalian Protein Metabolism (Academic, New York), pp. 21-132.
- 53.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 54.Kumar, S., Tamura, K. & Nei, M. (2004) Brief Bioinform. 5, 150-163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.