Abstract
Human polyomavirus JC virus (JCV) is a causative agent of progressive multifocal leukoencephalopathy which results from lytic infection of glial cells. Although significant progress has been made in understanding the regulation of JCV gene transcription, the mechanism(s) underlying the viral lytic cycle remains largely unknown. We recently reported that the JCV late auxiliary Agnoprotein may have a regulatory role in JCV gene transcription and replication. Here, we investigated its regulatory function in viral gene transcription through its physical and functional interaction with YB-1, a cellular transcription factor which contributes to JCV gene expression in glial cells. Time course studies revealed that Agnoprotein is first detected at day 3 postinfection and that its level increased during the late stage of the infection cycle. Agnoprotein is mainly localized to the cytoplasmic compartment of the infected cell, with high concentrations found in the perinuclear region. While the position of Agnoprotein throughout the infection cycle remained relatively unaltered, the subcellular distribution of YB-1 between the cytoplasm and nucleus changed. Results from coimmunoprecipitation and glutathione S-transferase pull-down experiments revealed that Agnoprotein physically interacts with YB-1 and that the amino-terminal region of Agnoprotein, between residues 1 and 36, is critical for this association. Further investigation of this interaction by functional assays demonstrated that Agnoprotein negatively regulates YB-1-mediated gene transcription and that the region corresponding to residues 1 to 36 of Agnoprotein is important for the observed regulatory event. Taken together, these data demonstrate that the interaction of the viral late regulatory Agnoprotein and cellular Y-box binding factor YB-1 modulates transcriptional activity of JCV promoters.
The human polyomavirus JC virus (JCV) is the etiologic agent of progressive multifocal leukoencephalopathy, which is a fatal demyelinating disease of the central nervous system observed primarily in immunocompromised patients (4, 9, 25). JCV is a small DNA virus, and its genome is composed of three functional domains including viral early and late coding regions and viral noncoding regulatory elements (8). JCV is closely related to other polyomaviruses, including simian vacuolating virus 40 (SV40) and the human BK virus (BKV), all of which have significant sequence homology in their coding sequences. The JCV late genome, like those of SV40 and BKV, contains an open reading frame for a small auxiliary protein designated Agnoprotein, whose function in the JCV lytic cycle remains unknown. Previous studies on both SV40 and BKV have established that the Agnoproteins for both viruses are expressed during the late phase of infection (18, 19, 37), and mutational analysis of SV40 Agnoprotein suggested that it may have a regulatory role in the viral lytic cycle including viral assembly (19, 27, 30, 31), transcription, translation, and maturation (1, 13, 14). However, the mechanisms involved in such implicated functions are currently unknown.
Transcriptional regulation of the JCV early and late promoters during the viral lytic cycle is a complicated event that requires participation of both viral and cellular factors. During the past 10 years we and others have identified and characterized a number of cellular transcription factors which are involved in JCV gene regulation including the Y-box binding protein YB-1 (6, 21, 39, 40), NF-κB, Sp-1, Tst-1, NF-1, and Purα (2, 6, 10, 15, 33-36, 41, 43). At the initial stages of infection, only host cellular factors are responsible for expression of early genes in the absence of the viral large T antigen. Cellular transcription factor YB-1 has been shown to have the ability to transactivate viral genes driven from JCV early and late promoters (6, 21, 40). Specifically, we recently demonstrated that YB-1 functionally interacts with the viral early regulatory protein T antigen and synergistically activates transcription from the JCV late promoter (39). YB-1 is a member of a large family of Y-box DNA binding proteins (50), whose members are conserved from bacteria to higher eukaryotes. YB-1 was originally cloned by virtue of its binding to CT-rich double-stranded oligonucleotide spanning the DRA X and Y elements found within the promoters of human major histocompatibility complex class I genes (7). Y-box binding proteins consist of three domains: a variable glycine-rich N terminus, a highly conserved central nucleic acid recognition domain, and a hydrophilic C terminus tail domain. The central nucleic acid recognition domain is also known as the cold shock domain and shows 43% homology to the bacterial cold shock protein (11). The tail domain, which is thought to stabilize protein-DNA interactions, contains alternating clusters of predominantly basic or acidic amino acids. Compiled experimental evidence indicates that Y-box binding proteins are involved in a wide variety of biological functions including regulation of gene expression at both transcriptional (20, 21, 28, 29, 39, 40) and translational levels (47, 50) and DNA repair (26). The family members were shown to be induced by stress-related stimuli including UV irradiation (22), anticancer agents (16, 23, 32), hypothermia (44), and small affecter molecules such as thrombin and interleukin-2 (38, 45, 46).
Like that for all other viruses, the regulation of JCV gene expression is not governed solely by the presence or absence of a particular viral or cellular transcription factor in a given cell; rather delicate interactions among host transcription factors and/or interactions between host and viral regulatory proteins determine pathways for a successful viral life cycle. Our recent efforts concentrated on revealing the mechanisms underlying the functional role of Agnoprotein in the viral life cycle through its interactions with both viral and cellular transcription factors. In this regard, we have demonstrated that Agnoprotein can functionally interact with the viral early regulatory protein large T antigen and can play a regulatory role in T-antigen-mediated viral DNA replication and gene transcription (42). In this report, we further examined its regulatory role in JCV gene expression through its interaction with cellular transcription factor YB-1. Our results show that JCV late Agnoprotein physically and functionally interacts with YB-1 and suppresses YB-1-mediated activation of transcription from both viral promoters.
MATERIALS AND METHODS
Cell lines and viruses.
U-87MG (ATCC HTB14), a human glioblastoma cell line, was grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (penicillin and streptomycin, 100 μg/ml). SVG-A is a subclonal population of a human glial cell line which was established by transformation of human fetal glial cells with an origin-defective SV40 mutant and which has been previously described (24). It should be noted that these cells are not considered infected but rather cells transformed by SV40 large T antigen. The growth conditions for SVG-A were the same as those described for U-87MG. Both cell lines were maintained at 37°C in a humidified atmosphere with 7% CO2. It is difficult to study the life cycle of JCV in tissue culture because of its restrictive replication in only primary human fetal glial cells. Established cell line SVG-A partially alleviates this requirement and offers a convenient cell system to study the JC virus infection cycle. Since the prototype JCV Mad-1 strain does not grow well on this established cell line, a hybrid virus (Mad-1/SVEΔ) was constructed using SV40 and JCV by insertion of the 72- and 21-bp repeats of SV40 into the regulatory region of the Mad-1 strain of JCV on the late side of the 98-bp repeats. DNA sequence analysis of a selected clone of the hybrid virus grown in human fetal glial cells indicated a 294-bp deletion from the original construction. This clone (kindly provided by E. O. Major, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, Md.) (49) retained the sequences from the JCV replication origin, 78 bp of one 98-bp repeat, 33 bp of one SV40 72-bp repeat, and one intact 72-bp repeat.
Plasmid constructs.
The pBLCAT3-Mad-1L and the pBLCAT3-Mad-1E reporter constructs containing the regulatory region of JCV Mad-1 strain in the late and early orientations, respectively, have been previously described (6). Plasmid pGEX1λT-Agno (1-71), encoding a glutathione S-transferase (GST) fusion protein construct of JCV Agnoprotein, and its deletion mutants pGEX1λT-Agno (1-54), pGEX1λT-Agno (1-36), pGEX1λT-Agno (18-71), pGEX1λT-Agno (18-54), pGEX1λT-Agno (37-71), and pGEX1λT-Agno (55-71) have been previously described (42). In addition, pGEX1λT-YB-1 (1-318), pGEX1λT-YB-1 (1-203), pGEX1λT-YB-1 (1-125), pGEX1λT-YB-1 (1-75), and pGEX1λT-YB-1 (1-37), encoding GST-YB-1 fusion protein constructs, have been described previously (40). Eukaryotic gene expression plasmid for full-length YB-1 pEBV-HisA-YB-1 and its deletion mutants pcDNA3-YB-1 (126-318), pcDNA3-YB-1 (204-318), and pcDNA3-YB-1 (250-318) have been described previously (40). Furthermore, eukaryotic expression plasmid for full-length JCV Agnoprotein pEBV-HisA-Agnoprotein and its deletion mutants pEBV-HisA-Agnoprotein (1-36) and pEBV-HisA-Agnoprotein (55-71) have been previously described (42). Baculovirus expression systems for both Agnoprotein and YB-1 were created with the commercially available pFastBact HTb expression vector (Gibco BRL). Plasmid pGEX1λT-Agno (1-71) was digested with restriction enzymes BamHI and EcoRI; the Agnoprotein fragment was gel purified, and its coding sequence was subsequently subcloned in-frame into the BamHI/EcoRI sites of the pFastBact HTb expression vector. The pGEX1λT-YB-1 (1-318) plasmid was digested with BamHI and EcoRI; the YB-1 fragment was gel purified, and then its coding sequence was subcloned in frame into BamHI/EcoRI sites of the pFastBact HTb expression vector. Recombinant baculoviruses for both Agnoprotein and YB-1 were prepared according to the manufacturer's recommendation.
Transient transfection assays.
U-87MG cells were transfected by the calcium phosphate precipitation method (12) with reporter constructs alone or in combination with Agnoprotein (pEBV-His Agno) and YB-1 (pEBV-His-YB-1) expression plasmids. Plasmid concentrations used in each transfection experiment are indicated below and/or in the figure legends. The total amount of DNA transfected into the cells was normalized with the respective empty vector. A glycerol shock was applied at 4 h posttransfection, and the medium was replenished. At 48 h posttransfection, cells were lysed by freeze-thaw cycles. After clearance of cell debris, the protein concentrations of the supernatants were normalized and the chloramphenicol acetyltransferase (CAT) activity of samples was determined by utilizing 100 μg of protein for each sample. Transfections were repeated more than three times with different plasmid preparations, and standard deviations were indicated by error bars.
Expression and purification of recombinant GST fusion proteins.
Overnight cultures (150 ml) of Escherichia coli DH5α, transformed with either pGEX1λT-Agno or pGEX1λT-YB-1 or their respective deletion mutant plasmids, were diluted 1:10 in fresh Luria-Bertani broth supplemented with ampicillin (100 μg/ml). Cultures were induced with 0.4 M isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density of 0.4 at a wavelength of 595 nm and were incubated for an additional 2 h at 37°C. Cells were collected by centrifugation and resuspended in 10 ml of lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 supplemented with 1 mM phenylmethylsulfonyl fluoride, 2 mM pepstatin A, 0.6 mM leupeptin, and 2 mM benzamidine. After sonication, lysates were cleared by centrifugation at 10,000 × g and incubated with 150 μl of 50% glutathione-Sepharose beads (Pharmacia, Piscataway, N.J.) overnight at 4°C. GST fusion proteins were purified by three cycles of washing and centrifugation with 5 ml of lysis buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
GST affinity chromatography assays (GST pull-down).
For the GST pull-down assay, 2 μg of either GST alone or GST-Agnoprotein or its deletion mutants immobilized on glutathione-Sepharose beads was incubated with 0.5 mg of whole-cell extract prepared from U-87MG cells transfected with pEBV-HisA-YB-1 expression plasmid overnight at 4°C in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% Nonidet P-40 and supplemented with a cocktail of proteinase inhibitors (Sigma). Formed complexes bound to Sepharose beads were washed extensively with lysis buffer and resolved by SDS-10% PAGE, followed by Western blot analysis using an anti-T7 antibody (Invitrogen) directed against the histidine tag of YB-1. In reciprocal GST pull-down assays, 0.5 mg of whole-cell extract from U-87MG cells transfected with pEBV-His-Agnoprotein expression plasmid was incubated with either GST or GST-YB-1 (2 μg each) and bound complexes were resolved by SDS-15% PAGE and analyzed by Western blotting using an anti-T7 antibody directed against the histidine tag of Agnoprotein. Additionally, whole-cell extracts from U-87MG cells transfected with the pEBV-His-YB-1 expression plasmid were treated with ethidium bromide (100 ng/ml) or DNase I (0.2 U/μg of protein) or RNase I (0.5 U/32 μg of protein) prior to incubation with GST-Agnoprotein to examine whether the observed interaction between Agnoprotein and YB-1 is mediated by a DNA or RNA molecule.
For mapping studies, 0.3 mg of whole-cell extract from U-87MG cells transfected with pEBV-His-YB-1 expression plasmid was incubated with GST, GST-Agnoprotein, or GST-Agnoprotein amino- and carboxy-terminal deletion mutants immobilized on glutathione-Sepharose beads. Bound complexes were analyzed by Western blotting using an anti-T7 antibody for the detection of histidine-tagged YB-1. In reciprocal-mapping studies, 4 μl of 35S-labeled in vitro-translated Agnoprotein was incubated with 2 μg of GST, GST-YB-1, or GST-YB-1 amino-terminal deletion mutants. Alternatively, 4 μl of 35S-labeled in vitro-translated amino-terminal YB-1 deletion mutants was incubated with full-length GST-Agnoprotein fusion proteins immobilized on glutathione-Sepharose beads. All reactions were performed in a total reaction volume of 400 μl in lysis buffer overnight at 4°C with continuous rocking. After incubation, the beads were washed extensively with lysis buffer. In both cases, complexes were resolved by SDS-15% PAGE and the presence of Agnoprotein or YB-1 amino-terminal deletion mutants was determined by autoradiography.
In vitro transcription and translation assay.
Agnoprotein (42) and YB-1 amino-terminal deletion mutants [YB-1(126-318), YB-1(204-318), and YB-1(250-318)] (40) were radiolabeled with [35S]methionine by using a TNT coupled in vitro transcription-translation system (Promega, Madison, Wis.) according to the recommendations of the manufacturer.
Coimmunoprecipitation and Western blot analysis.
For coimmunoprecipitation studies, SF9 insect cells were coinfected with recombinant baculoviruses expressing Agnoprotein and YB-1. On the third day postinfection, cells were collected and whole-cell lysates were prepared in lysis buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and a cocktail of proteinase inhibitors. Two micrograms of anti-YB-1 antibody (a polyclonal rabbit anti-YB-1 antibody raised against a peptide of YB-1 amino acids 242 to 267) was incubated with 0.5 mg of whole-cell extract overnight at 4°C with continuous rocking. Immunocomplexes were precipitated by the addition of protein A-Sepharose beads (20 μl of 50% slurry) (Pharmacia) for an additional 2 h and washed extensively with lysis buffer. Immunocomplexes were resolved by SDS-15% PAGE and transferred onto an immunoblotting membrane for 15 min at 250 mA. The membrane was probed with an anti-Agnoprotein antibody raised against a mixture of Agnoprotein peptides (peptides spanning amino acids 1 to 36, 20 to 53, and 36 to 70), developed with an ECL detection kit (Amersham, Arlington Heights) according to the manufacturer's recommendations, and analyzed for the presence of Agnoprotein. For the analysis of Agnoprotein expression during the course of the JCV infection cycle, SVG-A cells were infected with Mad-1/SVEΔ hybrid virus at 10 PFU/cell. Infections were terminated at various time points as indicated in the respective figure legends, and whole-cell lysate for each sample was prepared, resolved by SDS-15% PAGE, blotted onto a nitrocellulose membrane (250 mA for 10 min), and analyzed by Western blotting using an anti-Agnoprotein rabbit polyclonal antibody. For detection of expressed JCV capsid proteins, whole-cell lysates prepared from infected cells at different time points were analyzed by Western blotting using a rabbit polyclonal antibody raised against the SV40 capsid protein (Lee Biomolecular Research, San Diego, Calif.), which is cross-reactive with JCV viral proteins.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence microscopy studies were conducted as previously described (3). Briefly, SVG-A cells were seeded on polylysine-coated glass chamber slides in low density. Next day, cells were washed twice with phosphate-buffered saline (PBS) and infected with Mad-1/SVEΔ virus at 10 PFU per cell. Cells were then fixed with cold acetone at various time points of infection as indicated in the respective figure legends. Fixed cells were blocked with 5% bovine serum albumin in PBS for 2 h and incubated either with preimmune rabbit polyclonal antibodies or with anti-Agnoprotein rabbit polyclonal primary antibodies for 1 h. Cells were subsequently washed three times with PBS-0.01% Tween 20 for 10-min intervals and incubated with an anti-rabbit fluorescein isothiocyanate (FITC)-conjugated goat secondary antibody for 45 min. Finally, slides were washed three times with PBS, mounted, and examined by fluorescence microscopy for expressed Agnoprotein. In parallel assays, the subcellular distribution of the endogenous YB-1 transcription factor during the course of infection was also examined by indirect immunofluorescence microscopy utilizing a primary polyclonal anti-YB-1 rabbit antibody [anti-YB-1(242-267); a kind gift from R. Kelm, Mayo Clinic/Foundation, Rochester, Minn] and an anti-rabbit FITC-conjugated secondary goat antibody.
RESULTS
Expression of Agnoprotein during infection.
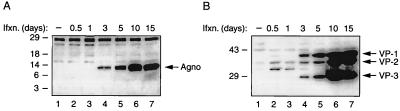
To examine the expression pattern of Agnoprotein during the course of infection, SVG-A cells were infected with JCV and, at various time points after infection, whole-cell lysates were prepared and analyzed by Western blotting. As shown in Fig. 1A, Agnoprotein is detectable as early as the third day postinfection. The levels of Agnoprotein increased in a time-dependent manner between 3 and 10 days postinfection and stabilized or slightly declined beyond 10 days after infection. Parallel examination of viral capsid proteins VP-1, VP-2, and VP-3 in infected cells revealed that the time of the initial expression of these proteins during the infection cycle coincides with that of Agnoprotein, suggesting that late transcripts of JCV are translated simultaneously to generate all late proteins. Of note, SV40 Agnoprotein and viral capsid proteins are not expressed in uninfected SVG-A cells (Fig. 1A and B, respectively, lanes 1). This eliminates the possibility that cross-reactivity of the antibodies may contribute to the experimental outcome.
FIG. 1.
Analysis of Agnoprotein expression by Western blotting during the course of infection. (A) SVG-A cells were infected with Mad-1/SVEΔ hybrid virus at multiplicity of infection of 10, and whole-cell lysates were prepared at various time points after infection (Ifxn.) as indicated. Twenty micrograms of whole-cell lysate for each time point was analyzed by Western blotting using an anti-Agnoprotein polyclonal rabbit antibody. Agno, band corresponding to the Agnoprotein. (B) Expression of viral capsid proteins. Whole-cell lysates prepared for panel A were also analyzed for the presence of viral capsid proteins by Western blotting using polyclonal rabbit antibodies raised against SV40 capsid proteins, which are cross-reactive with JCV capsid proteins. VP-1, VP-2, and VP-3 are indicated. Lanes 1, whole-cell lysate from uninfected cells loaded as a negative control.
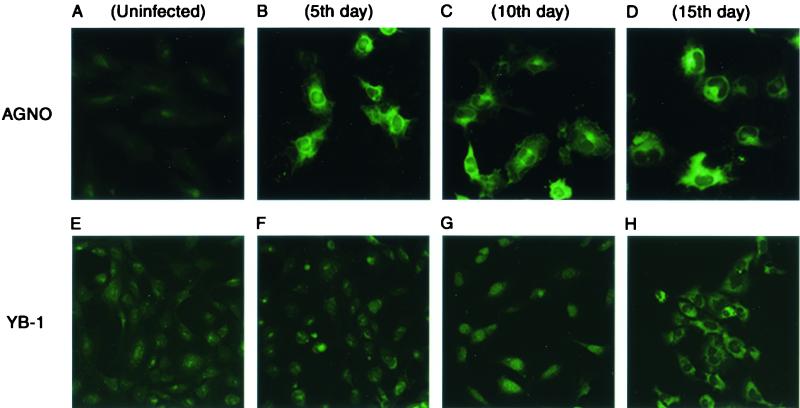
To examine the subcellular distribution of Agnoprotein during the course of infection, SVG-A cells seeded on glass chamber slides were infected with Mad-1/SVEΔ, fixed at various time points following infection, and, after incubation with primary (anti-Agnoprotein rabbit polyclonal) and secondary (anti-rabbit FITC-conjugated goat polyclonal) antibodies, were examined by fluorescence microscopy. As shown in Fig. 2B to D, Agnoprotein is mainly localized to the cytoplasmic compartment of the cell, with high concentrations found in the perinuclear area particularly at the late stages of infection. It is also apparent that Agnoprotein can be moderately distributed to the nuclear compartment. This is in agreement with previous observations in which SV40 and BKV Agnoproteins were detected in the cytoplasmic and perinuclear areas of the infected cells (5, 17, 37). To ensure specificity of the anti-Agnoprotein antibody, infected and uninfected samples were also reacted with preimmune antisera and no staining was observed (data not shown).
FIG. 2.
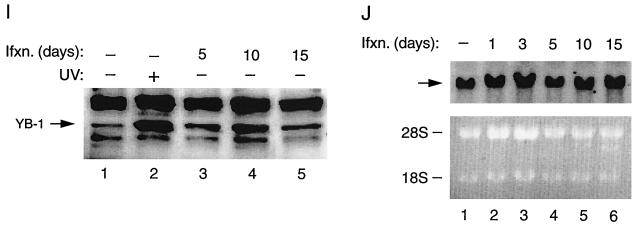
Subcellular distributions of both Agnoprotein and YB-1 during the infection cycle were examined by indirect immunofluorescence microscopy. SVG-A cells were grown on tissue culture glass slides and infected with Mad-1/SVEΔ at multiplicity of infection (MOI) of 10. Infected cells were fixed with cold acetone on days 5, 10, and 15 postinfection as indicated (B to D, Agnoprotein; F to H, YB-1). Samples were incubated with either polyclonal rabbit anti-Agnoprotein (A to D) or polyclonal rabbit anti-YB-1(242-267) (E to H) primary antibodies followed by incubation with a FITC-conjugated goat anti-rabbit secondary antibody. Samples were examined by fluorescence microscopy for the detection of Agnoprotein and YB-1. (A and E) Negative controls for Agnoprotein and YB-1, respectively. (I) Western blot analysis of nuclear YB-1 levels in infected cells. SVG-A cells were infected with hybrid virus (Mad-1/SVEΔ) at an MOI of 10. Nuclear extracts (40 μg of protein/lane) prepared on days 5, 10, and 15 postinfection were subjected to SDS-10% PAGE and analyzed by Western blotting using an anti-YB-1(242-267) antibody. Lane 1, nuclear extracts from uninfected cells loaded as a negative control; lane 2, nuclear extracts from UV-treated cells loaded as a positive control. (J) (Top) Northern blot analysis of YB-1 message in infected cells. In parallel to Western blot analysis of the nuclear proteins for YB-1, total RNA (20 μg of RNA/lane) prepared from uninfected (lane 1) and infected cells (lanes 2 to 6) was analyzed by Northern blotting for the presence of the YB-1 transcript. Arrow, hybridized YB-1 message. (Bottom) Analysis of total RNA prepared from both uninfected (lane 1) and infected cells (lanes 2 to 6) by ethidium bromide staining. Both 18S and 28S rRNA bands indicate the integrity of total RNA analyzed for the YB-1 message. Ifxn, infection.
We have previously demonstrated that cellular transcription factor YB-1 is involved in regulation of transcription from JCV gene promoters (6, 21, 40). Results from several laboratories indicated that YB-1 can be induced by a variety of stress-related stimuli, including UV irradiation (22), anticancer agents (16, 23, 32), and hypothermia (44). Viral infection can also create a stressful environment for host cells and lead to the induction of several important regulatory proteins. Examples of such an inducible protein are members of the NF-κB family of transcription factors, which, under normal conditions, are retained in the cytoplasmic compartment; upon induction by a variety of extracellular stimuli, including viral infection, they translocate to the nucleus and stimulate stress-responsive genes (48). As recent studies pointed to the stress inducibility of YB-1, we sought to determine whether viral infection plays a role in its subcellular distribution. To test this possibility, we performed indirect immunofluorescence microscopy studies on JCV-infected cells utilizing primary (anti-YB-1 polyclonal rabbit) and secondary (anti-rabbit FITC-conjugated polyclonal goat) antibodies. As shown in Fig. 2E to H, in accord with earlier data (50), endogenous YB-1 is mainly localized to the cytoplasmic compartment in uninfected cells (E). During early infection, no noticeable change in the subcellular distribution of YB-1 was observed (F). However, as the infection cycle progressed, the difference became more apparent, as YB-1 was mainly localized to the nucleus (G). Further, as the infection cycle approached its termination point, the level of YB-1 seemed to increase and its subcellular localization shifted back to the cytoplasmic compartment. In accord with previous observations, treatment of SVG-A cells with UV irradiation caused translocation of YB-1 from nuclei to cytoplasm (data not shown) (22). We repeated these experiments two additional times with similar results. Immunostaining of noninfected and infected cells with preimmune antisera (negative control for anti-YB-1 antibody) did not show any noticeable changes with respect to YB-1 induction, indicating the specificity of immunostaining (data not shown). Of note, although the subcellular distribution of YB-1 is affected by viral infection, it is not influenced by the expression of SV40 T antigen alone. We also examined YB-1 induction at the level of transcription and translation. As shown in Fig. 2I, Western blot analysis of nuclear extracts from infected (lanes 3 to 5) versus uninfected (lane 1) cells clearly indicates a noticeable increase in the level of YB-1 in infected cells, suggesting that nuclear translocation of YB-1 is induced by viral infection. Interestingly, a relatively high accumulation of YB-1 at day 10 postinfection correlates with our observations from immunostaining studies with the same data point (Fig. 2G). As expected, YB-1 levels in the nucleus are increased upon UV treatment (Fig. 2I, lane 2, positive control). In parallel, we also analyzed total RNA from the same data points by Northern blotting for the induced level of YB-1 mRNA. The data indicated that YB-1 mRNA levels remained relatively unaltered throughout the infection cycle, implying that YB-1 induction occurs at the level of translation rather than transcription. Taken together, these observations demonstrate that, although the subcellular distribution of Agnoprotein throughout the infection cycle remained relatively unaltered, YB-1 oscillates between nucleus and cytoplasm during the late phase of infection.
Functional interaction between Agnoprotein and YB-1 on the JCV promoters.
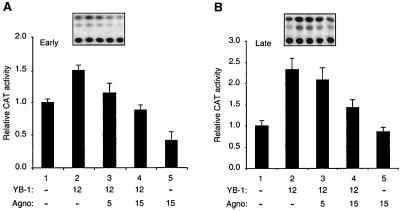
We previously reported that Agnoprotein and YB-1 negatively and positively regulate the transcriptional activity of JCV promoters, respectively (21, 39, 40, 42). In addition, the above immunofluorescence studies demonstrated that the subcellular distribution of YB-1 is modulated during the infection cycle, further suggesting the involvement of YB-1 in virus growth. Hence, we sought to determine whether Agnoprotein exhibits any influence on YB-1-mediated transcription of the viral promoters. To that end, cotransfection experiments were performed with glial cell line U-87MG by using reporter constructs containing the JCV early (Fig. 3A) and late (Fig. 3B) gene promoters alone or together with expression plasmids for Agnoprotein and YB-1. As shown in Fig. 3A, cotransfection of the reporter construct with a YB-1 expression plasmid resulted in an increase in the transcriptional activity of the viral early promoter (compare lane 2 to lane 1). Expression of Agnoprotein in the transfected cells decreased the level of viral promoter activation by YB-1 (compare lane 2 to lanes 3 and 4). In agreement with previous observations (42) overexpression of Agnoprotein alone suppressed the basal transcription of the early promoter (compare lanes 1 and 5).
FIG. 3.
Effect of Agnoprotein on YB-1-mediated activation of JCV promoters. (A and B) Agnoprotein suppresses YB-1-mediated transactivation from JCV promoters. Reporter plasmid constructs containing the JCV early (pBLCAT3-Mad-1E) and late (pBLCAT3-Mad-1L) gene promoters were transfected into U-87MG cells alone or in combination with Agnoprotein (pEBV-His-Agnoprotein) and YB-1 (pEBV-His-YB-1) expression plasmids. Expression plasmid DNA concentrations used in the transfections are indicated at the bottom (in micrograms per 60-mm-diameter plate). At 48 h posttransfection, cells were lysed by freeze-thaw cycles. After clearance of cell debris, protein concentrations of the supernatants were normalized and CAT activity of samples was determined by utilizing 100 μg of protein for each sample. The data are represented as CAT activity relative to basal-level expression of the promoter. The results of a representative CAT assay are shown at the top. All results are means of three independent experiments; bars, standard deviations.
Similar results were observed when the effect of Agnoprotein and YB-1 on viral late promoter activity was examined. As shown in Fig. 3B, production of Agnoprotein was accompanied by suppression of YB-1-induced transcription of the viral late promoter in glial cells. Additionally, we have statistically compared CAT values for the data points of Fig. 3A and B separately by pairwise analysis of variance with the Bonferroni correction for multiple comparisons. P values less than or equal to 0.05 from such comparisons indicated a statistical difference between data points, implying that the negative effect of Agnoprotein on YB-1-induced activation of JCV promoters is statistically significant. In conclusion, these findings demonstrate that Agnoprotein negatively influences YB-1-induced transactivation from JCV promoters.
Physical interaction of Agnoprotein with YB-1.
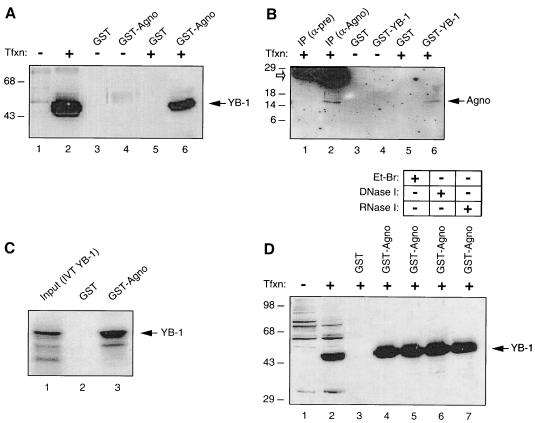
Our observations from functional studies suggested that Agnoprotein may physically interact with YB-1. To test this possibility, we first performed in vitro GST pull-down experiments. Bacterially expressed GST or GST-Agnoprotein was immobilized on glutathione-Sepharose beads and incubated with whole-cell extracts prepared from transfected U-87MG cells expressing histidine-tagged YB-1, His-YB-1. Proteins bound to GST or GST-Agnoprotein were analyzed by Western blotting using an anti-histidine tag antibody. As shown in Fig. 4A, His-YB-1 was retained on the Sepharose column containing GST-Agnoprotein (lane 6). An interaction of this type between YB-1 and GST alone was not observed (lane 5), indicating the specificity of association of YB-1 and Agnoprotein. Incubation of protein extracts from untransfected cells with GST alone (lane 3) or GST-Agnoprotein (lane 4) showed no protein band corresponding to His-YB-1, further verifying the specificity of this observation. Under our experimental conditions and based on densitometric scanning of our autoradiograms, 5.6% of the input of YB-1 was retained in the GST-Agnoprotein column.
FIG. 4.
In vitro interaction of Agnoprotein with YB-1. (A and B) Agnoprotein and YB-1 interact in GST pull-down assays. (A) Whole-cell extracts (0.5 mg) prepared from untransfected U-87MG cells (lanes 1, 3, and 4) and from U-87MG cells transfected with YB-1 expression plasmid pEBV-His-YB-1 (lanes 2, 5, and 6) were incubated with either GST alone (lanes 2 and 5) or GST-Agnoprotein (lanes 4 and 6). After being washed, proteins interacting with GST or GST-Agnoprotein were analyzed by Western blotting using an anti-T7 antibody for detection of histidine-tagged YB-1. Whole-cell extracts from U-87MG cells either untransfected (lane 1) or transfected with pEBV-His-YB-1 expression plasmid (lane 2) were loaded as negative and positive controls, respectively (50 μg/lane). Tfxn, transfection. (B) In reciprocal GST pull-down assays, whole-cell extracts (0.5 mg) prepared from untransfected U-87MG cells and from U-87MG cells transfected with Agnoprotein expression vector pEBV-His-Agnoprotein were incubated with GST alone or GST-YB-1 as indicated. Bound proteins were analyzed by Western blotting as described for panel A using an anti-T7 antibody to detect histidine-tagged Agnoprotein. Whole-cell extracts (50 μg/lane) from transfected cells were immunoprecipitated (IP) with preimmune (α-pre; lane 1) and anti-Agnoprotein (α-Agno; lane 2) antibodies and loaded as negative and positive controls, respectively. Open arrow, migration position of a small fragment of immunoglobulin of the antibodies used in immunoprecipitation. (C) Full-length in vitro-translated and [35S]methionine-labeled YB-1 (IVT YB-1) interacts with GST-Agnoprotein in a cell-free system. In vitro-labeled full-length YB-1 was incubated with either GST (lane 2) or GST-Agnoprotein fusion protein (lane 3), both of which were already immobilized on GST beads. (D) After extensive washing of the column with binding buffer, bound proteins were resolved by SDS-10% PAGE and analyzed by autoradiography. Whole-cell extracts prepared from U-87MG cells transfected with the YB-1 expression plasmid were treated with either ethidium bromide (Et-Br; 100 ng/ml; lane 5) or DNase I (0.2 U/μg of protein; lane 6) or RNase I (0.5 U/μg of protein; lane 7) (42) prior to incubation with GST-Agnoprotein, and bound proteins were analyzed by Western blotting as described for panel A.
In reciprocal GST pull-down assays, protein extracts prepared from U-87MG cells transfected with an Agnoprotein expression plasmid were incubated with either GST (lane 3) or GST-YB-1 (lane 4) and bound proteins were analyzed by Western blotting using antibodies against the histidine tag of Agnoprotein. As demonstrated in Fig. 4B, Agnoprotein produced in transfected cells was retained in the GST-YB-1 glutathione column (lane 6) but not in the GST column (lane 5), demonstrating the specificity of interaction between Agnoprotein and YB-1. As expected, no band was detected in protein extracts from untransfected cells incubated either with GST (lane 3) or GST-YB-1 (lane 4). Under conditions similar to those stated above, 7.9% of input Agnoprotein was retained in the GST-YB-1 column.
In addition to examining protein-protein interaction between YB-1 and Agnoprotein by utilizing cellular extracts, we investigated this interaction using a cell-free system. As shown in Fig. 4C, in vitro-transcribed and translated full-length YB-1 specifically and strongly interacts with GST-Agnoprotein (lane 3) but not GST alone (lane 2) in a GST pull-down assay; this result further confirms the observed interaction between YB-1 and Agnoprotein.
To determine whether the interaction between Agnoprotein and YB-1 is mediated through nucleic acids, the GST-Agnoprotein fusion protein and the protein extract from U-87MG cells expressing His-YB-1 were pretreated with either ethidium bromide, DNase I, or RNase prior to the pull-down step. As shown in Fig. 4D, neither ethidium bromide (lane 5) nor DNase I (lane 6) nor RNase (lane 7) treatment significantly affected the efficiency of binding between GST-Agnoprotein and YB-1, suggesting that the interaction of Agnoprotein and YB-1 is independent of nucleic acids.
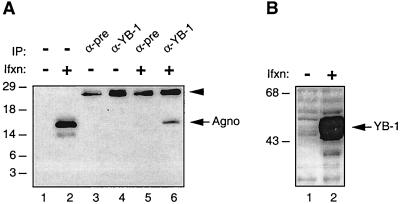
Next, to further examine the association of Agnoprotein with YB-1, we performed coimmunoprecipitation experiments. Protein extracts prepared from SF9 cells coinfected with recombinant baculoviruses expressing YB-1 and Agnoprotein were immunoprecipitated either with preimmune serum (control) or an anti-YB-1(242-267) antibody, and immunocomplexes were analyzed by Western blotting for the presence of Agnoprotein with an anti-Agnoprotein antibody. As shown in Fig. 5A, Agnoprotein was coimmunoprecipitated with YB-1 and was detected by the anti-Agnoprotein antibody (lane 6). Under conditions similar to those stated above, 3.12% of Agnoprotein was coimmunoprecipitated with the antibody to YB-1. The specificity of this coimmunoprecipitation was verified by the absence of a band corresponding to Agnoprotein when protein extracts from either uninfected or infected cells were incubated with normal mouse serum (lanes 3 and 5, respectively). Furthermore, the anti-YB-1 antibody showed no cross-reactivity with cellular proteins from uninfected cells (lane 4). Figure 5B demonstrates the detection of expressed YB-1 in infected cells by Western blot analysis. Altogether, results from both in vitro GST pull-down and coimmunoprecipitation experiments demonstrate that Agnoprotein associates with YB-1.
FIG. 5.
(A) Coimmunoprecipitation of Agnoprotein with YB-1. Coimmunoprecipitation experiments were performed as described in Materials and Methods. Antibodies used for the respective lanes are shown at the top. α-pre is as defined for Fig. 4. Whole-cell extracts prepared from uninfected (lane 1) and infected (lane 2) Sf9 cells were loaded as negative and positive controls, respectively (50 μg/lane). Arrowhead, position of immunoglobulin light chain detected by the secondary antibody. IP, immunoprecipitation; Ifxn, infection. (B) Western blot analysis of recombinant YB-1 expressed in Sf9 insect cells.
Localization of Agnoprotein sequences important for interaction with YB-1.
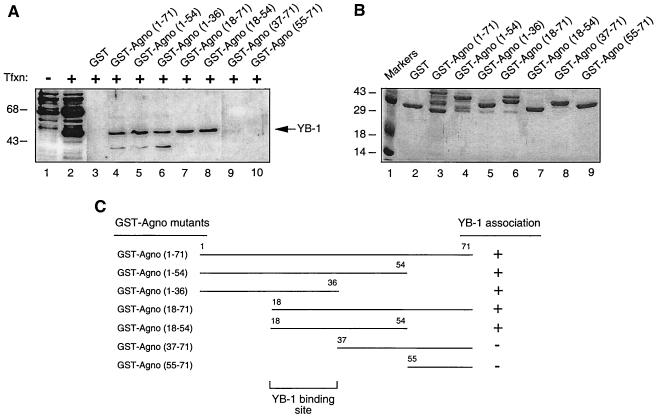
In the next series of experiments, we mapped the region of Agnoprotein which interacts with YB-1. A series of bacterially expressed Agnoprotein deletion mutants fused to GST were incubated with protein extract from U-87MG cells overexpressing YB-1. Protein complexes were resolved by SDS-PAGE and analyzed by Western blotting using an anti-Agnoprotein antibody. As shown in Fig. 6A, removal of the regions of Agnoprotein positioned between residues 55 and 71 (lane 5), 37 and 71 (lane 6), and 1 and 17 (lane 7), as well as the simultaneous deletion of residues 1 to 17 and 55 to 71, had no effect on the ability of Agnoprotein to bind YB-1. Deletion of residues 1 to 36 and 1 to 54 completely inhibited association of Agnoprotein with YB-1, indicating that the residues between 18 and 36 of Agnoprotein are important for its association with YB-1. Figure 6B illustrates the Coomassie blue-stained SDS-PAGE gel of the full-length and mutant Agnoproteins which were used in this study and verifies the integrity of the protein preparations. A schematic of the GST pull-down assay depicting the regions of Agnoprotein which bind to YB-1 is presented in Fig. 6C.
FIG. 6.
Mapping of the domain of the interaction between Agnoprotein and YB-1. (A) Whole-cell extracts from U-87MG cells transfected with YB-1 expression plasmid pEBV-His-YB-1 were incubated with either GST alone (lane 3) or GST-Agnoprotein (lane 4) or Agnoprotein deletion mutants fused to GST (lanes 5 to 10) immobilized on glutathione-Sepharose beads. Bound complexes were analyzed by Western blotting using an anti-T7 antibody. Whole-cell extracts prepared from untransfected (lane 1) and transfected (lane 2) U-87MG cells were loaded as negative and positive controls, respectively. Tfxn, transfection. (B) Analysis of GST and GST-Agnoprotein and its deletion mutants by SDS-PAGE. (C) Summary of the results obtained from in vitro mapping assays. +, ability of Agnoprotein and its deletion mutants to interact with YB-1; −, no interaction.
Mapping of the interaction domain of YB-1 with Agnoprotein.
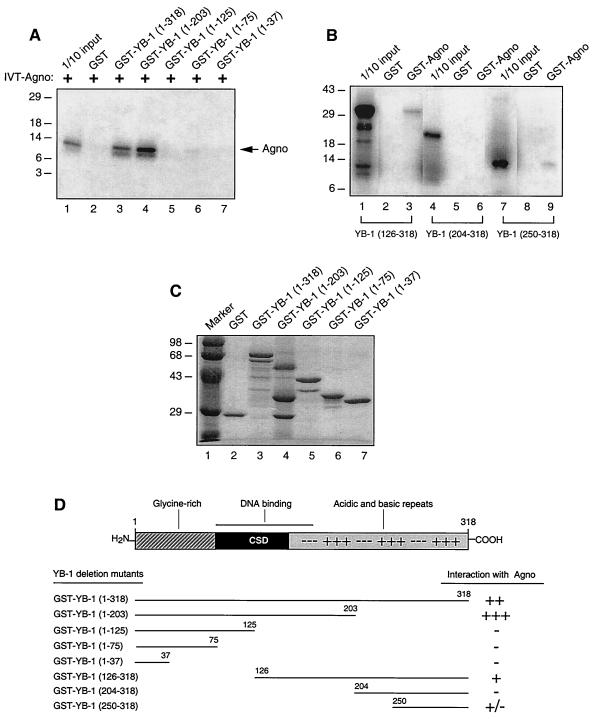
To identify the protein domains within YB-1 which confer the interaction with Agnoprotein, a series of YB-1 carboxy-terminal (Fig. 7A) deletion mutants fused with GST were prepared and incubated with in vitro-synthesized [35S]methionine-labeled Agnoprotein. In agreement with the results shown in Fig. 4A, GST-YB-1 but not GST bound to Agnoprotein (Fig. 7, lanes 2 and 3). A YB-1 carboxy-terminal deletion mutant retaining residues 1 to 203 showed a slight increase in Agnoprotein binding activity, suggesting that a region in the carboxy terminus of YB-1 may exert a negative effect on the interaction between YB-1 and Agnoprotein. The remaining three carboxy-terminal deletion mutants retaining residues 1 to 125, 1 to 75, or 1 to 37 showed no YB-1 binding activity.
FIG. 7.
Localization of the domain of the interaction between YB-1 and Agnoprotein. (A) GST (lane 2), GST-YB-1 (lane 3), and GST-YB-1 C-terminal deletion mutants (lanes 4 to 7) were immobilized on glutathione-Sepharose beads and incubated with in vitro-translated [35S]methionine-labeled Agnoprotein. The Sepharose beads were washed extensively, and bound proteins were resolved by SDS-PAGE and analyzed by autoradiography. Lane 1, 1/10 of the input Agnoprotein loaded for migration control. Agno, position of in vitro-translated [35S]methionine-labeled Agnoprotein; IVT-Agno, in vitro translation of Agnoprotein. (B) Three different [35S]methionine-labeled in vitro-translated YB-1 amino-terminal deletion mutants, i.e., YB-1(126-318), YB-1 (204-318), and YB-1 (250-318), were incubated with GST (lanes 2, 5, and 8) or GST-Agnoprotein (lanes 3, 6, and 9). Bound proteins were analyzed as described for panel A. Lanes 1, 4, and 7 contain 1/10 of the amount used in the pull-down experiments with YB-1(126-318), YB-1(204-318), and YB-1 (250-318), respectively. (C) Analysis of GST, GST-YB-1, GST-YB-1 C-terminal deletion mutants by SDS-PAGE. (D) Summary of the results obtained from in vitro mapping assays. A schematic representation of YB-1 is shown at the top (not shown to scale). The abilities of YB-1 and its deletion mutants to interact with Agnoprotein are shown on the right (+++, very strong interaction; ++, strong interaction; +, minimal interaction; +/−, weak interaction; −, no interaction). CSD, cold shock domain.
We also examined the Agnoprotein binding activities of three in vitro-synthesized YB-1 amino-terminal deletion mutants, including YB-1(126-318), YB-1(204-318), and YB-1(250-318), by GST pull-down assays. For these mutants we utilized in-vitro synthesized proteins, as our initial studies showed that GST fusion proteins corresponding to these three YB-1 deletion mutants are unstable in bacteria. Each deletion mutant was incubated with either GST or GST-Agnoprotein, and bound proteins were visualized by autoradiography. As demonstrated in Fig. 7B, the YB-1 deletion mutant lacking the residues between 1 and 125 (lane 3) showed a moderate binding ability. Removal of amino acids 1 to 249 resulted in very weak Agnoprotein binding activity of YB-1 (lane 9). However, the deletion mutant that retains amino acid residues 204 to 318 completely abolished the interaction of YB-1 with Agnoprotein (lane 6), implying that YB-1 residues 204 to 250 negatively affect the interaction between Agnoprotein and YB-1. This finding is consistent with our observation in Fig. 7A, where we demonstrated that the Agnoprotein binding activity of YB-1 was significantly increased when carboxy-terminal residues 204 to 318 were deleted (lane 4). Taken together, results from mapping experiments demonstrate that the domain(s) of the interaction of YB-1 with Agnoprotein lies within the carboxy-terminal region of the protein between amino acids 126 and 318, where residues 204 to 249 negatively regulate this interaction. Figure 7C illustrates the Coomassie blue-stained SDS-PAGE gel of the full-length YB-1 and YB-1 mutants which were used in this study. A scheme for these observations is shown in Fig. 7D.
Effect of mutant Agnoprotein on YB-1-mediated JCV gene transcription.
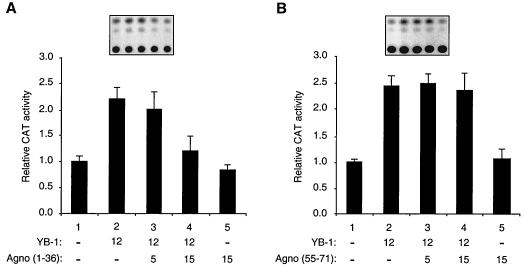
To further assess the functional interaction between Agnoprotein and YB-1, we examined the ability of mutant Agnoproteins which have retained or lost their YB-1 binding activity in GST pull-down assays. We chose mutant Agnoprotein(1-36), which had the ability to interact with YB-1 in in vitro GST pull-down assays, and mutant Agnoprotein(55-71), which displayed no YB-1 binding activity. Transient transfection assays were performed utilizing a reporter JCV late CAT construct and expression plasmids for mutants Agnoprotein(1-36) and -(55-71). As shown in Fig. 8A, mutant Agnoprotein(1-36), which binds to YB-1 with the same efficiency as the full-length protein (Fig. 6A, lane 6), suppressed YB-1-mediated transcriptional activation from the JCV late promoter while mutant Agnoprotein(55-71) had no effect on the level of transcriptional activation of the JCV promoter by YB-1 (Fig. 8B). The findings from functional studies, where we used mutant Agnoproteins to further verify the interaction between Agnoprotein and YB-1, corroborate our observations from in vitro GST pull-down assays (Fig. 6A). Of note, the stable expression of Agnoprotein mutants used in transfection assays has been reported previously (42), and therefore the absence of a functional interaction between YB-1 and mutant Agnoprotein(55-71) may not be attributed to its lack of expression in transfected cells. In addition, we also statistically compared CAT data points for Fig. 8A and B in a pairwise manner to demonstrate that the effect of mutant Agnoprotein(1-36) on YB-1-induced CAT expression is statistically significant. We used analysis of variance with the Bonferroni correction for multiple comparisons for this analysis. The obtained P values (less than or equal to 0.05) suggested that the effect of mutant Agnoprotein(1-36) on YB-1-induced CAT expression of the JCV late promoter is statistically significant. As expected, a similar comparison of data points for mutant Agnoprotein(55-71) did not show any statistically significant difference. Taken together, the data from functional assays utilizing Agnoprotein mutants, one of which physically and functionally interacts with YB-1, mutant YB-1(1-36), and the other of which lacks such characteristics, mutant YB-1(55-71), further confirm the significance of the physical and functional interactions between Agnoprotein and YB-1 in the regulation of JCV gene expression.
FIG. 8.
Two Agnoprotein deletion mutants confirm the interaction between Agnoprotein and YB-1. Shown is the effect of Agnoprotein deletion mutants on YB-1-mediated transcriptional activation of the JCV late promoter. A CAT reporter plasmid (pBLCAT3-Mad-1L; 7 μg/60-mm-diameter dish) containing the JCV late promoter was transfected into U-87MG cells alone or in combination with expression plasmids for deletion mutant constructs Agnoprotein(1-36) (A) and Agnoprotein(55-71) (B) and YB-1. The concentrations of expression plasmids in micrograms are indicated at the bottom. Representative CAT assays are shown at the top. The results represent the means of three independent experiments. Error bars indicate standard deviations.
DISCUSSION
In this report, we examined the molecular mechanism(s) involved in the regulation of JCV gene transcription by JCV late Agnoprotein via an interaction with cellular transcription factor YB-1 and showed that Agnoprotein negatively regulates YB-1-mediated gene transcription. These findings support our previous observations (42) indicating that Agnoprotein, similar to large T antigen, has regulatory functions in the JCV life cycle.
Examination of Agnoprotein expression during the course of infection by Western blot analysis and immunofluorescence microscopy demonstrated that its expression is detectable by both techniques as early as the third day postinfection. Further, its expression coincides with that of the viral capsid proteins. As observed for Agnoproteins of the other closely related polyomaviruses including BKV and SV40, JCV Agnoprotein is also mainly localized to the cytoplasmic and perinuclear compartments of the infected cells. To a lesser extent, however, its nuclear localization is also apparent. At this moment, it is not clear what function(s) might be associated with its subcellular distribution. Studies with SV40 Agnoprotein have suggested that it may have regulatory roles in the viral lytic cycle including viral assembly (19, 27, 30, 31), transcription and translation, and maturation (1, 13, 14).
Results from in vitro protein-protein interaction studies reported in this paper have clearly shown a direct association between Agnoprotein and YB-1. Such an interaction does not appear to be mediated by a nucleic acid molecule, since the treatment of cell extracts with either DNase I, RNase I, or ethidium bromide did not result in any noticeable change in the band intensities corresponding to YB-1. Results from in vitro mapping experiments utilizing various deletion mutant constructs demonstrated that the Agnoprotein interaction domain of YB-1 is localized to its carboxy-terminal half. This region of the protein contains alternating basic and acidic amino acid islands, which are thought to stabilize protein-DNA interactions, and perhaps acts as a transactivation domain of the protein (50). Consistent with our observations from transfection assays, when Agnoprotein is coexpressed with YB-1 in cells, the suppressive effect of Agnoprotein on YB-1-mediated activation of transcription from both JCV promoters was observed, suggesting that Agnoprotein, by binding to the carboxy-terminal half of YB-1, may diminish its transcriptional activity.
Many reports indicate that viral infection induces stress in host cells that can alter the activity of regulatory proteins including the NF-κB family of transcription factors (48). Such factors under normal conditions are retained in the cytoplasm and, upon induction, translocate into the nucleus and thereby stimulate stress-responsive genes. YB-1 is also a cytoplasmic stress-inducible factor and can be induced by a variety of stress-related stimuli including anticancer agents (16, 23, 32), UV irradiation (22), and hypothermia (44). These observations suggested that perhaps viral infection may also be an important factor in its induction. We explored such a possibility by immunofluorescence microscopy studies with infected cells. We observed that, although early during infection the subcellular distribution of YB-1 was not noticeably altered, as the infection cycle progressed, its subcellular localization oscillated between the cytoplasm and the nucleus. This finding strongly suggests that YB-1 can be induced by viral infection and thereby may play an important role in JCV gene regulation. In fact, we have previously demonstrated that YB-1 is able to upregulate JCV transcription from the viral early promoter in the absence of the viral early regulatory protein large T antigen in transfection assays. Together with T antigen, YB-1 is then able to synergistically transactivate viral late genes (39).
One of the interesting characteristics of viruses is their ability to use the host machinery to maximize the efficiency of the viral lytic cycle. It was interesting to observe that Agnoprotein, which is expressed at the late phase of lytic infection, negatively regulates both viral DNA replication and transcription (42), suggesting that negative regulation mediated by Agnoprotein on both processes may have physiological consequences for a successful viral life cycle which includes capsid formation. Experiments are under way to further analyze the role of Agnoprotein in the JCV life cycle by using JCV Agno mutant viruses.
Acknowledgments
We thank E. O. Major and W. Atwood for providing the cells and viruses which were used in this study, Robert Kelm for kind gifts of anti-YB-1 antibodies, and past and present members of the Center for Neurovirology and Cancer Biology for their insightful discussion, sharing of ideas, and reagents. We thank Cynthia Schriver for editorial assistance.
This work was made possible by grants awarded by NIH to K.K.
REFERENCES
- 1.Alwine, J. 1982. Evidence for simian virus 40 late transcriptional control: mixed infections of wild-type simian virus 40 and a leader deletion mutant exhibit trans effects on late viral RNA synthesis. J. Virol. 42:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amemiya, K., R. Traub, L. Durham, and E. O. Major. 1989. Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J. Biol. Chem. 264:7025-7032. [PubMed] [Google Scholar]
- 3.Ansari, S. A., M. Safak, L. Del Valle, S. Enam, S. Amini, and K. Khalili. 2001. Cell cycle regulation of NF-kappa B-binding activity in cells from human glioblastomas. Exp. Cell Res. 265:221-233. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J. R., and M. Concha. 1995. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J. Neurovirol. 1:5-18. [DOI] [PubMed] [Google Scholar]
- 5.Carswell, S., J. Resnick, and J. C. Alwine. 1986. Construction and characterization of CV-1P cell lines which constitutively express the simian virus 40 agnoprotein: alteration of plaquing phenotype of viral agnogene mutants. J. Virol. 60:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, N. N., and K. Khalili. 1995. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Purα in glial cells. J. Virol. 69:5843-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didier, D. K., J. Schiffenbauer, S. L. Woulfe, M. Zacheis, and B. D. Schwartz. 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA 85:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisque, R. J., G. L. Bream, and M. T. Cannella. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisque, R. J., and F. A. White III. 1992. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy, p. 25-158. In E. R. P. Roos (ed.), Molecular neurovirology. Humana Press Inc., Totowa, N.J.
- 10.Gallia, G. L., M. Safak, and K. Khalili. 1998. Interaction of the single-stranded DNA-binding protein Purα with the human polyomavirus JC virus early protein T-antigen. J. Biol. Chem. 273:32662-32669. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 13.Haggerty, S., D. L. Walker, and R. J. Frisque. 1989. JC virus-simian virus 40 genomes containing heterologous regulatory signals and chimeric early regions: identification of regions restricting transformation by JC virus. J. Virol. 63:2180-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay, N., D. H. Skolnik, and Y. Aloni. 1982. Attenuation in the control of SV40 gene expression. Cell 29:183-193. [DOI] [PubMed] [Google Scholar]
- 15.Henson, J., J. Saffer, and H. Furneaux. 1992. The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann. Neurol. 32:72-77. [DOI] [PubMed] [Google Scholar]
- 16.Ise, T., G. Nagatanni, T. Imamura, K. Kato, M. Nomoto, H. Izumi, H. Ohmari, T. Okamota, T. Ohga, T. Uchiumi, M. Kuwano, and K. Kohno. 1999. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 59:342-346. [PubMed] [Google Scholar]
- 17.Ishii, N., N. Minami, E. Y. Chen, A. L. Medina, M. M. Chico, and H. Kasamutsu. 1996. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J. Virol. 70:1317-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, V., and R. Chalkley. 1981. Use of whole-cell fixation to visualize replicating and maturing simian virus 40: identification of a new viral gene product. Proc. Natl. Acad. Sci. USA 78:6082-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jay, G., S. Nomura, C. W. Anderson, and G. Khoury. 1981. Identification of the SV40 agnogene product: a DNA binding protein. Nature 291:346-349. [DOI] [PubMed] [Google Scholar]
- 20.Kelm, R. J., P. K. Elder, and M. J. Getz. 1999. The single-stranded DNA-binding proteins Purα, Purβ and MSY1 specifically interact with an exon 3-derived mouse vascular smooth muscle a-actin messenger RNA sequence. J. Biol. Chem. 274:38268-38275. [DOI] [PubMed] [Google Scholar]
- 21.Kerr, D., C. F. Chang, N. Chen, G. L. Gallia, G. Raj, B. Schwartz, and K. Khalili. 1994. Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J. Virol. 68:7637-7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koike, K., T. Uchiumi, T. Ohga, S. Toh, M. Wada, K. Kohno, and M. Kuwano. 1997. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 417:390-394. [DOI] [PubMed] [Google Scholar]
- 23.Li, W. W., Y. Hsiung, V. Wong, K. Galvin, Y. Zhou, Y. Shi, and A. S. Lee. 1997. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol. Cell. Biol. 17:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Major, E. O., A. E. Miller, P. Mourrain, R. G. Traub, D. de With, and J. Sever. 1985. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl. Acad. Sci. USA 82:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Major, E. O., K. Amemiya, C. Tornatore, S. A. Houff, and J. B. Berger. 1992. Pathogenesis and molecular biology of progressive multifocal encephalophathy. The JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 5:49-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marenstein, D. R., M. T. A. Ocampo, M. K. Chan, A. Altamirano, A. K. Basu, R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 2001. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem. 276:21242-21249. [DOI] [PubMed] [Google Scholar]
- 27.Margolskee, R. F., and D. Nathans. 1983. Suppression of a VP1 mutant of simian virus 40 by missense mutations in serine codons of the viral agnogene. J. Virol. 48:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mertens, P. R., S. Harendza, A. S. Pollock, and D. H. Lovett. 1997. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J. Biol. Chem. 272:22905-22912. [DOI] [PubMed] [Google Scholar]
- 29.Mertens, P. R., M. A. Alfonso-Jaume, K. Steinmann, and D. H. Lovett. 1998. A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J. Biol. Chem. 273:32957-32965. [DOI] [PubMed] [Google Scholar]
- 30.Mertz, J. E., A. Murphy, and A. Barkan. 1983. Mutants deleted in the agnogene of simian virus 40 define a new complementation group. J. Virol. 45:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng, S. C., J. E. Mertz, S. Sanden-Will, and M. Bina. 1985. Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. J. Biol. Chem. 260:1127-1132. [PubMed] [Google Scholar]
- 32.Ohga, T., T. Uchiumi, Y. Makina, K. Koike, M. Wada, M. Kuwano, and K. Kohna. 1998. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem. 273:5997-6000. [DOI] [PubMed] [Google Scholar]
- 33.Raj, G. V., and K. Khalili. 1994. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol. Cell. Biol. 14:7770-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raj, G. V., and K. Khalili. 1995. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology 213:283-291. [DOI] [PubMed] [Google Scholar]
- 35.Ranganathan, P. N., and K. Khalili. 1993. The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res. 21:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renner, K., H. Leger, and M. Wegner. 1994. The POU doamin protein Tst-1 and Papovaviral large tumor antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc. Natl. Acad. Sci. USA 91:6433-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinaldo, C. H., T. Traavik, and A. Hey. 1998. The agnogene of the human polyomavirus BK is expressed. J. Virol. 72:6233-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabath, D. E., P. L. Podolin, P. G. Comber, and M. B. Prystowky. 1990. cDNA cloning and characterization of interleukin 2-induced genes in a cloned T helper lymphocyte. J. Biol. Chem. 265:12671-12678. [PubMed] [Google Scholar]
- 39.Safak, M., G. L. Gallia, S. A. Ansari, and K. Khalili. 1999. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J. Virol. 73:10146-10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safak, M., G. L. Gallia, and K. Khalili. 1999. Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol. Cell. Biol. 19:2712-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safak, M., G. L. Gallia, and K. Khalili. 1999. A 23-bp sequence element from human neurotropic JC virus is responsive to NF-κB subunits. Virology 262:178-189. [DOI] [PubMed] [Google Scholar]
- 42.Safak, M., R. Barrucco, A. Darbinyan, Y. Okada, K. Nagashima, and K. Khalili. 2001. Interaction of JC virus Agno protein with T-antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 75:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safak, M., and K. Khalili. 2001. Physical and functional interaction between viral and cellular proteins modulate JCV gene transcription. J. Neurovirol. 7:288-292. [DOI] [PubMed] [Google Scholar]
- 44.Stein, U., K. Jurchott, W. Walther, S. Bergmann, P. M. Schlag, and H.-D. Royer. 2001. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J. Biol. Chem. 276:28562-28569. [DOI] [PubMed] [Google Scholar]
- 45.Stenina, O. I., E. J. Poptic, and P. E. DiCorleto. 2000. Thrombin activates a Y box-binding protein (DNA-binding protein B) in endothelial cells. J. Clin. Investig. 106:579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenina, O. I., K. M. Shaneyfelt, and P. E. DiCarleto. 2001. Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation. Proc. Natl. Acad. Sci. USA 98:7277-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tafuri, S. R., and A. P. Wolffe. 1993. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4). J. Biol. Chem. 268:24255-24261. [PubMed] [Google Scholar]
- 48.Thanos, D., and T. Maniatis. 1995. NF-κB: a lesson in family values. Cell 80:529-532. [DOI] [PubMed] [Google Scholar]
- 49.Vacante, D. A., R. Traub, and E. O. Major. 1989. Extension of JC virus host range to monkey cells by insertion of a simian virus SV40 enhancer into the JC virus regulatory region. Virology 170:353-361 [DOI] [PubMed] [Google Scholar]
- 50.Wolffe, A. P., S. Tafuri, M. Ranjan, and M. Familari. 1994. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 4:290-298. [PubMed] [Google Scholar]