Abstract
Borna disease virus (BDV) establishes a persistent infection in the central nervous system of vertebrate animal species as well as in tissue cultures. In an attempt to characterize the life cycle of BDV in persistently infected cultured cells, we developed 30 clones by single-cell cloning from a human oligodendroglioma (OL) cell line after infection with BDV. According to the percentage of cells expressing the BDV major proteins, p40 (nucleoprotein) and p24 (phosphoprotein), the clones were classified into two types: type I (>20%) and type II (<20%). mRNAs corresponding to both proteins were detected by in situ hybridization (ISH) in a percentage of cells consistent with that for the protein expression in the two types. Surprisingly, ISH for the detection of the genomic RNA, mainly in type II, revealed a significantly larger cell population harboring the genomic RNA than that with the protein as well as the mRNA expression. By recloning from type II primary cell clones, the same phenotype was confirmed in the secondary cell clones obtained: i.e., low percentage of protein-positive cells and higher percentage of cells harboring the genomic RNA. After nerve growth factor treatment, the two types of clones showed increases in the percentage of cells expressing BDV-specific proteins that reached 80% in type II clones, in addition to increased expression levels per cell. Such enhancement might have been mediated by the activation of the mitogen-activated protein kinase in the clones as revealed by the detection of activated ERK1/2. Thus, our findings show that BDV may have established a persistent infection at low levels of viral expression in OL cells with the possibility of a latent infection.
Borna disease virus (BDV) causes progressive meningoencephalomyelitis in horses and sheep, in addition to several other vertebrate species (4, 26, 41). BDV is a single nonsegmented, negative-stranded (NNS) RNA virus (7, 11, 14, 42) that is unique among members of the order Mononegaverales, in that it uses the nucleus as a site for replication and transcription (10, 43). BDV is a highly neurotropic virus that persists in the central nervous system (CNS) of infected animals. In adult rats, BDV infection is associated with massive brain inflammation and extensive neuronal damage (33). Neonatal infection, by contrast, results in life-long persistence without encephalitis (2, 3, 8, 16, 33). However, infected rats of all ages express a variety of behavioral abnormalities. Therefore, BDV provides an important model for the investigation of the mechanisms and consequences of viral persistence in the CNS.
Generally, the CNS is known to be unique in its response to viral infections. This may be because of the lack of specialized lymphatic drainage, potentially limiting and delaying viral antigen recognition, in addition to the lack of dendritic cells and the use of microglia for the initial recognition and presentation of viral antigens, as well as the lack of expression of major histocompatibility molecules by normal neurons and glial cells (36). Thus, these factors result in immune limitations, which could contribute to the fate of viral persistence in the CNS, in general and for BDV as well. In addition, persistent BDV infection in the CNS is due to its noncytolytic strategy for replication (25).
Persistent infection with BDV has been established in a variety of neuronal and glial cell lines during which viral antigens are continuously detected (9). The choice of a cell line for the in vitro characterization of BDV appears to depend on the in vivo tropism of the virus, as well as on the functional importance and abundance of cells in the CNS such as neurons and glial cells (astrocytes and oligodendrocytes). The rat astrocytoma cell line (C6) is frequently used for such a purpose (9, 17), in addition to the neuronal cell lines SK-N-SH and SK-N-SHEP (9).
The cell-specific variations in BDV replication in vitro have been studied. BDV persistence in the C6 cell line is characterized by high-level expression of BDV-specific proteins and RNA and low-level production of infectious virions (9). Such an expression is related in part to the secretion of several neurotrophic factors into the culture medium by the cells (9, 35). Although BDV is characterized by high genomic stability, a recent report demonstrated the possibility of isolating stable virus variants from a persistently infected C6 cell line (17).
Although oligodendrocytes are a major cellular component of the white matter in the CNS and have a pivotal role in neuronal cell functions, they have not been well studied in terms of BDV characterization. Oligodendrocytes have been shown to support experimental and natural infection with neurotropic NNS RNA viruses, such as canine distemper virus (31, 49) and measles virus (1, 20), revealing the important role of these cells as a target for establishing persistent infection and participating in the pathogenesis of such viruses. Even more, they may be targets for hosting the viral genome for life-long persistence at undetectable levels.
In this study, we characterized persistent BDV infection in a human oligodendroglioma (OL) cell line. To test for possible variations in BDV replicative phenotype, we developed a total of 30 biological cell clones after infection with BDV. We examined the differences in BDV replication patterns among the clones in terms of the expression of BDV-specific major proteins and their corresponding mRNAs. According to the percentage of BDV protein-expressing cells, clones were classified into two types, i.e., type I (>20%) and type II (<20%). However, the cell population harboring the genomic RNA was significantly larger than that expressing viral protein or mRNA especially in type II cell clones. This persistent infection of BDV exhibited by type II clones showed a shift to a phenotype similar to that of type I on treatment with nerve growth factor (NGF). Such activation might possibly be mediated by the activation of the mitogen-activated protein (MAP) kinases (ERK1/2).
MATERIALS AND METHODS
Cells and virus.
A human OL cell line and an OL cell line persistently infected with BDV, HuP2br (OL/BDV), isolated from the brain of a schizophrenic patient (32), were maintained in Dulbecco’s modified Eagle’s medium with high glucose (4.5 g/liter) containing penicillin, streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) (complete medium). A C6 cell line persistently infected with horse BDV, He-80 (C6/BV) (9), was also used.
Single cell cloning.
OL/BDV and C6/BV were grown to confluence, washed with phosphate-buffered saline (PBS) and harvested by trypsinization. Thereafter, the cells were suspended in complete medium and disrupted by three 10-s ultrasonic bursts, followed by centrifugation at 10,000 rpm for 5 min. The clarified samples containing the cell-free viruses from each cell line were separately incubated with 200 uninfected OL cells. After 1 h of incubation at 37°C in a 5% CO2 incubator, the cells were washed and resuspended in complete medium. Cells were seeded as a single cell per well in 96-well microplates and then incubated for 14 days without medium change. Cell growth per well was monitored daily under the microscope, and single microcolonies growing out in individual wells were picked up on the 14th day and expanded for optimal division rates in 24- and 6-well microplates and then in T-25 tissue culture flasks. For recloning, selected cell clones were grown to complete confluence and then trypsinized and suspended in complete medium. Cells were seeded to a single cell per well in 96-well microplates, and the subsequent steps were performed as described above.
Preparation of polyclonal anti-BDV antibodies.
For the preparation of the polyclonal anti-BDV p40 and p24 antibodies, rabbits were immunized with the purified proteins from glutathione S-transferase (GST)-p40 and GST-p24, respectively, expressed as fusion proteins in Escherichia coli, as described previously (48).
Immunofluorescence (IF) assay.
Selected cell clones, OL/BDV, and uninfected OL cells growing to complete confluence were collected after being washed and treated with trypsin and then suspended in PBS. Cells were smeared onto eight-well glass slides, dried, fixed in 4% paraformaldehyde solution for 20 min at room temperature, and permeabilized for 5 min in 0.4% Triton X-100 (23). For BDV-specific protein detection, the slides were incubated with p40 and p24 polyclonal rabbit anti-BDV antibodies (100 μl; dilution 1:250) in PBS for 30 min at 37°C, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (IgG; 100 μl; dilution 1:200) in PBS for 30 min at 37°C.
Antigen-capture ELISA.
Cells were grown until complete confluence in 12-well plates. The culture media were collected and centrifuged. The supernatant was mixed with Nonidet P-40 at a final concentration of 0.5%. The cells were washed with PBS and then collected in 500 μl of 0.5% Nonidet P-40. The cells were sonicated, and the supernatant was collected after centrifugation. Antigen-capture enzyme-linked immunosorbent assay (ELISA) to quantify BDV-p40 within the cell fraction, as well as in the culture media, was performed as described previously (48). Briefly, microtiter plates were coated with 100 μl of PBS containing 1 μg of anti-p40 monoclonal antibody overnight at 4°C. The plates were blocked in 5% skim milk in PBS for 2 h at 37°C. After being washed with 0.05% Tween 20 in PBS (PBST), 100 μl of the sample was added per well (three wells per sample) for 1 h at 37°C. After another wash with PBST, the plates were incubated with 100 μl of 5 μg of rabbit anti-p40 polyclonal antibody/ml for 1 h at 37°C. After five washes with PBST, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (50 μl; dilution, 1:2,000) was added for 1 h at 37°C. Then, 100 μl of a solution of 0.4 mg of o-phenylenediamine in citrate-phosphate buffer (pH 5.4) with 0.012% hydrogen peroxide/ml was added. Finally, 100 μl of stop solution (3 N H2SO4) was added after color development for 1 min. The absorbance was measured at 492 nm.
BDV infectivity assay.
The infectivity of BDV in the culture media of the OL cell clones was determined for uninfected OL cells. Briefly, the culture medium was collected and then centrifuged to remove cellular debris. The supernatant was reacted with 105 uninfected OL cells, and the cells were incubated for 1 h at 37°C in a CO2 incubator. The cells were then washed and plated in 12-well plates. The cells were monitored for BDV-p40 production every third day, for four passages, by IF assay.
Western blot analysis.
Monolayers of representative OL cell clones, OL and OL/BDV, were collected after being washed and mixed with an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, as described previously (46). After cell lysis by sonication, followed by boiling, the proteins were separated by using a 12% gel and transferred to polyvinylidene difluoride membranes. The nonspecific binding sites were blocked with a solution of 5% skim milk in PBST. Polyclonal rabbit anti-BDV-p40 (1:1,000) was incubated with the membrane for 1 h at room temperature. After being washed with PBST, the membranes were incubated with the HRP-conjugated anti-rabbit IgG. Specific binding was detected by using an ECL Western blotting kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
Total RNA extraction and Northern blot analysis.
Total RNA was extracted from representative cell clones and OL and OL/BDV cells by using Isogen (Nippon Gene Co., Tokyo, Japan). Aliquots, each of 5 μg of RNA, were separated on 1% agarose gel containing formaldehyde and transferred to GeneScreen Plus Hybridization Transfer Membranes (NEN Life Sciences, Boston, Mass.). After being baked at 80°C, the membranes were prehybridized in hybridization solution (7% SDS; 50% deionized formamide; 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]; 50 mM phosphate buffer, pH 7.0; 0.1% [wt/vol] N-lauroylsarcosine; 2% blocking reagent) for 2 h at 68°C. This step was followed by overnight hybridization with digoxigenin (DIG)-labeled RNA probes for the detection of p40 genomic RNA and mRNA at a concentration of 50 ng/ml. The hybridized probe was detected with alkaline phosphatase-conjugated anti-DIG Fab fragment (Boehringer Mannheim) and visualized by using the chemiluminescence CSPD substrate (Boehringer Mannheim). The signals were visualized by exposure to X-ray film.
The RNA p40 sense and antisense probes used for hybridization were labeled with DIG by in vitro transcription with T7 RNA polymerase from the plasmids containing the cDNAs of BDV-p40, which was linearized with NotI and BamHI for the generation of the sense and antisense probes, respectively, as described previously (47). The DIG-labeled riboprobe was synthesized by using a DIG RNA labeling kit (Boehringer Mannheim).
ISH.
In situ hybridization (ISH) was carried out as described previously with minor modifications (47). Briefly, cells grown in eight-chamber slides until 80% confluence were washed with PBS, dried, fixed in 4% paraformaldehyde solution in PBS for 20 min at room temperature, and then denatured in 0.2 N HCl and in 2× SSC at 70°C. After acetylation in 0.1 M triethanolamine and 0.5% acetic anhydride, the slides were prehybridized in hybridization solution (50% formamide; 3× SSC; 50 mM HEPES, pH 7.0; 5× Denhardt's solution; 250 μg of salmon sperm DNA/ml) for 30 min at 37°C, followed by overnight hybridization at 37°C with the DIG-labeled RNA probes at a concentration of 10 ng/ml. After being washed, the slides were incubated with FITC-conjugated rabbit anti-DIG antibodies (50 μl; dilution 1:10; Boehringer Mannheim) for 45 min at 37°C. The slides were washed, counterstained with propidium iodide, and mounted for examination under the microscope.
The RNA probes (sense and antisense RNAs) used for ISH were labeled with DIG by in vitro transcription with T7 RNA polymerase from plasmids containing the cDNAs of BDV-p40 and BDV-p24, which were linearized with BamHI for the generation of the antisense probes and with NotI for the p40 sense probe. The DIG-labeled riboprobes were synthesized by using a DIG RNA labeling kit. Probes 300 bp in length were then generated by alkaline hydrolysis.
Serum starvation.
Cells grown in 12-well plates were washed with PBS and cultured in FBS-free medium (30). At each time point 0, 2, 3, and 5 days after starvation, IF assay was carried out.
NGF-β treatment.
Cells growing in monolayers were treated with NGF-β (Sigma) at a concentration of 50 ng/ml in FBS-free medium every other day (9). At each time point, 0, 2, and 4 days after the treatment, samples for IF assay and antigen-capture ELISA were collected.
NGF signaling pathway.
Cells were cultured in eight-chamber slides in medium containing 0.5% FBS for 2 days. This was followed by serum starvation for 2 h. Next, 100 ng of NGF-β/ml was added to the cells in FBS-free medium for given time points (19). At each time point (0, 5, 10, 15, 30, and 60 min), the slides were washed and fixed in 3% paraformaldehyde solution in PBS for 30 min at room temperature. The subsequent steps were performed according to the protocol supplied with the phospho-p44/42 MAP kinase (Thr202/Tyr204) E10 monoclonal antibody (ERK1/2) used (Cell Signaling Technology; New England Biolabs).
RESULTS
Isolation of OL cell clones persistently infected with BDV.
The cell-free virus derived from OL/BDV was incubated with OL cells for 1 h at 37°C. After being washed, the infected OL cells were subjected to single-cell cloning by limiting dilution. A total of 30 clones were obtained, each of a single-cell origin. These clones were expanded in 24-well plates, then in 6-well plates, and then in T-25 tissue culture flasks.
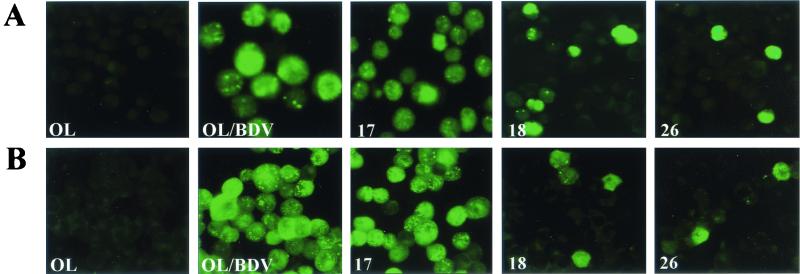
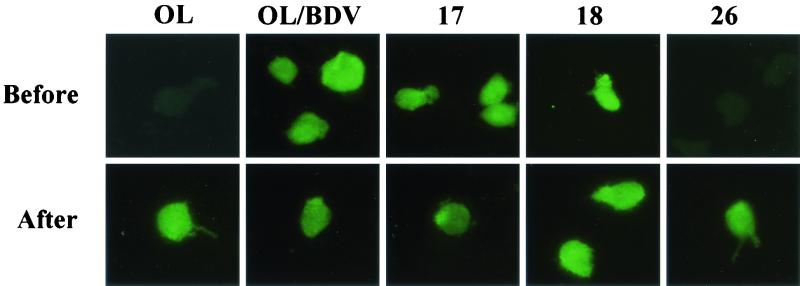
The cell clones were examined for BDV protein expression by IF assay. The slides were incubated with polyclonal rabbit antiserum against BDV-p40 or BDV-p24 and then with the secondary antibody (Fig. 1). The cell clones showed the characteristic punctate nuclear staining of the corresponding proteins in addition to diffused staining in the cytoplasm. According to the percentage of positive cells that expressed either protein, the clones were classified into three types: I (>20%), II (<20%), and III (0%) (Table 1). Type III showed no viral protein expression among the whole-cell population and was considered negative and thus was excluded from further characterization. Similarly, the cell clones obtained after infection of OL cells with cell-free BDV derived from C6/BV were also divided into three types, as summarized in Table 1. These data revealed no apparent difference in the infection of OL cells with the cell-free virus derived from either BDV isolate. Therefore, representative OL/BDV-derived cell clones from type I (clones 17 and 18) and type II (clone 26) were selected for more detailed characterization, together with the parental OL/BDV and uninfected OL cells as controls.
FIG. 1.
Percentage of BDV protein-expressing cells among cell clones persistently infected with BDV. The cell clones were analyzed for the BDV proteins, p40 and p24, by IF. Representative cell clones belonging to types I (clones 17 and 18) and II (clone 26), as well as parental OL/BDV and uninfected OL cells, were smeared on glass slides and incubated with polyclonal rabbit antiserum against BDV-p40 (A) and BDV-p24 (B), followed by the addition of FITC-conjugated secondary antibody.
TABLE 1.
Classification of persistently BDV-infected OL cell clones
| Type | % BDV-expressing cellsa | Cell clonesc detected in BDV strain:
|
|
|---|---|---|---|
| HuP2br | He/80 | ||
| Ib | >80 | 1, 10, 16, 17, 24, 38, 41, 45, 47 | 2, 10, 16, 19, 20, 22, 27, 39, 41 |
| 20-50 | 8, 18, 28, 35, 65, 69, 86 | 12, 30, 32, 37 | |
| II | <20 | 21, 26, 30, 57 | 3, 13, 20, 26, 29 |
| III | 0 | 2, 7, 9, 22, 31, 33, 37, 39, 42 | 4, 8, 9, 15, 18 |
% BDV p40-expressing cells in at least five microscopic fields as determined by IF assay.
Type I comprises cell clones with a wide range of protein expression: from those at >80% to others at 20 to 50%. However, there were no clear differences between them as detected by ELISA, Western blotting, and Northern blotting. From type I cell clones obtained after infection with HuP2br, underlined clones 17 and 18 were picked up for characterization, together with clone 26 from type II.
Cell clone designations are listed.
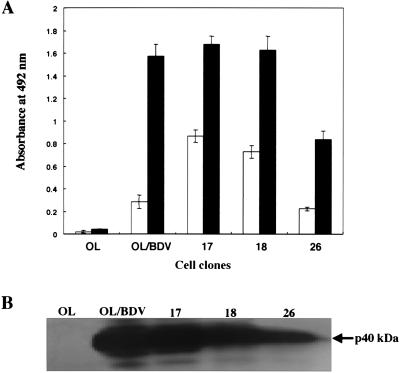
Quantification of BDV-p40 protein expression among the cell clones.
As shown in Fig. 2A, the production level of BDV-p40 within the cells among the two types of cell clones showed some variability by ELISA. Such variability in the protein contents, confirmed by Western blotting as shown in Fig. 2B, is consistent with the difference in the percentage of BDV protein-producing cells between types I and II as detected by the IF assay (Fig. 1).
FIG. 2.
Quantitative detection of BDV-p40 in persistently BDV-infected cell clones. (A), Representative cell clones (clones 17 and 18 in type I and clone 26 in type II), as well as OL/BDV and OL cells, were harvested and fractionated into cells (▪) and culture medium (□). The concentration of BDV-p40 protein in both fractions was estimated by antigen-capture ELISA. (B) Western blot analysis of cell lysates from representative cell clones. Cell lysates were separated by SDS-PAGE (12% gel). The proteins were transferred to polyvinylidene difluoride membranes, which where then blocked with 5% skim milk in PBST. The membranes were probed with rabbit anti-BDV-p40 antibodies. Bound antibodies were detected by using HRP-conjugated anti-rabbit IgG.
A significant amount of BDV-p40 was detected in the culture media of both types of cell clones (Fig. 2A). Thus, we infected OL cells with the supernatant from both types of clones in addition to the parental OL/BDV, as described in Materials and Methods. Along four passages, we detected no p40-positive cells in the newly infected cells, indicating that such a level of protein in the supernatant was not due to release of infectious virus particles into the medium but rather due to cell death and the release of their protein contents.
Quantification of RNA signals within the cell clones.
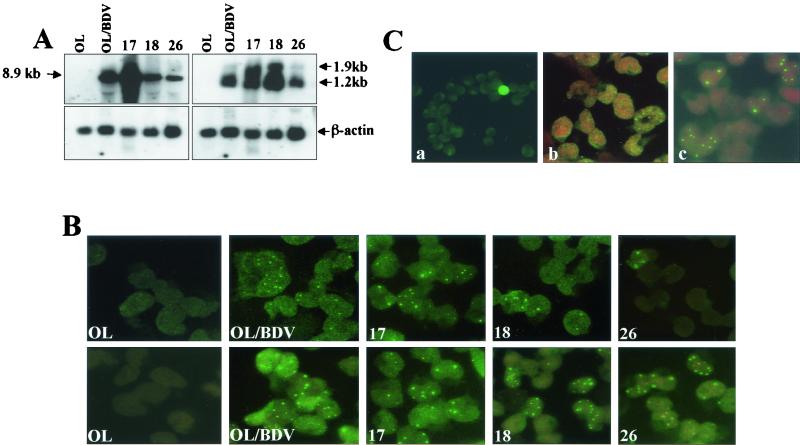
Next, we characterized representative clones of each type for the expression of the viral mRNA by Northern blot analysis with p40 antisense probe (Fig. 3A, right panel). The expression level of the 1.2- and 1.9-kb transcripts was lower in type II than in type I cells, as well as in the parental OL/BDV, and this appears to be consistent with the low production of p40 protein by this type as shown in Fig. 2.
FIG. 3.
Quantification of the viral RNA (genomic and mRNA) in the cell clones. (A) RNA was extracted from representative cell clones (clones 17 and 18 in type I and clone 26 in type II), as well as OL/BDV and OL cells, and analyzed by Northern blot hybridization for the expression of BDV-genomic RNA (left panel) and mRNA (right panel) with the p40 sense and antisense probes, respectively. (B) The same representative cell clones, as well as OL/BDV and OL cells, growing in eight-chamber slides were characterized by ISH with the p40 antisense (upper panel) and sense (lower panel) probes as described in Materials and Methods. (C) The type II reclone II-26/3 was characterized by IF with polyclonal rabbit antiserum against BDV-p40 (a), ISH with p40 antisense probe (b), and ISH with p40 sense probe (c).
By using the p40 sense probe to quantify the expression level of the 8.9-kb BDV-genomic RNA among the cell clones, we found that the expression level in type I cell clones was higher than that in type II, as expected (Fig. 3A, left panel).
Detection of BDV RNA in cells of the two types of cell clones.
We then tried to quantify the cell population positive for viral RNAs (mRNA and genomic RNA) among the two types of clones by ISH. Using p40 antisense probe, it was found that the mRNA expression (Fig. 3B, upper panel) was consistent with the expression of p40 protein (Fig. 1) among the two types of clones, >20% in type I and ca. 20% or less in type II. Similar results were obtained with p24 antisense probe (data not shown). The mRNA signals were detected in the nuclei of the infected cells.
To quantify the cell population harboring the genomic RNA within cells of the two types of clones, the cells were hybridized with the p40 sense probe. Types I and II as well as the parental OL/BDV showed the genomic RNA signals localized within the nuclei of infected cells (Fig. 3B, lower panel), the site for BDV transcription and replication (10, 43). The percentage of cells harboring the genomic RNA in type I (cell clone 17) and the parental OL/BDV was consistent with that producing the p40 protein and that expressing the mRNA. On the other hand and unexpectedly, in type II, the cell population harboring the genomic RNA was two- to threefold higher (40 to 60%) than that with the corresponding protein and mRNA. However, cell clone 18 belonging to type I had a percentage of cells harboring the genomic RNA higher than that with the protein and the mRNA expression; its other characteristics were the same as those of cell clone 17.
We then tried recloning from the type II cell clone 26 that showed low expression level of the protein. One of the new clones (reclones) obtained, designated II-26/3, had a level of viral protein expression that was much lower (<2%) than that of the original clone and was used for further characterization and for confirmation of the above mentioned phenotype, i.e., a larger cell population harboring the genomic RNA than that harboring the protein and the corresponding mRNA. ISH for the detection of the mRNA and for the viral genomic RNA with p40 antisense and sense probes, respectively, was carried out. As shown in Fig. 3C, the II-26/3 clone showed an increase in the cell population harboring the viral genomic RNA over that with the protein or mRNA expression. Moreover, more cells may have been carrying the genomic RNA in very few copies that fell below the detection threshold of ISH. Meanwhile, this replication pattern for BDV points to the development of a different BDV replication cycle within the cell population in the two types of clones.
Enhancement of replication and transcription in type II cell clones.
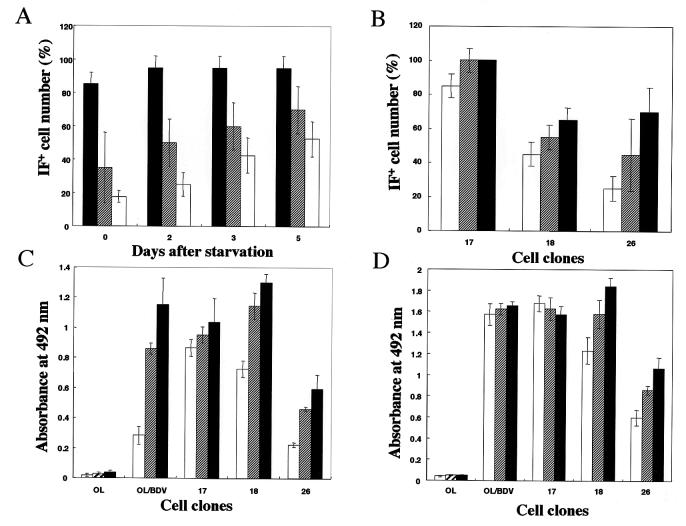
As previously reported (30), serum starvation of persistently BDV-infected cells results in cell growth arrest and enhancement of viral transcription. Thus, we tested the effects of serum starvation on the replication of BDV in OL cell clones, especially type II clones. At each time point (0, 2, 3, and 5 days) after starvation, IF assay was carried out with p40 polyclonal antibodies. The cell clones showed a gradual increase in the cell population positive for p40 expression with a very high intensity of staining within individual cells (Fig. 4A).
FIG. 4.
Enhancement of BDV replication in the cell clones by serum starvation and NGF-β treatment. (A) Representative cell clones (clones 17 and 18 in type I and clone 26 in type II) were serum starved for 0 to 5 days and examined for BDV antigen by IF with anti-p40 polyclonal antibodies. The percentages of cells positive for p40 expression by IF at 0, 2, 3, and 5 days after starvation are shown. Type I, clone 17 (▪) and clone 18 (▨); type II, clone 26 (□). (B to D) The same representative cell clones were treated with NGF-β for the indicated time points as described in Materials and Methods. At 0 days (□), 2 days (▨), and 4 days (▪) after treatment, the cells were subjected to IF assay by using polyclonal rabbit antiserum against BDV-p40 (B). The same cell cultures after treatment were fractionated into culture medium (C) and cells (D) and were separately analyzed for BDV-p40 production by antigen-capture ELISA.
We then examined whether other factors can enhance BDV replication in the clones, mainly in type II. NGF has been reported to enhance the replication of BDV in cultured cells (9). Thus, we applied NGF-β to representative cell clones of types I and II. We quantified the population of cells expressing BDV-p40 by IF assay (Fig. 4B). NGF-β treatment of the two types of clones resulted in a gradual increase in the percentage of cells expressing BDV-p40 in addition to increased intensity per cell. The increase was prominent for type II clones compared to the start point before stimulation. Further, antigen-capture ELISA confirmed the gradual increase in BDV protein production within the culture media as well as in the cells (Fig. 4C and D, respectively). However, the significant amount of p40 protein detected in the culture media of the cell clones was found, by the infectivity assay (as described in Materials and Methods), to not be due to infectious virus particle release into the media, as revealed by the complete absence of infected cells along four passages. Nevertheless, it may be due to the overgrowth of the cells under the effect of NGF and their subsequent lysis releasing their protein contents into the medium.
As shown here, there was no apparent difference between reactivation levels as determined by serum starvation and NGF-β application. However, the only difference was in the direct effect of NGF on the proliferation and cell activation where the cell growth and proliferation as well as differentiation in the cell clones were highly stimulated after NGF application, a result that was not noticed in the case of serum starvation, where cell proliferation was arrested and many cells died. This may indicate the involvement of intrinsic cellular factors for BDV activation in type II cell clones, which directed us to proceed with the following experiment.
Activation of MAP kinase in OL cell clones by NGF.
In the culture system with PC12 cells infected with herpes simplex virus, it was shown that the viral expression level was significantly amplified by NGF through the Ras/Raf signaling pathway (18). Recently, BDV was also shown to cause sustained activation of MAP kinase (ERK1/2) in the PC12 cell line (19). In addition, a very recent report showed that the Raf/MEK/ERK signaling cascade is activated upon BDV infection in several cell lines, including OL cells (37). Therefore, we next examined whether the NGF-β-enhanced replication in the cell clones may have occured due to the activation of MAP kinase. IF assay was carried out with antibodies against the activated form of MAP kinases (ERK1/2) before and after NGF-β treatment. As shown in Fig. 5, before the NGF-β was applied, types I and parental OL/BDV expressed ERK1/2, whereas type II as well as the uninfected OL cells did not. After NGF-β treatment, the two types of clones, as well as the uninfected OL cells, expressed ERK1/2 that was continuously detected in the BDV-infected cell clones with continuous application of NGF-β for the tested time points (data not shown).
FIG. 5.
Activation of ERK1/2 in OL cell clones after NGF-β treatment. Representative cell clones (clones 17 and 18 in type I and clone 26 in type II), as well as OL/BDV and OL, were cultured in eight-chamber slides. After 2 h of serum starvation, the cells were untreated (upper panels) and treated with NGF-β (lower panel) and then processed for IF assay as described in Materials and Methods.
DISCUSSION
Host cell-virus interactions have been described for many viruses. However, the consequences of such interactions vary, either resulting in host cell death or complete clearance of the virus. Other viruses adopt different mechanisms to maintain their genome within the cells by developing either a persistent or a latent infection. In the present study, we obtained cell clones (type II) in which <20% of the population positively expressed BDV-specific proteins. However, by ISH, the genomic RNA was found to be harbored in a significantly higher proportion of cells within individual cell clones. Moreover, serum starvation and NGF potently enhanced BDV replication in type II, as well as in type I. Thus, we postulate that BDV established a restricted persistent infection in type II cell clones which could represent an in vitro state of latency for BDV in OL cells. Furthermore, this latent pattern may provide some understanding about the possible host cell-virus interactions.
Host cells were found to affect the replication of measles virus during persistent infection (44). Moreover, a defective replication cycle for measles virus has been reported in astroglial cell cultures with restricted gene expression (44). Also, neuroblastoma cells have been shown to modify measles virus RNA during replication (39). For BDV, cell-specific variations in the replication pattern affecting the production level of the virus as well as that of the antigen have been reported (9). In addition, virus variants have been shown to evolute as a result of cell cloning, with <20% of the cell population positive for the protein expression (17). Thus, we can hypothesize that the clonal selection of OL cells resulted in the varied replication phenotypes of BDV, as seen in types I and II, which may be derived from selection of viral variants during cell cloning.
A similar pattern of infection with restricted expression of viral proteins, as exhibited by type II cell clones, has been described for other NNS RNA viruses such as canine distemper virus that showed restricted infection of OL cells in vitro (50) and in vivo (49). Similarly, restricted infections with measles virus (1, 20, 21, 38), Sendai virus (24, 45), and influenza C virus (28, 29) have been demonstrated in vivo as well as in vitro. To date, in vitro latent infection with BDV has not been described. However, in vivo latent infection of BDV was suggested in healthy human subjects with low antigen expression in addition to frequent reactivation in chronically ill patients (5). Latent infection for DNA viruses such as Epstein-Barr virus is associated with a downregulation of gene expression (6, 27). On the other hand, persistent infection is the most common mechanism thus far adapted by RNA viruses for their maintenance within infected cells over periods that can extend for the life of the host. In type II cell clones, the downregulation was not only in the protein production level but also in the corresponding viral transcripts compared with type I as well as with the parental clone. Thus, such downregulating factors, to be outlined in the future, might contribute to such restricted infection of BDV in OL cells.
NGF is an important factor for the growth, differentiation, and survival of neurons as well as glia. Furthermore, it was also found to be important for the pathogenesis of some viral infections. For example, NGF was found to be involved in the pathogenesis of reovirus T3C9 infection (15). In addition, reactivation of herpes simplex virus type 1 in latently infected cells was found to be mediated by NGF (22). Human immunodeficiency virus type 1 long terminal repeat was also strongly activated by NGF (40). Moreover, NGF has been shown to activate the Ras/Raf signal transduction pathways in PC12 cells that resulted in the activation of the herpes simplex virus type 1 latency-associated transcripts (18).
In the case of BDV, the viral replication is favored in the hippocampus. Further, NGF and other neurotrophic factors exist in the hippocampus at high concentrations (34), suggesting a relation between BDV replication and such neurotrophic factors. Moreover, BDV replication was enhanced in several persistently infected cell lines after application of NGF (9). This directed us to examine the effect of this neurotrophic factor on BDV replication in our cell clones. We observed that BDV replication was significantly enhanced by NGF in type II cell clones as well as in type I clones. This enhancement exerted by NGF might be due to an indirect effect(s) on viral replication through the activation of the cellular transcription machinery and the activation of the MAP kinase signaling cascade.
A variety of DNA and RNA viruses induce signaling through MAP kinase cascade in infected cells. In addition, the MAP kinase signaling cascade is implicated in the response of cells to several growth factors. Further, Hans et al. (19) reported that BDV infection constitutively activated ERK1/2 proteins in PC12 cells but blocked the neurite extension response of the PC12 cells to NGF. In our BDV-infected cell clones, the same can be applied to both parental and type I clones, where the ERK1/2 were already expressed before NGF application, and this also explains the effect of serum starvation to activate BDV expression in type I cell clones. Moreover, there was no effect for BDV on the OL cell morphology before or after NGF application (data not shown). On the contrary, type II cell clones did not follow the same assumption, where ERK1/2 was not expressed except after NGF application. The activation effect of serum starvation on type II cell clones may be due to viral intrinsic factors to survive the diverse effects of cell death.
Whether BDV or MAP kinases are the first to be triggered by NGF in type II cell clones is not yet known. Which one resulted in the activation of the other and how this bidirectional activation (BDV and MAP kinases) occurred and changed the phenotype of type II cell clones is also not yet known. However, this suggests that BDV may be able to adopt a new mechanism to persist, inactively, in infected cells.
During the preparation of this study, Planz et al. (37) reported that MEK-specific inhibitors blocked virus spread to neighboring cells. Further, it was shown that the spread of BDV infection among the cells is due to cell-to-cell transmission and not to virus released into the culture medium (12, 13), suggesting that the increase in the percentage of BDV protein-expressing cells in type II cell clones, after NGF treatment, may be due to the enhancement of cell-to-cell transmission. However, hypothesizing that activated ERK1/2, in type II cell clones, enhanced virus spread to neighboring cells might not apply, as we determined by ISH that the number of cells harboring the genomic RNA was greater than those expressing the protein. In addition, a wider cell population might have had the genomic RNA in copies that fell under the detection limit of our ISH technique. Moreover, infected but viral-protein-nonproducing cells may harbor the virus in a latent state, which upon NGF stimulation resumed activity and started replication.
Although BDV, as shown here, may produce an in vitro latent infection that can change to an active one under the influence of NGF, the in vivo interaction of BDV with NGF and the possible consequences for viral replication are still unknown.
Viral persistence or latency is a mechanism adapted by the virus for the maintenance of the viral genome in infected cells. In dividing cells, there must be a mechanism for maintaining the viral genome so that it is not cleared by continuous cell division, as in retroviruses where the viral genome is integrated into the host chromosome as a provirus. Although it is unknown how BDV can maintain its genomic RNA with no or only low levels of expression in dividing cultured cells, the in vivo model with cell-free virus from type II cell clones may contribute to the understanding of the pathogenesis of this pattern of BDV persistence or even latency in the nondividing neuronal cells in vivo. In addition, the low level of BDV-p40, the target for the T-cell-mediated immune response, can allow viral escape from immune recognition with consequent persistence of the virus and the possibility of establishing an in vivo latent infection. Thus, BDV may be regarded as not always establishing a persistent infection with high levels of viral expression but may under certain conditions establish a latent infection with low levels of viral expression. Since BDV is one of the slow viruses, studying the factors controlling its gene expression in cells of neuronal origin will be important in understanding the biology of persistent or latent BVD infection in the CNS.
Acknowledgments
We thank J. C. de la Torre, The Scripps Research Institute, La Jolla, Calif., for providing C6/BV and Hanns Ludwig, Free University of Berlin, for providing the OL cell line.
This work was supported by a Grant-in-Aid for BDV Research from the Ministry of Health and Welfare; Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture; and a Special Coordination Fund for Science and Technology from the Science and Technology Agency of Japan.
REFERENCES
- 1.Baczko, K., U. G. Liebert, R. Cattaneo, M. A. Billeter, R. P. Roos, and V. ter Meulen. 1988. Restriction of measles virus gene expression in measles inclusion body encephalitis. J. Infect. Dis. 158:144-150. [DOI] [PubMed] [Google Scholar]
- 2.Bautista, J. R., S. A. Rubin, T. H. Moran, G. J. Schwartz, and K. M. Carbone. 1995. Developmental injury to the cerebellum following perinatal Borna disease virus infection. Brain Res. Dev. Brain Res. 90:45-53. [DOI] [PubMed] [Google Scholar]
- 3.Bautista, J. R., G. J. Schwartz, J. C. de la Torre, T. H. Moran, and K. M. Carbone. 1994. Early and persistent abnormalities in rats with neonatally acquired Borna disease virus infection. Brain Res. Bull. 34:31-40. [DOI] [PubMed] [Google Scholar]
- 4.Bode, L., R. Ferszt, and G. Czech. 1993. Borna disease virus infection and affective disorders in man. Arch. Virol. Suppl. 7:159-167. [DOI] [PubMed] [Google Scholar]
- 5.Bode, L., S. Riegel, W. Lange, and H. Ludwig. 1992. Human infections with Borna disease virus: seroprevalence in patients with chronic diseases and healthy individuals. J. Med. Virol. 36:309-315. [DOI] [PubMed] [Google Scholar]
- 6.Bogedain, C., P. Alliger, F. Schwarzmann, M. Marschall, H. Wolf, and W. Jilg. 1994. Different activation of Epstein-Barr virus immediate-early and early genes in Burkitt lymphoma cells and lymphoblastoid cell lines. J. Virol. 68:1200-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briese, T., A. Schneemann, A. J. Lewis, Y. S. Park, S. Kim, H. Ludwig, and W. I. Lipkin. 1994. Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. USA 91:4362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone, K. M., S. W. Park, S. A. Rubin, R. W. Waltrip II, and G. B. Vogelsang. 1991. Borna disease: association with a maturation defect in the cellular immune response. J. Virol. 65:6154-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbone, K. M., S. A. Rubin, A. M. Sierra-Honigmann, and H. M. Lederman. 1993. Characterization of a glial cell line persistently infected with Borna disease virus (BDV): influence of neurotrophic factors on BDV protein and RNA expression. J. Virol. 67:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cubitt, B., and J. C. de la Torre. 1994. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J. Virol. 68:1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cubitt, B., C. Oldstone, and J. C. de la Torre. 1994. Sequence and genome organization of Borna disease virus. J. Virol. 68:1382-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danner, K., K. Luthgen, M. Herlyn, and A. Mayr. 1978. Comparative studies on the demonstration and formation of serum antibodies against the Borna virus. Zentbl. Veterinarmed. B 25:345-355. [PubMed] [Google Scholar]
- 13.Danner, K., and A. Mayr. 1979. In vitro studies on Borna virus. II. Properties of the virus. Arch. Virol. 61:261-271. [DOI] [PubMed] [Google Scholar]
- 14.de la Torre, J. C. 1994. Molecular biology of Borna disease virus: prototype of a new group of animal viruses. J. Virol. 68:7669-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien, M., and B. N. Fields. 2000. Anti-interleukin-3 and anti-nerve growth factor increase neonatal mice survival to reovirus type 3 clone 9 per oral challenge. J. Neuroimmunol. 110:209-213. [DOI] [PubMed] [Google Scholar]
- 16.Dittrich, W., L. Bode, H. Ludwig, M. Kao, and K. Schneider. 1989. Learning deficiencies in Borna disease virus-infected but clinically healthy rats. Biol. Psychiatry 26:818-828. [DOI] [PubMed] [Google Scholar]
- 17.Formella, S., C. Jehle, C. Sauder, P. Staeheli, and M. Schwemmle. 2000. Sequence variability of Borna disease virus: resistance to superinfection may contribute to high genome stability in persistently infected cells. J. Virol. 74:7878-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazier, D. P., D. Cox, E. M. Godshalk, and P. A. Schaffer. 1996. The herpes simplex virus type I latency-associated transcript promoter is activated through Ras and Raf by nerve growth factor and sodium butyrate in PC12 cells. J. Virol. 70:7424-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hans, A., S. Syan, C. Crosio, P. Sassone-Corsi, M. Brahic, and D. Gonzalez-Dunia. 2001. Borna disease virus persistent infection activates mitogen-activated protein kinase and blocks neuronal differentiation of PC12 cells. J. Biol. Chem. 276:7258-7265. [DOI] [PubMed] [Google Scholar]
- 20.Isaacson, S. H., D. M. Asher, M. S. Godec, C. J. Gibbs, Jr., and D. C. Gajdusek. 1996. Widespread, restricted low-level measles virus infection of brain in a case of subacute sclerosing panencephalitis. Acta Neuropathol. 91:135-139. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, S., and H. F. Macfarland. 1982. Measles virus persistence in human lymphocytes: a role for virus-induced interferon. J. Gen. Virol. 63:351-357. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, S. 1996. Effects of nerve growth factor and phorbol derivative on reactivation of herpes simplex virus type 1 in cultured cells of latently infected adult mouse trigeminal ganglia. Microbiol. Immunol. 40:645-650. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, T., W. Kamitani, Q. Zhang, M. Watanabe, K. Tomonaga, and K. Ikuta. 2001. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J. Virol. 75:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch, E. M., W. J. Neubert, and P. H. Hofschneider. 1984. Lifelong persistence of paramyxovirus Sendai-6/94 in C129 mice: detection of a latent viral RNA by hybridization with a cloned genomic cDNA probe. Virology 15:78-88. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, H., L. Bode, and G. Gosztonyi. 1988. Borna disease: a persistent virus infection of the central nervous system. Prog. Med. Virol. 35:107-151. [PubMed] [Google Scholar]
- 26.Lundgren, A. L., G. Czech, L. Bode, and H. Ludwig. 1993. Natural Borna disease in domestic animals other than horses and sheep. Zentbl. Veterinarmed. B 40:298-303. [DOI] [PubMed] [Google Scholar]
- 27.Marschall, M., F. Schwarzmann, U. Leser, B. Oker, P. Alliger, H. Mairhofer, and H. Wolf. 1991. The BI'LF4 trans-activator of Epstein-Barr virus is modulated by type and differentiation of the host cell. Virology 181:172-179. [DOI] [PubMed] [Google Scholar]
- 28.Marschall, M., C. Boswald, A. Schuler, E. Youzbashi, and H. Meier-Ewert. 1993. Productive and non-productive phases during long-term persistence of influenza C virus. J. Gen. Virol. 74:2019-2023. [DOI] [PubMed] [Google Scholar]
- 29.Marschall, M., A. Schuler, and H. Meier-Ewert. 1996. Influenza C virus RNA is uniquely stabilized in a steady state during primary and secondary persistent infections. J. Gen. Virol. 77:681-686. [DOI] [PubMed] [Google Scholar]
- 30.Mizutani, T., H. Inagaki, D. Hayasaka, H. Kariwa, and I. Takashima. 1999. Enhancement of Borna disease virus transcription in persistently infected cells by serum starvation. J. Vet. Med. Sci. 61:831-834. [DOI] [PubMed] [Google Scholar]
- 31.Muller, C. F., R. S. Fatzer, K. Beck, M. Vandevelde, and A. Zurbriggen. 1995. Studies on canine distemper virus persistence in the central nervous system. Acta Neuropathol. 89:438-445. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura, Y., H. Takahashi, Y. Shoya, T. Nakaya, M. Watanabe, K. Tomonaga, K. Iwahashi, K. Ameno, N. Momiyama, H. Taniyama, T. Sata, T. Kurata, J. C. de la Torre, and K. Ikuta. 2000. Isolation of Borna disease virus from human brain tissue. J. Virol. 74:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayan, O., S. Herzog, K. Frese, H. Scheefers, and R. Rott. 1983. Behavioral disease in rats caused by immunopathological responses to persistent Borna virus in the brain. Science 220:1401-1403. [DOI] [PubMed] [Google Scholar]
- 34.Nieto-Sampedro, M., and P. Bovolenta. 1990. Growth factors and growth factor receptors in the hippocampus. Role in plasticity and response to injury. Prog. Brain Res. 83:341-355. [DOI] [PubMed] [Google Scholar]
- 35.Notter, M. F., J. T. Hansen, S. Okawara, and D. M. Gash. 1989. Rodent and primate adrenal medullary cells in vitro: phenotypic plasticity in response to coculture with C6 glioma cells or NGF. Exp. Brain Res. 76:38-46. [DOI] [PubMed] [Google Scholar]
- 36.Parra, B., D. R. Hinton, N. W. Marten, C. C. Bergmann, M. T. Lin, C. S. Yang, and S. A. Stohlman. 1999. IFN-γ is required for viral clearance from central nervous system oligodendroglia. J. Immunol. 162:1641-1647. [PubMed] [Google Scholar]
- 37.Planz, O., S. Pleschka, and S. Ludwig. 2001. MEK-specific inhibitor U0126 blocks spread of Borna disease virus in cultured cells. J. Virol. 75:4871-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rapp, F., and S. J. Robbins. 1981. Evolution of a human cell line persistently infected with measles virus. Intervirology 16:149-159. [DOI] [PubMed] [Google Scholar]
- 39.Rataul, S. M., A. Hirano, and T. C. Wong. 1992. Irreversible modification of measles virus RNA in vitro by nuclear RNA-unwinding activity in human neuroblastoma cells. J. Virol. 66:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recio, J. A., and A. Aranda. 1997. Activation of the HIV-1 long terminal repeat by nerve growth factor. J. Biol. Chem. 272:26807-26810. [DOI] [PubMed] [Google Scholar]
- 41.Rott, R., and H. Becht. 1995. Natural and experimental Borna disease in animals. Curr. Top. Microbiol. Immunol. 190:17-30. [DOI] [PubMed] [Google Scholar]
- 42.Schneemann, A., P. A. Schneider, R. A. Lamb, and W. I. Lipkin. 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology 210:1-8. [DOI] [PubMed] [Google Scholar]
- 43.Schneider, P. A., A. Schneemann, and W. I. Lipkin. 1994. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J. Virol. 68:5007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider-Schaulies, S., U. G. Liebert, K. Baczko, and V. ter Meulen. 1990. Restricted expression of measles virus in primary rat astroglial cells. Virology 177:802-806. [DOI] [PubMed] [Google Scholar]
- 45.Shimokata, K., Y. Nishiyama, Y. Ito, Y. Kimuta, and I. Nagata. 1976. Pathogenesis of Sendai virus infection in the central nervous system of mice. Infect. Immun. 13:1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, M., T. Kobayashi, K. Tomonaga, and K. Ikuta. 2000. Antibodies to Borna disease virus in infected adult rats: an early appearance of anti-p10 antibody and recognition of novel virus-specific proteins in infected animal brain cells. J. Vet. Med. Sci. 62:775-778. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe, M., B.-J. Lee, W. Kamitani, T. Kobayashi, H. Taniyama, K. Tomonaga, and K. Ikuta. 2001. Neurological diseases and viral dynamics in the brains of neonatally Borna disease virus-infected gerbils. Virology 282:65-76. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, M., Q. Zhong, T. Kobayashi, W. Kamitani, K. Tomonaga, and K. Ikuta. 2000. Molecular ratio between Borna disease viral-p40 and -p24 proteins in infected cells determined by quantitative antigen-capture ELISA. Microbiol. Immunol. 44:765-772. [DOI] [PubMed] [Google Scholar]
- 49.Zurbriggen, A., I. Schmid, H. U. Graber, and M. Vandevelde. 1998. Oligodendroglial pathology in canine distemper. Acta Neuropathol. 95:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zurbriggen, A., M. Yamawaki, and M. Vandevelde. 1993. Restricted canine distemper virus infection of oligodendrocytes. Lab. Investig. 68:277-284. [PubMed] [Google Scholar]