Abstract
Herpes simplex virus (HSV) is characterized by its ability to establish a latent infection in sensory neurons, from which it can periodically reactivate. The mechanisms of latency, however, remain unclear. The HSV genome is quiescent during latency except for the expression of the latency-associated transcripts (LATs). Although the exact function of the LATs remains obscure, current evidence suggests they are multifunctional and are involved in both establishment of latency and reactivation from latency. The LATs contain several open reading frames (ORFs). One or more of the functions of the LATs could therefore be protein mediated. We have previously reported that deregulated expression of the largest of the HSV type 1 (HSV-1) LAT ORFs (∼274 amino acids) greatly enhances virus growth in cell types that are normally relatively nonpermissive for HSV replication and also that it complements mutations to the immediate-early (IE) gene ICP0 (S. K. Thomas, G. Gough, D. S. Latchman, and R. S. Coffin, J. Virol. 73:6618-6625, 1999). Here we show that LAT ORF expression overcomes the repression of expression from exogenous promoters introduced into the HSV-1 genome which normally occurs in the absence of IE gene expression. To further explore LAT ORF function, we have generated an epitope-tagged LAT ORF, LATmycHis, which forms punctate structures in the infected-cell nucleus reminiscent of the structures formed by ICP0. These are associated with the appearance of a phosphorylated form of the protein and are formed adjacent to, or around the edges of, viral replication compartments. These results provide further evidence that the HSV-1 LAT ORF protein is biologically functional and that the tightly regulated expression of this protein may be important in the wild-type latency phenotype in vivo.
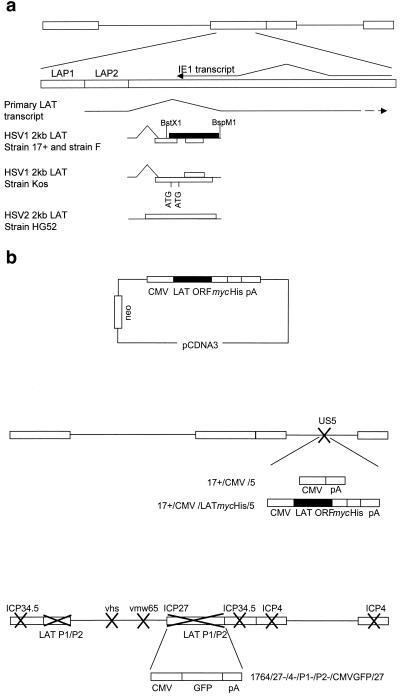
As is characteristic of all members of the family Herpesviridae, herpes simplex virus types 1 and 2 (HSV-1 and -2) establish lifelong latent infections in specific cell types in the infected host. Following an initial productive infection, HSV-1 and -2 enter and latently infect the sensory ganglia that innervate the infection site, from which they can then periodically reactivate. During latency the only abundant viral RNAs are the latency-associated transcripts (LATs) (56) (Fig. 1a), although very low levels of lytic gene RNAs have been detected by sensitive reverse transcription-PCR techniques (33). The ∼8.3-kb polyadenylated primary LAT, which is transcribed from the LAP1 promoter within the long repeat regions of the genome (56), is spliced to yield an abundant nuclear 2.0-kb LAT, which is further spliced to produce a smaller, 1.5-kb transcript in neurons (22, 55). The 2.0-kb LAT is thought to be an intron and as such is not capped or polyadenylated (20, 69). Recent work has shown that the 2.0-kb LAT is in a lariat structure (51, 67). While the function of the LATs has remained elusive, all strains of HSV-1 and -2 sequenced to date contain several open reading frames (ORFs) within the 2.0-kb LAT which may contribute to LAT function (8, 12, 65).
FIG. 1.
The HSV genome, the LAT region, and constructs used in this study. (a) The LATs are transcribed from the long terminal repeats of the genome (larger rectangles), resulting in production of an 8.3-kb primary transcript that is spliced to release smaller LATs, which are abundant during latency. The IE1 transcript and the positions of the LAP1 and LAP2 promoters are shown. Open boxes, potential ORFs within the HSV-1 and HSV-2 2.0-kb LAT; solid box, the largest ORF, which is the focus of this study. (b) Constructs and viruses used. Further details are given in Materials and Methods.
The molecular mechanisms of the establishment of HSV latency and the reactivation of HSV from latency are not well understood, including the role of the LATs. HSV-1 LAT-negative mutants have a reduced potential to establish latency (47, 54, 60, 62) and reactivate from latency (2, 3, 26, 45, 54, 60, 63), and a region within the first 1.5 kb of the 8.3-kb primary transcript, which has been partially localized to the first 811 nucleotides, has been shown to be essential for spontaneous reactivation in a particular rabbit model (13, 46). Various mechanisms have been suggested to explain these observations, including the possibility that the LATs reduce the expression of ICP0 during latency by antisense effects (21, 41, 55) or that they act by keeping the genome in a transcriptionally competent state from which reactivation is possible. However, while this could be relevant to the phenotype of LAT-negative viruses, it is not known how mutations in the first 1.5 kb of the primary LAT, which would not be anticipated to affect 2.0-kb LAT production or the level of ICP0 antisense RNA, exert their phenotype. This is particularly the case because it has been shown that inserting the deleted 1.5-kb sequence elsewhere in the genome restores the wild-type reactivation phenotype (46). More recently, it has been suggested that this region within the primary LAT provides an antiapoptotic function both in vitro and in vivo, which may contribute to neuron survival during establishment of and/or reactivation from latency (28, 48, 62), although others have questioned the interpretation of these data (61).
The potential role of a functional LAT-encoded protein during HSV latency, first suggested by Wilcox and coworkers (11), has largely been discounted, not least due to the lack of a consistent phenotype when the LAT ORFs are mutated (19, 21) and the fact that no LAT-encoded proteins have been reliably detected in latently infected neurons (11, 38). However, experimentally induced HSV-1 reactivation in animal models may not be representative of spontaneous reactivation in humans, which may account for the lack of identification of a phenotype associated with LAT ORF mutation so far. The HSV-1 2.0-kb LAT contains three potential ORFs that are highly conserved among strains of HSV-1 (see Fig. 1) (8). The largest of these ORFs is positioned in the 1.5-kb neuronal LAT such that its translation might be expected if the RNA were not retained in the nucleus, as is usually the case (8). However, since the 2.0-kb LAT is normally retained within the nucleus as a lariat (51, 67), translation of even this ORF would not normally be expected. This suggests that if 2-kb LAT-encoded ORFs are translated during the HSV life cycle, translation is likely to occur in a tightly regulated and probably transient fashion, which may explain the difficulty in detecting the expression of such proteins so far. Recently it has been reported that introduction directly into the 2.0-kb LAT coding sequence of the gene encoding green fluorescent protein (GFP) did not result in reporter gene expression in either neuronal or nonneuronal cells in culture, or in acutely or latently infected trigeminal ganglia in the mouse model (37), suggesting that LAT ORFs are not expressed during the HSV life cycle. However, in view of the likely tightly regulated nature of LAT ORF expression if LAT ORFs are expressed, such a conclusion would seem premature. Moreover, because the GFP gene was not inserted in frame with the major LAT ORF, and because it was preceded by out-of-frame ATG sequences in the 2.0- or 1.5-kb LAT, expression would not have been expected in any case.
We have previously reported that deregulated expression of the largest HSV-1 2.0-kb LAT-encoded ORF (∼274 amino acids) (referred to below as the LAT ORF) greatly enhances HSV-1 replication in both epithelial and neuronal cells, most obviously in cells which are normally nonpermissive for virus growth (57). The effects observed were shown to be mediated at the protein level, rather than as RNA or DNA. Furthermore, LAT ORF expression could fully complement for mutations in ICP0 or vmw65, which otherwise significantly reduce virus growth. Interestingly, the effects of LAT ORF expression were specific to HSV-1, as evidenced by the fact that expression of the HSV-1 LAT ORF had no effect on the growth of HSV-2. Taken together, these data suggested a role for the LAT ORF in virus reactivation as the LAT ORF has not reliably been found to be expressed in lytic infection or latently infected ganglia and as it can functionally complement for ICP0, regulated or transient expression during latency might be anticipated to result in reactivation of the virus, since ICP0 is known to be important for this process (24).
Here we report that the LAT ORF, like ICP0 (27, 53), can prevent the silencing of exogenous promoters inserted into the HSV-1 genome in the absence of immediate-early (IE) gene expression, again demonstrating that the LAT ORF protein can perform functions similar to those of ICP0. In order to aid the study of the mechanism by which the LAT ORF functions, we have constructed a biologically active epitope-tagged version of the LAT ORF protein which has allowed us to localize the LAT ORF protein in the cell by immunological techniques. This has shown the LAT ORF protein to be localized to distinct punctate structures in the infected-cell nucleus which are associated with a phosphorylated form of the protein that appears at this time. These punctate structures, which are reminiscent of structures formed by ICP0 in HSV-infected cells, occur adjacent to viral replication compartments. The relevance of these observations in view of the phenotype associated with the expression of the LAT ORF protein is discussed.
MATERIALS AND METHODS
Plasmid and virus construction.
The LAT ORF was subcloned from a 3.5-kb NotI HSV-1 genomic fragment (nucleotides 120412 to 121285) by using BstXI and BspMI (Fig. 1a) and was inserted into pSP72 (Promega) at the HindIII and EcoRI sites to yield pSP72LAT. In order to delete the stop codon for generation of a fusion protein, a 275-bp fragment (from the NarI site to the last base before the stop codon, nucleotides 120963 to 121238) was generated by PCR using primers 5′ TGCCGAGACCGCAGGCTGCG 3′ and 5′ CCCTGTCGGAATTCGTGAACAAGAC 3′ and was then used to replace the 3′ section of the LAT ORF sequence in pSP72LAT to yield pSP72LATstop. The modified LAT ORFstop was then subcloned into pVP22mycHis (Invitrogen) to ensure that the epitope tags were in frame with the LAT ORF coding sequence. This was confirmed by sequencing. The VP22 coding sequence was then excised from pVP22LATmycHis to yield pcDNA3LATmycHis (Fig. 1b). Viruses 17+/CMV/5 and 17+/CMV/LATmycHis were produced by inserting an Nru/PvuII fragment from pCDNA3 (containing the cytomegalovirus [CMV] promoter and poly(A) sequences; Invitrogen) or pcDNA3LATmycHis (containing the CMV promoter, LATmycHis, and poly(A) sequences) into a shuttle vector containing the US5 coding sequences at nucleotide 137945 to yield pCMV/5 and pCMV/LATmycHis/5, respectively. pCMV/5 and pCMV/LATmycHis/5 were then recombined into the HSV-1 strain 17+ genome by standard techniques (59) to yield 17+/CMV/5 and 17+/CMV/LATmycHis/5, respectively (Fig. 1b). Virus 1764/27-/4-/P2-/pR20.5/vhs/LAT was produced by insertion of the LAT ORF in place of the lacZ gene into the pR20.5/vhs shuttle vector (35) to yield pR20.5/vhs/LAT, which was recombined into an HSV-1 1764-/27-/4-/P2- (35) genome (Fig. 1b). HSV-1 1764/27-/4-/P1-/P2-/CMVGFP/27 is like 1764/27-/4-/P2- (described in reference 35) with the additional deletion of LAP1 sequences (nucleotides 6151 to 8366 and 113273 to 120470) and the insertion of a CMV GFP cassette so as to replace ICP27 and one LAT region between nucleotides 113273 and 120470. The DNA structures of all the viruses were confirmed by Southern blot analysis.
Cell culture and viruses.
Vero cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, and BHK and ND7 cells were cultured as previously described (57). The ND7 and BHK cell lines expressing the LAT ORF and LATmut (a mutant LAT ORF in which a frameshift mutation has been inserted at amino acid 163) have been described previously (57); LAT ORF RNA expression was confirmed by Northern blotting using RNA extracted by standard methods (59). Cell lines expressing VP22mycHis and LATmycHis were produced by using standard calcium phosphate transfection of ND7 cells with pVP22mycHis or pcDNA3LATmycHis, followed by selection of clones with 800 μg of neomycin/ml. Primary adult rat dorsal root ganglion (DRG) cultures were prepared from Lewis rats as previously described (10) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% horse serum. The medium was changed every third day, and infections were carried out after 5 to 7 days in culture. All disabled viruses were propagated on BHK-derived 27/12/M:4 complementing cell lines as previously described (58). Growth curves were performed as previously described (57).
Western blot analysis.
Samples were prepared from cells infected with the appropriate virus at a multiplicity of infection (MOI) of 5, run on a discontinuous sodium dodecyl sulfate-polyacrylamide gel (34) (15% resolving gel), and then transferred to a nitrocellulose membrane (Hybond C; Amersham) (25). Extracts from 2 × 105 cells per lane were used. Equal loading of samples was confirmed by Coomassie staining of duplicate gels. The LATmycHis protein was detected by probing membranes with an antibody to the myc tag (Invitrogen). Detection was performed with the enhanced chemiluminescence (ECL) system (Amersham).
Indirect immunofluorescence and antibodies.
Cells were grown on glass coverslips in 24-well dishes. At the times indicated, the cells were washed twice with phosphate-buffered saline (PBS), then fixed either with ice-cold ethanol (for the anti-ICP0 antibody; 4°C for 20 min) or with formaldehyde (for the anti-tubulin, anti-promyelocytic leukemia protein [anti-PML], anti-cyclin D3, and anti-ICP8 antibodies; 1% formaldehyde in PBS, room temperature for 10 min; 1% Triton X-100 in PBS, 4°C for 20 min), and again washed twice with PBS. Both fixation methods are suitable for use with the anti-myc antibody. Cells were then blocked with 10% fetal calf serum at room temperature for 15 min, incubated with the appropriate primary antibodies for 60 min at 37°C, washed twice with PBS, incubated with the appropriate secondary antibody for 60 min at 37°C, and then washed twice again with PBS. Coverslips were mounted by using the ProLong Antifade kit (Molecular Probes, Eugene, Oreg.) to reduce photobleaching. All images were obtained using a Zeiss Axiophot2 microscope and the software supplied. The anti-myc monoclonal antibody (MAb) (immunoglobulin G1 [IgG1]) (Invitrogen) was used at a 1:500 dilution. The anti-neuron-specific tubulin (β111) MAb TUJI (IgG2a) (Covance) was used at a 1:500 dilution. The anti-ICP0 (exon 2) rabbit polyclonal antibody, a gift from Bernard Roizman (University of Chicago) (32), was used at a 1:200 dilution. The anti-PML rabbit polyclonal antibody (Chemicon) was used at a 1:200 dilution. The anti-cyclin D3 rabbit polyclonal antibody (Santa Cruz) was used at a 1:50 dilution. The anti-ICP8 rabbit polyclonal antibody 383, a gift from David Knipe (Harvard Medical School, Boston, Mass.) (9), was used at a 1:200 dilution. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and rhodamine (tetramethyl rhodamine isocyanate [TRITC])-conjugated swine anti-rabbit antibodies (DAKO) were used at a 1:200 dilution. TRITC-conjugated goat anti-mouse IgG2a and FITC-conjugated goat anti-mouse IgG1 (Southern Biotechnology Associates) were used at a 1:100 dilution.
PNGase F and λ-PPase treatment of cell lysates.
Samples were prepared from Vero cells infected at an MOI of 5. For peptide-N-glycosidase F (PNGase F) treatment, infected-cell monolayers were washed twice with PBS, harvested into the denaturing buffer supplied (New England Biolabs) at a concentration of 2.5 × 107 cells/ml, and incubated at 95°C for 10 min. Aliquots (20 μl) were treated with 1,000 U of PNGase F (New England Biolabs) for 1 h at 37°C according to the manufacturer's instructions and then analyzed by Western blotting. For lambda phosphatase (λ-PPase) treatment, cells were washed twice with PBS and then harvested into 100 mM Tris HCl (pH 7.5) at a concentration of 2.5 × 107 cells/ml. The cells were disrupted by three freeze-thaw cycles followed by homogenization. Aliquots (20 μl) were treated with 800 U of λ-PPase (New England Biolabs) for 2 h at 30°C according to the manufacturer's instructions and then analyzed by Western blotting.
RESULTS
LAT ORF expression prevents repression of exogenous promoters in the HSV-1 genome in the absence of IE gene expression.
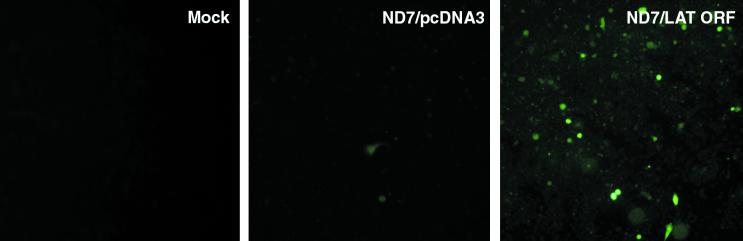
It has previously been shown that in the absence of IE gene expression, both HSV promoters and exogenous promoters inserted into the HSV-1 genome are repressed immediately after infection, even though the viral genome persists in a potentially functional state within the nuclei of infected cells (49, 53). In the absence of ICP4, the activity of these exogenous promoters has been found to be dependent on the expression of ICP0 (27, 53). In order to examine whether the LAT ORF could affect the activity of exogenous promoters in such viruses similarly to ICP0, ND7 cells stably transfected with pcDNA3 or with pcDNA3/LAT ORF were infected at an MOI of 5 with a nonreplicating virus with ICP4 and ICP27 deleted, with an inactivating mutation in vmw65, and containing GFP under the control of a CMV promoter (1764/27-/4-/P1-/P2-/CMVGFP/27) (Fig. 1b). This virus behaves similarly in cultured cells to the previously reported virus constructed by Samaniego et al. in which all IE genes are deleted (53) such that the CMV promoter is repressed immediately after infection of noncomplementing cells. Infected cells were examined 24 h postinfection (p.i.) by fluorescence microscopy. Infection of control ND7 cells resulted in GFP expression in <0.1% of the cells (Fig. 2), as we have previously found with viruses of this type (35; unpublished data). Superinfection of these cells with a less disabled virus which expressed ICP0 but no marker gene (virus strain 17+/27-, with only ICP27 deleted [35]) demonstrated that >50% of the cells contained viral genomes, as GFP was then expressed in >50% of the cells (data not shown). Thus, as previously found, the CMV promoter is repressed in these cells in the absence of IE gene expression, which can then be activated by infection with a virus expressing ICP0. However, when the LAT ORF-containing cells were examined 24 h after infection with the 1764/27-/4-/P1-/P2-/CMVGFP/27 virus, ∼10% of the cells expressed GFP (i.e., a 100-fold increase in GFP-positive cell numbers over those in control cells not expressing the LAT ORF) (Fig. 2), demonstrating that the LAT ORF can prevent the repression of exogenous promoters in the HSV genome which otherwise occurs in the absence of ICP4, ICP0, and vmw65. The difference between the numbers of GFP-positive cells in the presence of the LAT ORF and with 17+/27- superinfection in this experiment may be due to only low-level LAT ORF expression at the time of infection, as constitutive expression of LAT ORF RNA in these cells has previously been shown to be low (57). However, this finding does demonstrate that the LAT ORF can function similarly to ICP0 in overcoming the repression of gene expression that occurs with HSV when IE genes are not expressed.
FIG. 2.
Deregulated expression of the LAT ORF prevents repression of exogenous gene promoters inserted into the HSV-1 genome in the absence of IE gene expression. ND7 cells stably transfected with pcDNA3 and ND7 cells constitutively expressing the wild-type LAT ORF were infected (at an MOI of 5) with a nonreplicating virus deficient in the IE genes ICP4 and ICP27 and in the virion transactivator vmw65 and containing GFP under the control of a CMV promoter (1764/27-/4-/P1-/P2-/CMVGFP/27). Cells were examined by fluorescence microscopy 24 h p.i. Mock-infected ND7 cells stably transfected with pcDNA3 were used as a negative control.
Construction of cell lines and viruses expressing a biologically active epitope-tagged LAT ORF.
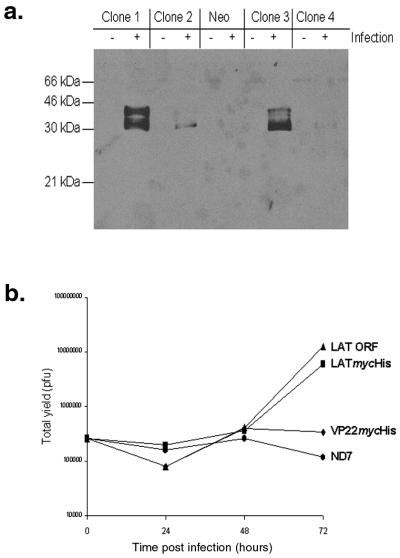
In order to examine the intracellular localization of the LAT ORF protein, and any potential interactions with viral and/or cellular factors that may be involved in LAT ORF function, we generated a LAT ORF mycHis epitope-tagged fusion protein (LATmycHis). Generation of such a protein allowed easy detection by Western blotting and immunofluorescence microscopy using a commercially available anti-myc antibody. Initially the LATmycHis protein under the control of a CMV promoter was expressed in stably transfected ND7 cell lines. Control stably transfected ND7 cell lines expressing VP22mycHis were also generated. Clones expressing either LATmycHis or VP22 mycHis were initially identified by Western blot analysis of wild-type virus (17+)-infected lysates. A representative screen of LATmycHis-expressing clones is shown in Fig. 3a. LATmycHis expression levels in these cell lines were found to be very low in the absence of virus infection. However, following wild-type HSV-1 infection, LATmycHis expression from the CMV promoter was increased such that it was easily detected, as we have previously found to be the case for proteins expressed from some other CMV promoter-containing stable cell lines (58). Infection with a replication-deficient mutant such as 1764/27-/4-/P2-/20.5/vhs did not, however, result in up-regulation of LATmycHis expression (data not shown), most likely due to lack of ICP0 expression.
FIG. 3.
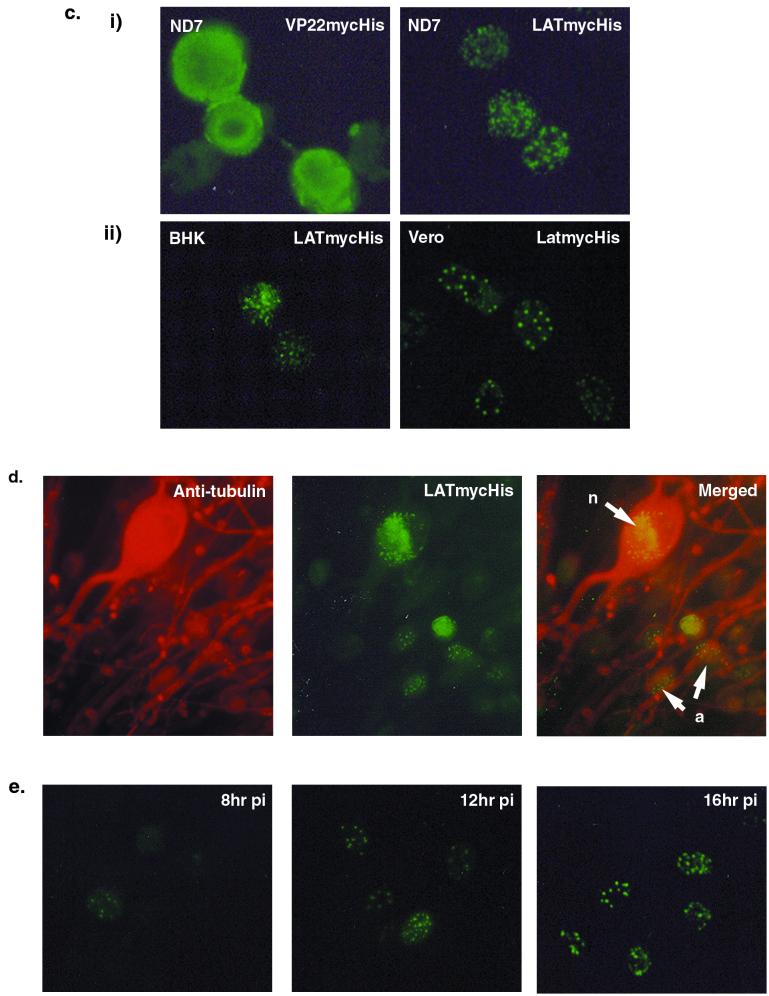
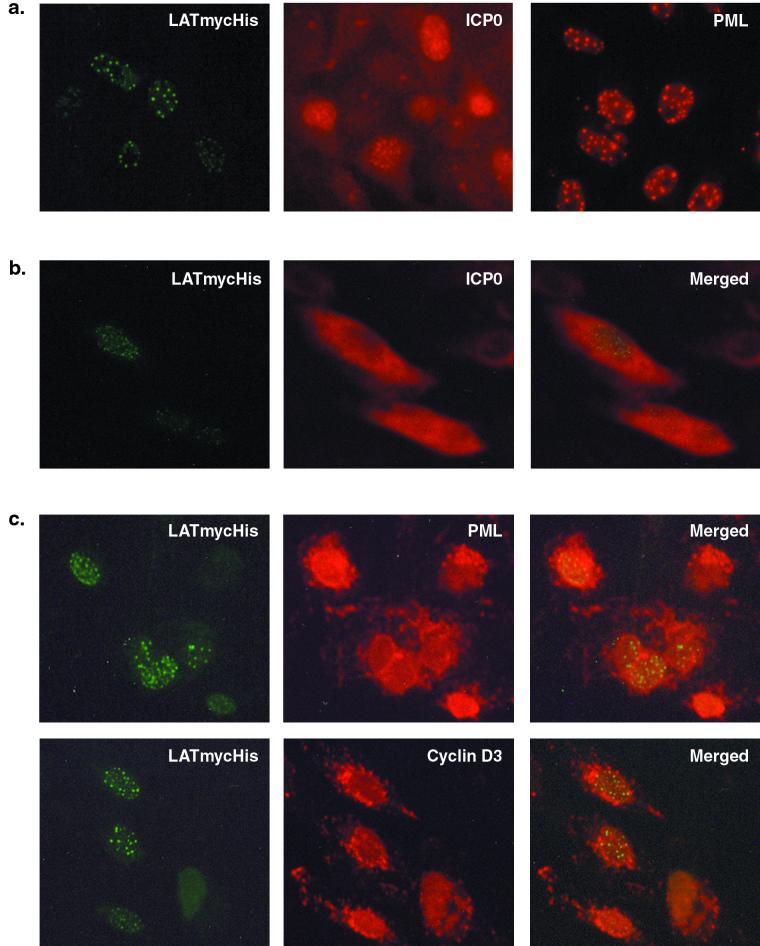
An epitope-tagged LAT ORF, LATmycHis, is biologically active and forms punctate structures in the nuclei of infected cells. (a) Western blot screening of infected and uninfected (with 17+ at an MOI of 5) stably transfected ND7 cell lines expressing LATmycHis probed with an anti-myc MAb. (b) Growth curves using wild-type (17+) virus performed on pools of six ND7 cell lines expressing either LAT ORF, LATmycHis, or VP22myc together with parental ND7 cells. (c) ND7 cell lines expressing either LATmycHis or VP22mycHis infected with wild type (17+) HSV-1 (at an MOI of 0.1), and BHK and Vero cells infected with a wild-type virus constitutively expressing LATmycHis (17+/CMV/LATmycHis/5), were stained with an anti-myc MAb at16 h p.i. The secondary antibody used was an FITC-conjugated goat anti-mouse IgG. (d) Primary adult rat DRG cells infected with 17+/CMV/LATmycHis/5 at 16 h p.i. Cells are dually labeled with an anti-myc MAb (IgG1) and an anti-neuron-specific tubulin MAb (IgG2a). The secondary antibodies used were FITC-conjugated goat anti-mouse IgG1 and TRITC-conjugated goat anti-mouse IgG2a. Punctate structures are formed both in neurons (n) and accessory cells (a). (e) Kinetics of punctate structure formation in Vero cells infected with 17+/CMV/LATmycHis (at an MOI of 0.1) and stained with an anti-myc MAb and an FITC-conjugated goat anti-mouse IgG.
In order to confirm that the LATmycHis protein was biologically active, growth curves using wild-type (17+) virus were performed on pools of six ND7 cell lines expressing either the LAT ORF, LATmycHis, or VP22myc together with parental ND7 cells (Fig. 3b). These demonstrated that LATmycHis enhanced HSV-1 growth in the normally semipermissive ND7 cells to a level similar to that with the wild-type LAT ORF (∼50- to 100-fold). In contrast, the constitutive expression of VP22mycHis in the cells did not affect virus growth.
Nondisabled viruses constitutively expressing LATmycHis were then also constructed by inserting the fusion protein-encoding gene under the control of the CMV promoter together with bovine growth hormone (BGH) poly(A) into the US5 gene of HSV-1 strain 17+ (to produce virus strain 17+/CMV/LATmycHis/5) (Fig. 1b). Gene insertions into US5 have previously been shown not to affect virus growth in vitro, the virulence of the virus, or the latency phenotype (1). LATmycHis expression from these viruses was confirmed by Western blot analysis (see Fig. 6). Control viruses in which the CMV promoter and BGH poly(A) were inserted into US5 without the LATmycHis ORF were also generated.
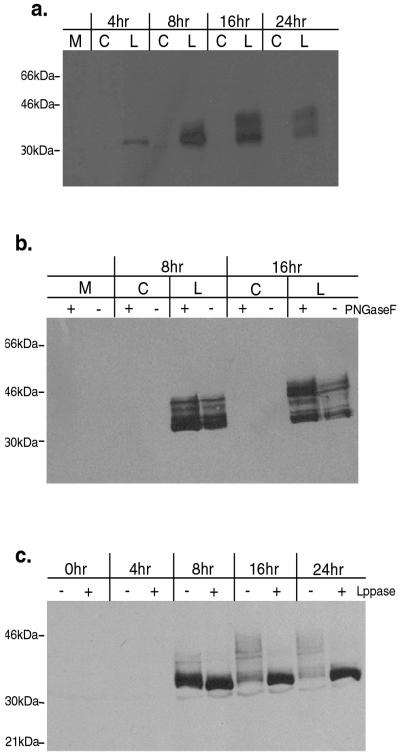
FIG. 6.
The LATmycHis protein is modified by phosphorylation 8 to 16 h p.i. (a) Kinetics of modification. Extracts were prepared from Vero cells infected with either 17+/CMV/LATmycHis/5 or the control virus 17+/CMV/5 (at an MOI of 5) at the times indicated, and expression of the LATmycHis protein was analyzed by probing a Western blot with an anti-myc antibody. (b) The LATmycHis protein is not modified by N-linked glycosylation. Extracts were prepared from Vero cells infected with either 17+/CMV/LATmycHis/5 or the control virus 17+/CMV/5 at the times indicated, treated with PNGase F according to the manufacturer's instructions (see Materials and Methods), and analyzed by Western blotting as before. (c) The LATmycHis protein is modified by phosphorylation. Extracts were prepared from Vero cells infected with 17+/CMV/LATmycHis/5 at the times indicated, treated with λ-PPase according to the manufacturer's instructions (see Materials and Methods), and analyzed by Western blotting as before.
The LATmycHis protein forms punctate structures in the nuclei of infected cells.
In order to determine the intracellular localization of the LATmycHis protein, ND7 cell lines expressing either LATmycHis or VP22mycHis were infected with wild-type HSV-1 (at an MOI of 0.1) to induce expression and fixed at 16 h p.i., and LATmycHis expression was visualized by immunofluorescence microscopy using an anti-myc antibody (Fig. 3c). A low MOI was used for the immunohistochemical studies in order to ensure that there were noninfected cells within each field of view to act as internal negative controls. The LATmycHis protein was shown to form punctate structures within the nuclei of infected cells. In contrast, VP22mycHis, which was used as a control, was distributed throughout the cell (Fig. 3c).
We next examined the intracellular localization of LATmycHis in permissive BHK and Vero cells as well as in nonpermissive ND7 cells following infection with the recombinant virus 17+/CMV/LATmycHis/5 (which constitutively expresses the LATmycHis protein). In all cases LATmycHis formed punctate structures in the nuclei of infected cells at 8 to 10 h p.i. Further analysis of these cells showed that the punctate structures formed by LATmycHis were first visible at 8 to 10 h p.i. and then increased in intensity up to 16 h p.i. (Fig. 3e). We also used this virus to examine the intracellular localization of LATmycHis in primary adult rat DRG neurons. In this case dual-labeling immunofluorescence microscopy using anti-myc together with an anti-neuron-specific tubulin antibody was used to demonstrate that the protein forms punctate structures within infected neurons, as well as in accessory cells (Fig. 3d).
LATmycHis punctate structures do not colocalize with ICP0, PML, or cyclin D3 in the nuclei of infected cells.
As discussed previously, the LAT ORF protein is able to complement mutations in ICP0 during lytic infection and to prevent the repression of exogenous promoters inserted into the HSV-1 genome that occurs in the absence of IE gene expression. We noted that the punctate structures formed in the cells by the LATmycHis protein bore a striking resemblance to those formed by ICP0 during the early stages of HSV-1 infection (42) (Fig. 4a). However, the previously published kinetics of the formation of ICP0 punctate structures are different from those we have found here with the LATmycHis protein. Thus, following infection, ICP0 is rapidly expressed and transported to the nucleus, giving a punctate appearance. At later times during infection (5 to 9 h p.i.), ICP0 is translocated back to the cytoplasm (32). Therefore, at 8 to 10 h p.i., the time when the LATmycHis punctate structures are first visible, ICP0 is mainly located in the cytoplasm. Unsurprisingly, therefore, we do not observe colocalization of LATmycHis and ICP0. This can be clearly seen following dual-labeling immunofluorescence microscopy to visualize LATmycHis and ICP0 in infected Vero cells at 8 h p.i. (Fig. 4b).
FIG. 4.
(a) LATmycHis, ICP0, and ND10 structures show similar intracellular localization. (Left) Vero cells infected with 17+/CMV/LATmycHis/5 (at an MOI of 0.1) and stained with anti-myc at 16 h p.i. (Center) Vero cells infected with 17+ and stained with an anti-ICP0 MAb at 5 h p.i. (Right) ND10 structures in uninfected Vero cells stained with a polyclonal anti-PML antibody. (b) LATmycHis structures do not colocalize with ICP0 in the nuclei of infected cells. Vero cells infected with 17+/CMV/LATmycHis (at an MOI of 0.1) were fixed at 8 h p.i. and stained with an anti-myc MAb and polyclonal anti-ICP0 antibodies. (c) LATmycHis structures do not colocalize with the ND10 protein PML or with cyclin D3. Vero cells infected with 17+/CMV/LATmycHis (at an MOI of 0.1) were fixed at 8 h p.i. and stained with an anti-myc MAb and a polyclonal anti-PML or anti-cyclin D3 antibody. Secondary antibodies used were FITC-conjugated goat anti-mouse IgG and TRITC-conjugated swine anti-rabbit IgG.
We next examined whether the LATmycHis protein colocalizes with cellular proteins that are known to interact with ICP0. Several cellular proteins, both nuclear and cytoplasmic, have been shown to interact with ICP0, including forms of PML and Sp100 modified by the ubiquitin-like protein SUMO-1 (or PIC1) (7, 16), a ubiquitin-specific protease (USP7, formerly known as HAUSP) (15), cellular DNA-dependent protein kinase (DNA-PK) (44), centromeric protein C (CENP-C) (17), cyclin D3 (32, 64), and elongation factor 1δ (31). The most extensively studied interaction is that of ICP0 with small punctate nuclear substructures known as ND10 or PML bodies (42). The exact function of these structures is unclear; one possibility is that they represent nuclear protein accumulations and function as nuclear deposits of particular proteins until they are required (42). The two best-characterized ND10 components, PML and Sp100, are both modified by covalent linkage to the small ubiquitin-like protein SUMO-1 (reviewed in reference 52). ICP0 has been shown to induce the rapid proteasome-dependent degradation of the modified forms of both PML and Sp100, leading to the destruction of ND10s, and colocalizes with PML at early times after infection (7, 16). Again, there is a striking resemblance between the distribution of the LATmycHis structures in infected cell nuclei and the distribution of ND10s in uninfected cells as visualized by immunofluorescence microscopy using an anti-PML antibody (average number of structures, 11.36 ± 4.09 and 12.48 ± 5.52, respectively) (Fig. 4a). However, LATmycHis structures form at later times after infection, after degradation of the SUMO-1-modified forms of PML has occurred and when PML is found in the cytoplasm. This can be seen clearly in dual-labeling immunofluorescence microscopy used to visualize the LATmycHis protein and PML in Vero cells at 8 h p.i. (Fig. 4c). The same result is obtained if ND10 structures are visualized by immunofluorescence using an anti-Sp100 antibody (data not shown). The LATmycHis structures therefore do not colocalize with ND10 structures as defined by visualization of PML and Sp100, although it is possible that LATmycHis is interacting with other components of the ND10 structures that are not targeted for degradation following interaction with ICP0. It is also possible that if LATmycHis were expressed at a time when PML was still retained in the nucleus, colocalization might be observed. All these immunohistochemical studies (and those described below) were carried out in Vero cells because the anti-PML and anti-Sp100 antibodies do not recognize the rodent forms of these proteins, as would be necessary for work with BHK, ND7, or primary DRG cultures.
ICP0 has also been shown to colocalize with cyclin D3 in structures similar to ND10s at early times in infection, leading to the stabilization of cyclin D3 and its translocation to ND10s (32, 64). We therefore used dual-labeling immunofluorescence microscopy to visualize the intracellular localization of LATmycHis and cyclin D3 in Vero cells at 8 h p.i. (Fig. 4c). However, since cyclin D3 is also translocated from the nucleus to the cytoplasm at later times after infection, the LATmycHis structures, unsurprisingly, could not be colocalized with cyclin D3, because the LATmycHis structures are not formed until after cyclin D3 translocation to the cytoplasm has occurred.
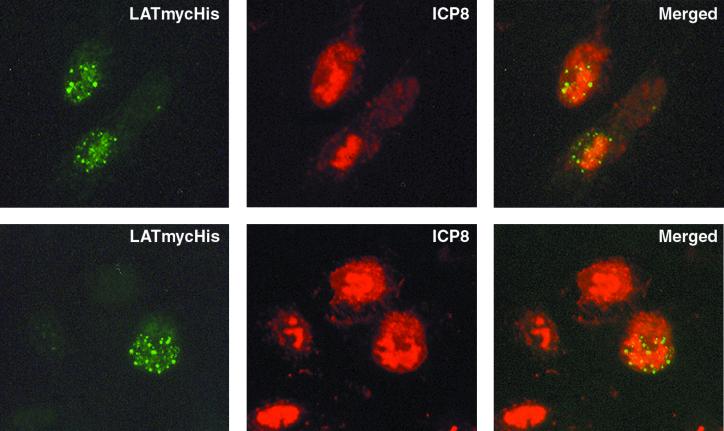
LATmycHis punctate structures localize adjacent to viral replication compartments.
The kinetics of LATmycHis punctate structure formation are similar to those of the formation of viral DNA replication compartments. These were originally defined by immunofluorescence visualization of the single-stranded DNA-binding protein ICP8 (50), and they also contain six other viral replication proteins, the viral DNA polymerase (UL30 and UL42) (4, 23), the UL9 origin-binding protein (43), and the helicase-primase complex proteins UL5, UL8, and UL52 (36, 39, 40, 43). In order to determine whether the LATmycHis structures colocalized with the replication compartments, we used dual-labeling immunofluorescence microscopy with anti-myc and anti-ICP8 antibodies to examine Vero cells infected with 17+/CMV/LATmycHis/5 virus (at an MOI of 0.5) at 10 h p.i. (Fig. 5). This demonstrated that in many cells the LATmycHis punctate structures form adjacent to or around the edges of early viral replication compartments in a striking and reproducible fashion. Interestingly, ND10 or sites close to ND10 have been proposed to play an important role in the establishment of productive infection, since viral replication compartments have been shown to form at sites adjacent to ND10 (29, 43). It is therefore possible that the LATmycHis punctate structures may be formed at the same subnuclear sites that were previously occupied by ND10s, even though PML and Sp100 have already been translocated to the cytoplasm following degradation by ICP0 by the time these structures are visible.
FIG. 5.
LATmycHis punctate structures form adjacent to early viral replication compartments. Vero cells infected with 17+/CMV/LATmycHis/5 (at an MOI of 0.1) were fixed 9 h p.i. and stained with an anti-myc MAb and polyclonal anti-ICP8. Secondary antibodies used were FITC-conjugated goat anti-mouse IgG and TRITC-conjugated swine anti-rabbit IgG.
The LATmycHis protein is modified by phosphorylation 8 to 16 h p.i.
In order to further explore the kinetics of punctate structure formation and the relationship of punctate structure formation to LATmycHis expression per se, Western blot analysis was performed following infection of Vero cells with 17+/CMV/LATmycHis/5. We were particularly interested in examining the levels of protein expression between 8 h p.i., when the LATmycHis structures were first visible, and 16 h p.i., when the structures appeared to be fully formed. Infected-cell lysates were prepared from Vero cells infected with either 17+/CMV/LATmycHis/5 or the control virus 17+/CMV/5 (at an MOI of 5) at 4, 8, 16, and 24 h p.i. and were analyzed by probing a Western blot with an anti-myc antibody (Fig. 6a). This revealed that a single LATmycHis protein species of the expected size could be detected at 4 h p.i. and that expression increased by 8 h p.i. After this time, modified forms of the protein with higher molecular sizes (∼34 to 45 kDa) were detectable, and the lower-molecular-size, ∼32-kDa form was less abundant. The kinetics of this modification suggested that the punctate structures might in fact be composed of these larger forms of the protein, since these are the most abundant LATmycHis protein forms at this time. A similar pattern of LATmycHis protein modification was also seen following infection of ND7 and BHK cells (data not shown).
In order to determine the nature of this modification, which was suggestive of glycosylation or phosphorylation, infected-cell lysates were prepared from Vero cells infected with 17+/CMV/LATmycHis at 4, 8, 16, and 24 h p.i. These lysates were then treated with either PNGase F, which removes N-linked oligosaccharides from glycoproteins, or λ-PPase, which removes phosphate groups from serine, threonine, and tyrosine residues, and were analyzed by Western blotting. PNGase treatment of infected lysates had no effect on the molecular weights of the larger LATmycHis forms, indicating that the protein is not modified by N-linked glycosylation (Fig. 6b). In contrast, treatment with λ-PPase resulted in a single protein species of ∼32 kDa, indicating that LATmycHis is multiply phosphorylated 8 to 16 h p.i. (Fig. 6c.) Interestingly, the kinetics of this modification correspond to the kinetics of punctate structure formation in infected cells. It would thus appear likely that it is a phosphorylated form of the LATmycHis protein that forms the punctate structures.
DISCUSSION
The molecular mechanisms of the establishment of HSV latency and reactivation from latency are not well understood. It is clear from the available data that the LATs play a role in these processes, although to date their exact role remains obscure. There are strong indications that the LATs have a role in both establishment of latency (47, 54, 60) and reactivation from latency (2, 3, 26, 45, 60, 62, 63). They may also prevent apoptosis or otherwise aid survival in latently infected neurons (28, 48, 62). Multiple mechanisms may well contribute to these LAT-encoded functions, including an antisense effect on ICP0. The LATs encode a number of putative ORFs, and so one or more functions of the LATs may be mediated through encoded proteins. However, the potential role of a functional LAT-encoded protein during HSV latency has largely been discounted, because mutations within the LAT ORFs do not affect the latency phenotype in animal models (19, 21), and no LAT-encoded proteins have been reliably detected in latently infected neurons (11, 38). Due to the structure of the LAT RNA, it is unlikely that these ORFs could be constitutively expressed throughout latency; instead, it would seem likely that if they are expressed, their expression is tightly regulated. We have previously reported that deregulated expression of the largest HSV-1 LAT-encoded ORF (referred to here as the LAT ORF) can greatly enhance virus growth both in vitro and in vivo and can compensate for mutations of ICP0 during lytic replication (57). This enhancement in virus growth is particularly evident in cells such as the DRG neuron-neuroblastoma fusion cell line ND7, which is otherwise relatively nonpermissive for HSV-1 replication. This fact suggested that the LAT ORF protein might have a mechanism of action similar to that of ICP0. It also suggested that the LAT ORF protein might be able to substitute for a lack of ICP0 early in HSV-1 reactivation. Thus, if the LAT ORF protein was transiently expressed during latency in response to currently unknown stimuli, it might aid in the periodic reactivation of HSV which occurs in infected individuals by substituting for the usual lack of ICP0 at this time. Interestingly, the HSV-1 LAT ORF has no effect on the growth of HSV-2 (57). It is notable that while the HSV-1 and HSV-2 genomes are colinear and the vast majority of the protein-coding sequences are very similar, HSV-1 and -2 generally have different sites of latency and reactivation (trigeminal and sacral dorsal root ganglia, respectively) (66), and each reactivates from latency much less efficiently from the other virus's natural site (68). The HSV-2 2.0-kb LAT also contains an ORF potentially encoding a protein of significant size (419 amino acids) (8), but this has no sequence similarities with any of the LAT ORFs of HSV-1. It is interesting to speculate, therefore, that if the HSV LAT ORFs are important for the wild-type reactivation phenotype in individuals infected with HSV-1 or -2, differences in the sequences of these ORFs may facilitate reactivation from the different sites from which this occurs.
Here we report work aimed at beginning to understand the mechanisms by which the HSV-1 LAT ORF might function, and particularly how it can compensate for mutations of ICP0. ICP0 is an important regulator of viral gene expression (5). HSV-1 mutants deficient in ICP0 initiate lytic replication inefficiently at low MOIs and reactivate poorly in mouse models (6). Similarly, in the absence of IE gene expression, both HSV and exogenous promoters inserted into the genome are repressed soon after infection, and the genome remains in a quiescent state in the nuclei of infected cells (49, 53). ICP0 expression is sufficient to initiate gene expression from these persistent, quiescent genomes, suggesting that ICP0 can counteract the repression mechanism (53). Although the activation of gene expression by ICP0 has been shown to occur at the level of mRNA synthesis (30), ICP0 does not bind directly to DNA (14). However, it is known that ICP0 controls the stability of a number of cellular proteins during the early stages of viral replication, including SUMO-1 modified forms of PML and Sp100, which are components of nuclear substructures known as ND10 or PML nuclear bodies (42), DNA-PK (44), CENP-C (17), and cyclin D3 (32, 64). This and other activities of ICP0 are thought to result in the alteration of the intracellular environment such that virus-directed gene expression is favored (reviewed in reference 18).
In view of what is known about the function of ICP0, we set out to determine whether the LAT ORF protein could also overcome cell-mediated repression of exogenous promoters in the HSV genome, which it proved able to do.
The mechanism by which the LAT ORF is able to enhance virus replication and overcome the repression of exogenous promoters in the HSV genome is unclear. ICP0 is known to affect the stability of several cellular proteins by targeting them for proteasome-dependent degradation (18, 32, 42, 44). In order to test whether the LAT ORF is able to interact with these proteins similarly to ICP0, we generated a biologically active epitope-tagged LAT ORF, termed LATmycHis. This protein formed punctate structures in the nuclei of infected cells which are similar in number and distribution to ND10 structures (visualized by using anti-PML antibodies in noninfected nuclei) and also to the structures formed by ICP0 at early times p.i. However, the LATmycHis punctate structures formed at later times after infection (8 to 10 h p.i.), after PML and ICP0 had been translocated to the cytoplasm. The LATmycHis protein therefore did not colocalize with these proteins or with cyclin D3, which is also known to interact with ICP0 (32, 64). ND10 or sites close to ND10 have been proposed to play an important role in the establishment of productive infection, since viral replication compartments have been shown to form at sites adjacent to ND10 (29, 39). Interestingly, LATmycHis punctate structures were shown to localize adjacent to early viral replication compartments, visualized by using an anti-ICP8 antibody. It is therefore possible that the LATmycHis punctate structures may form at the same subnuclear sites that were previously occupied by ND10s, even though PML and Sp100 have already been translocated to the cytoplasm by the time these structures are visible. The formation of the LATmycHis punctate structures was also shown to correlate with the appearance of phosphorylated forms of the protein. Since the LAT ORF can compensate for ICP0 deficiencies, it might be expected that the protein would be active at early times during infection. The LATmycHis punctate structures may in fact, therefore, be inactivated protein that has then been translocated to nuclear deposit structures similar to ND10 following phosphorylation. The failure of the LAT ORF protein to colocalize with either ICP0, PML, or cyclin D3 could indicate that the LAT ORF protein acts by a mechanism different from that of ICP0, even though it can perform at least some similar functions in the infected cell. This is further indicated by the fact that the HSV-1 LAT ORF had no effect on the growth of HSV-2, which would not be expected if the LAT ORF functioned as a generalized transcription activator similar to ICP0. We are currently conducting further dual-labeling immunofluorescence analysis in order to further elucidate the nature of the LATmycHis protein structures. We are also employing coimmunoprecipitation and yeast-two-hybrid analysis to identify potential interactions with cellular proteins in order to gain an understanding of the mechanism by which the LAT ORF protein acts.
Acknowledgments
We are grateful to Bernard Roizman (University of Chicago) and David Knipe (Harvard Medical School) for providing anti-ICP0 and anti-ICP8 antibodies, respectively, and to James Palmer for help with preparation of primary DRG cultures.
This work was funded by GlaxoSmithKline and the Medical Research Council.
REFERENCES
- 1.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245-1258. [DOI] [PubMed] [Google Scholar]
- 2.Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618-630. [DOI] [PubMed] [Google Scholar]
- 3.Bloom, D. C., J. M. Hill, G. Devi-Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, M., D. R. Yager, M. Gao, K. Weisshardt, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 65:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelbi-Alix, M. K., and H. de Thé. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 8.Coffin, R. S., S. K. Thomas, and D. S. Latchman. 1998. The herpes simplex virus 2-kb latency associated transcript leader sequence allows efficient expression of downstream proteins which is enhanced in neuronal cells: possible functions of LAT ORFs. J. Gen. Virol. 79:3019-3026. [DOI] [PubMed] [Google Scholar]
- 9.Curtin, K. D., and D. M. Knipe. 1993. Altered properties of the herpes simplex virus ICP8 DNA-binding protein in cells infected with ICP27 mutant viruses. Virology 196:1-14. [DOI] [PubMed] [Google Scholar]
- 10.De Felipe, C., and S. P. Hunt. 1994. The differential control of c-jun expression in regenerating sensory neurons and their associated glial cells. J. Neurosci. 14:2911-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doerig, C., L. I. Pizer, and C. I. Wilcox. 1991. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J. Virol. 65:2724-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drolet, B. S., G.-C. Perng, R. J. Villosis, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1999. Expression of the first 811 nucleotides of the herpes simplex virus type 1 latency-associated transcript (LAT) partially restores wild-type spontaneous reactivation to a LAT-null mutant. Virology 253:96-106. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 19:6155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D. 1999. A surprising role for the proteasome in the regulation of herpesvirus infection. Trends Biochem. Sci. 8:293-295. [DOI] [PubMed] [Google Scholar]
- 19.Fareed, M. U., and J. G. Spivack. 1994. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J. Virol. 68:8071-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell, M. J., J. M. Hill, T. P. Margolis, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1993. The herpes simplex virus type 1 reactivation function lies outside the latency-associated transcript open reading frame ORF-2. J. Virol. 67:3653-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich, L. D., P. A. Schaffer, D. I. Dorsky, C. S. Crumpacker, and P. S. Parris. 1990. Localization of the herpes simplex virus type 1 65-kilodalton DNA-binding protein and DNA polymerase in the presence and absence of viral DNA synthesis. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 27.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inman, M., G. C. Perng, G. Henderson, H. Ghiasi, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as a site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan, R., and P. A. Schaffer. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaguchi, Y., B. Renato, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translation machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, M. F., and D. M. Coen. 1995. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 69:1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Lilley, C. E., F. Groutsi, Z. Han, J. A. Palmer, P. N. Anderson, D. S. Latchman, and R. S. Coffin. 2001. Multiple immediate-early gene-deficient herpes simplex virus vectors allowing efficient gene delivery to neurons in culture and widespread gene delivery to the central nervous system. J. Virol. 75:4343-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liptak, L., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lock, M., C. Miller, and N. W. Fraser. 2001. Analysis of protein expression from within the region encoding the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1. J. Virol. 75:3413-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luganoff, M., and B. Roizman. 1994. Expression of a herpes simplex virus type 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J. Virol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukonis, C. J., and S. K. Weller. 1996. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mador, N., D. Goldenberg, O. Cohen, A. Panet, and I. Steiner. 1998. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J. Virol. 72:5067-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maul, G. G., A. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 43.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1989. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J. Virol. 63:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perng, G.-C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perng, G.-C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J. Virol. 70:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perng, G.-C., S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J. Virol. 74:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perng, G.-C., C. Jones, J. Ciacci-Zarella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 49.Preston, C. M., and C. M. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 51.Rodahl, E., and L. Haar. 1997. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable nonlinear structure. J. Virol. 71:1703-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitoh, H., R. T. Pu, and M. Dasso. 1997. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22:374-376. [DOI] [PubMed] [Google Scholar]
- 53.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66:2157-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spivack, J. G., G. M. Woods, and N. W. Fraser. 1991. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency associated transcript (LAT): translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J. Virol. 65:6800-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens, J. G., E. K. Wagner, G. B. Devi-Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science 235:1056-1059. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, S. K., G. Gough, D. S. Latchman, and R. S. Coffin. 1999. Herpes simplex virus encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from latency. J. Virol. 73:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas, S. K., C. E. Lilley, D. S. Latchamn, and R. S. Coffin. 1999. Equine herpesvirus 1 gene 12 can substitute for vwm65 in the growth of herpes simplex virus (HSV) type 1, allowing the generation of optimized cell lines for the propagation of HSV vectors with multiple immediate-early gene defects. J. Virol. 73:7399-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas, S. K., and D. S. Latchman. 1999. Analysis of transcriptional control in DNA virus infections, p. 113-156. In A. J. Cann (ed.), DNA viruses: a practical approach. IRL Press, Oxford, England.
- 60.Thompson, R. L., and N. M. Sawtell. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J. Virol. 71:5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, R. L., and N. M. Sawtell. 2000. HSV latency-associated transcript and neuronal apoptosis. Science 289:1651-1652. [PubMed] [Google Scholar]
- 62.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trousdale, M. D., I. Steiner, J. G. Spivack, S. L. Deshmane, S. M. Brown, A. R. Maclean, J. H. Subak-Sharpe, and N. W. Fraser. 1991. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J. Virol. 65:6989-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus type 1 ICP0. J. Virol. 75:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wechsler, S. L., A. B. Nesburn, J. Zwaagstra, and H. Ghiasi. 1989. Sequence of the latency related gene of herpes simplex virus type 1. Virology 168:168-172. [DOI] [PubMed] [Google Scholar]
- 66.Whitely, R. J. 1996. Herpes simplex viruses, p. 2297-2342. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 67.Wu, T.-T., Y.-H. Su, T. M. Block, and J. M. Taylor. 1996. Evidence that two latency-associated transcripts of herpes simplex virus are nonlinear. J. Virol. 70:5962-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshikawa, T., J. M. Hill, L. R. Stanberry, N. Bourne, J. F. Kurawadwala, and P. R. Krause. 1996. The characteristic site-specific reactivation phenotypes of HSV-1 and HSV-2 depend upon the latency-associated transcript region. J. Exp. Med. 184:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zabolotny, J. M., C. Krummenacher, and N. W. Fraser. 1997. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J. Virol. 71:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]