Abstract
The development of surrogate markers capable of detecting residual ongoing human immunodeficiency virus type 1 (HIV-1) replication in patients receiving highly active antiretroviral therapy is an important step in understanding viral dynamics and in developing new treatment strategies. In this study, we evaluated the utility of circular forms of the viral genome for the detection of recent infection of cells by HIV-1. We measured the fate of both one-long terminal repeat (1-LTR) and 2-LTR circles following in vitro infection of logarithmically growing CD4+ T cells under conditions in which cell death was not a significant contributing factor. Circular forms of the viral genome were found to be highly stable and to decrease in concentration only as a function of dilution resulting from cell division. We conclude that these DNA circles are not intrinsically unstable in all cell types and suggest that the utility of 2-LTR circle assays in measuring recent HIV-1 infection of susceptible cells in vivo needs to be reevaluated.
In some human immunodeficiency virus type 1 (HIV-1)-infected individuals, treatment with highly active antiretroviral therapy (HAART) decreases the amount of virus in plasma to levels below the limit of detection of standard clinical assays (10, 11, 24). However, initial hopes that HAART might permit virus eradication have been dampened by two findings (reviewed in reference 26). First, in both children and adults receiving HAART, replication-competent HIV-1 persists in latently infected resting memory CD4+ T lymphocytes, providing the potential for lifetime persistence of the virus despite potent HAART regimens (1a, 2, 7, 8, 25, 32). Second, a low level of virus production continues in patients receiving HAART (9, 14, 19; V. Natarajan, M. Bosche, J. A. Metcalf, D. J. Ward, H. C. Lane, and J. A. Kovacs, Letter, Lancet 353:119-120, 1999). Patients who have plasma virus levels below the limit of detection of conventional ultrasensitive assays (50 copies/ml) often are found to be virus positive when even more sensitive assays for viral RNA are used (limit of detection = 5 to 50 copies/ml) (3). In addition, unintegrated forms of HIV-1 DNA can also be detected in patients receiving HAART (9, 29). If these forms are intrinsically unstable, their presence can only result from ongoing viral replication.
In considering studies of ongoing replication, it is important to make a distinction between continuous new cycles of infection and virus release by cells that were infected previously, for example, latently infected cells that become activated and produce virus. This distinction is important because evolution of drug resistance is dependent upon the errors made in reverse transcription during new cycles of infection. The detection of very low levels of viremia in patients with plasma HIV-1 RNA levels below 50 copies/ml does not prove that new cycles of infection are occurring. In principle, this extremely low level of viremia could result from the release of virus from chronically infected cells or latently infected cells that become activated (13). Therefore, there is an urgent need for an assay to detect new cycles of infection in patients receiving HAART who have plasma HIV-1 RNA levels below the limit of detection.
The molecular processes that mediate HIV-1 infection provide several potentially useful markers of recent infection, including circular DNA forms that arise when the reverse-transcribed HIV-1 DNA fails to integrate into the host genome and instead undergoes one of two possible circularization reactions. 1-LTR circles form through homologous recombination between the two LTRs that flank the linear genome (27, 28), while 2-LTR circles form through the intramolecular (end-to-end) ligation of the linear HIV-1 cDNA (15, 17, 30, 31).
In several recent studies, 2-LTR circles have been used as a marker of recent infection on the basis of the notion that these circles are unstable (9, 21-23, 29, 34). Sharkey et al. demonstrated the apparent rapid decay of 2-LTR circles in T-cell lines infected in vitro and in patients starting HAART (29). In contrast, several different types of DNA circles, both viral and cellular, are known to be stable (4, 35). Therefore, in this study the intrinsic stability of circular forms of the HIV-1 genome was evaluated.
The fate of DNA intermediates generated during HIV-1 replication is a complex function of the stability of the viral DNA, the life span of the infected cell, and the fate of the DNA intermediate following cell division. The number of 2-LTR circles in a population of infected cells might decline if the circles are intrinsically unstable or if the cells harboring the circles die. In addition, since circular forms of the viral genome lack an origin of replication, the average number of circles per cell will decrease if the cells are proliferating. Our approach was designed to measure the stability of circular forms of the viral genome in a system in which the contribution of cell death and cell division could be carefully measured. Primary T cells could not be used because of variable growth and poor survival in long-term culture. Therefore, studies were carried out with the transformed CD4+ T-cell line MT-2.
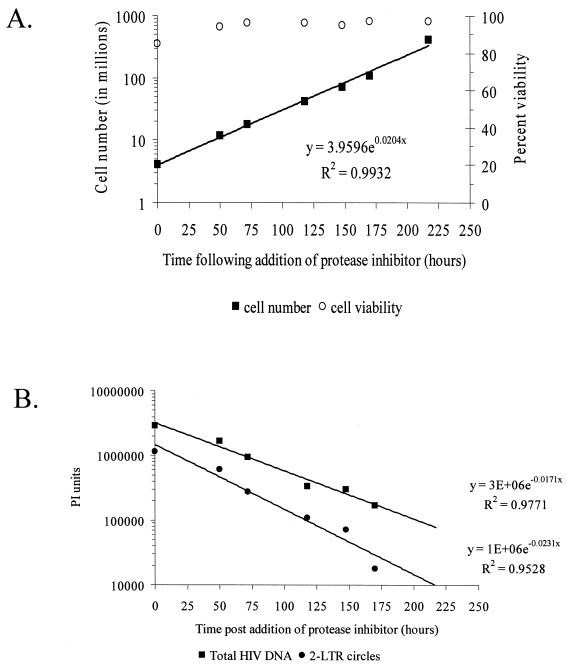
MT-2 cells were spin inoculated with DNase I-treated HIV-1 IIIb at a multiplicity of infection (MOI) of 0.05 as described recently (20). Cells were then incubated in minimal medium (MM) consisting of RPMI 1640 supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine. Forty-eight hours after infection, the infected cells were washed twice in MM containing 60 μM indinavir, an inhibitor of the HIV-1 protease (50% inhibitory concentration, 20 nM) (18). To prevent further rounds of viral replication, the cells were maintained in the presence of indinavir at a concentration of 0.5 × 106 cells per ml. Periodic sampling of culture supernatant was performed for analysis in a p24-specific antigen capture ELISA (limit of detection, 15 pg/ml; Beckman Coulter Electronics Ltd., Hialeah, Fla.). The efficacy of indinavir treatment in these experiments was confirmed by the absence of p24 release in the infected culture during the study period. Following infection, cell number and viability were carefully monitored by cell counting and vital dye exclusion (Fig. 1A). Analysis of cellular proliferation in culture demonstrated that logarithmic growth occurred throughout the period of study, with a doubling time of 34.0 h (95% confidence interval [CI], 31.5 to 36.9 h). Average cell viability during culture was 94.3%. At each time point, DNA was isolated and the relative amounts of 2-LTR circles and total HIV-1 DNA were measured.
FIG. 1.
Intrinsic stability of 2-LTR circles following infection of log-phase MT-2 cells. (A) Proliferation of MT-2 cells infected with HIV-1 IIIb. Cells were infected with HIV-1 IIIb (MOI = 0.05) and cultured for 48 h in MM. Indinavir was then added to the culture, and at the indicated times, cells were counted and viability was measured by trypan blue exclusion. Cells were maintained at a concentration of 0.5 million/ml after each count in order to sustain an exponential rate of growth. (B) Analysis of the decay of 2-LTR circles. At the indicated times, cells were harvested and assayed by 28 and 25 cycles of semiquantitative PCR for 2-LTR circles and total HIV-1 DNA, respectively. The observed decay rates of 2-LTR circles and total HIV DNA were measured by phosphorimager analysis. To normalize for DNA input, amplification of β-globin was performed (data not shown).
To measure 2-LTR circles, we developed a PCR assay based on a modification of a previously published strategy (15, 17, 30, 31). PCR primers were positioned at the terminus of the linear genome, allowing amplification across the junction of the two LTRs only after a covalently closed circle had formed (LPCR-L, TGGTACTAGCTTGAAGCACCATCCA; LPCR-R, GCCTGTACTGGGTCTCTCTGGTTAG). Specificity of this reaction was confirmed by sequence analysis of cloned products. Measurements of 1-LTR circles were made as previously described (1a). Total HIV-1 DNA was monitored using a previously described method employing primers that span the LTR and gag regions. This allows detection of reverse transcription products that have been largely or fully completed as well as integrated HIV-1 DNA and 1- and 2-LTR circles (33). Each PCR was performed on a mass of 500 ng of DNA using a 1 μM concentration of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 2.5 U of Platinum Taq polymerase in 1× proprietary reaction buffer (Invitrogen Corp., Carlsbad, Calif.). Reaction mixtures were incubated initially for 3 min of denaturation at 94°C, followed by cycling between 94 and 65°C for 30 s at each temperature. To ensure each reaction was performed with the same mass of DNA, each sample was assayed by PCR specific for β-globin as described previously (1a). Reaction products were run on a 2% agarose gel, transferred to nylon membranes, and Southern blotted with a 32P-end-labeled oligonucleotide probe (for 1- and 2-LTR circles, CACACACAAGGCTACTTCCCTGA; the probe for β-globin was described in reference 1a). The intensity of each band was quantified using phosphorimager analysis and is expressed as arbitrary phosphorimager units.
This study focused on the rate of decay of 2-LTR circles generated after an initial cycle of infection, which involved the analysis of a small number of HIV-1 DNA molecules per mass of DNA. To ensure that PCR measurements in this study maintained a linear relationship between band intensity and copy number, control experiments were performed to demonstrate the linearity of our PCR chemistry. Plasmids mimicking proviral DNA and 1-LTR and 2-LTR circles were diluted into uninfected human genomic DNA and analyzed as described above. Under the conditions described, the reaction mixtures used in this study maintained a linear relationship between copy number and band intensity up to 104 copies/500 ng of genomic DNA (data not shown). Due to the low MOI, DNA from infected cells contained only a small number of HIV-1 molecules (10 to 1,000 copies/500 ng), which was well within the linear range of the assays employed. Because the study focused on the rate of decay of 2-LTR circles generated in the initial cycle of infection, band intensities were compared to the initial time point, and no attempt was made to determine the absolute number of circles formed in a given experiment.
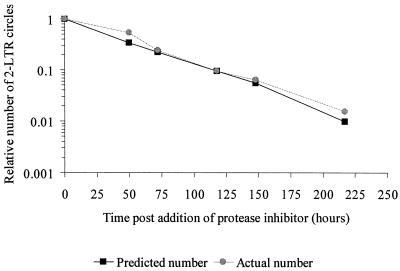
While both the 2-LTR circles and LTR-gag DNA were detected throughout the 10 days of culture, each decayed at a different rate after new infection was halted by the addition of indinavir. Phosphorimager analysis (Fig. 1B; Table 1) revealed that the decay of the 2-LTR circles (half life [t1/2], 35.4 h; 95% CI, 32.2 to 39.4 h) was very similar to the inverse of the doubling time of the host cells (34.0 h), suggesting that the decrease in the number of 2-LTR circles per microgram of DNA could be completely accounted for by dilution. Using a linear model with the combined data from cell division and 2-LTR circle experiments, there was no statistical evidence (P = 0.510) of any residual decay in the 2-LTR circles beyond that expected from dilution by cell division (Fig. 2). Decay of 2-LTR circles could be caused by either molecular decay of the circles or death of cells harboring the circles. Our results suggest that neither process occurred at a detectable rate over the course of the experiment. The decay of the LTR-gag signal was even slower (t1/2, 49.5 h; 95% CI, 41.0 to 62.3 h), presumably due to integration of HIV-1 into a fraction of the infected cells. In contrast to the episomal circles, integrated DNA does not decay by dilution. These results are representative of three replicate experiments, all of which demonstrated the stability of 2-LTR circles (Table 1). It should be noted that our analysis of decreases in 2-LTR circle numbers in a population of proliferating cells is not based on the assumption that the rate of division of cells containing circles is the same as the rate of division of cells that do not contain circles. Even if cells with circles divide more slowly, the decay in the number of circles per unit DNA would be slower, but this would be corrected for in our analysis, which is normalized for cell number.
TABLE 1.
Analysis of residual decay in LTR circles
| Expt. no. | Parameter | Phosphorimager results
|
Residual decay resultsc
|
|||
|---|---|---|---|---|---|---|
| t1/2 (h) | 95% CI | Pb | t1/2 (h) | 95% CI | ||
| 1d | Cells−1a | 33.97 | 30.00-39.16 | |||
| 2-LTR circles | 35.40 | 32.15-39.37 | 0.510 | NA | NA - 367.45 | |
| Total HIV | 49.45 | 41.01-62.26 | 0.005 | |||
| 2d | Cells−1 | 29.90 | 23.63-40.71 | |||
| 2-LTR circles | 26.12 | 24.31-28.22 | 0.025 | 206.53 | 1225.37 - 112.77 | |
| 1-LTR circles | 27.50 | 22.11-36.38 | 0.590 | 342.69 | NA - 64.66 | |
| 3e | Cells−1 | 27.74 | 25.66-30.19 | |||
| 2-LTR circles | 28.30 | 26.13-30.85 | 0.691 | NA | NA - 291.47 | |
| 1-LTR circles | 23.88 | 20.56-28.48 | 0.185 | 171.46 | NA - 64.99 | |
| Total HIV | 21.02 | 18.07-25.13 | 0.042 | |||
Cells−1, doubling time of the cells.
P values are for comparison of t1/2 and cells−1.
Residual decay relative to cell division. NA, not available.
Experiment with replication-competent virus.
Experiment with integrase mutant virus.
FIG. 2.
Comparison of the rate of observed decline of the 2-LTR circle signal to the decrease in signal predicted, assuming complete stability of the circles and a decrease in concentration as a consequence of dilution from cell division measured during the course of study. Data are normalized to the signal at time zero.
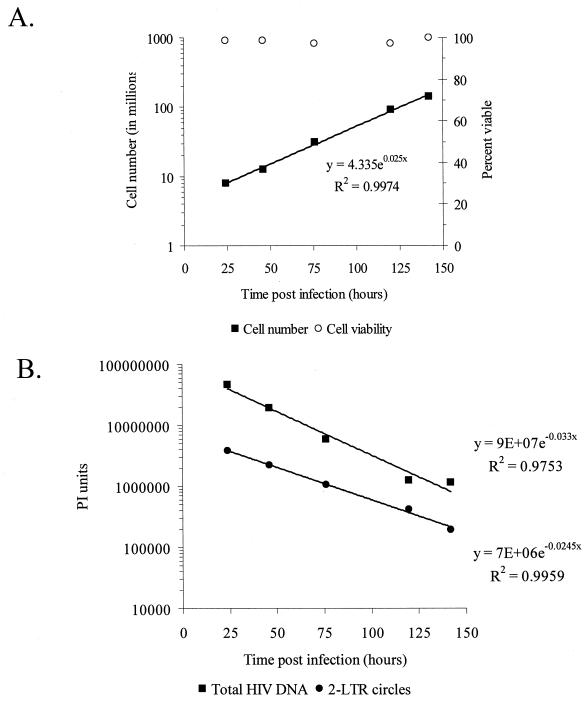
To confirm these observations, a second series of experiments was performed to study the decay of 2-LTR circles after infection of MT-2 cells with an HIV-1 IIIb strain bearing a mutation (D64N) in the first catalytic residue of the integrase protein. This well-characterized mutant is incapable of mediating either the terminal processing or integration functions of integrase (5, 6, 16). Previously published studies demonstrate that viruses deficient in integrase function produce larger quantities of 2-LTR circles compared to wild-type virus but with similar kinetics (12, 15). We reasoned that the increased number of circles generated by the D64N mutant virus would allow more accurate analysis of decay rates in a setting where none of the cells became productively infected. MT-2 cells were infected with DNase I-treated HIV-1 D64N mutant as described above. Total cell number and the proportion of dead cells were monitored over the course of infection. MT-2 cells infected with the HIV-1 D64N mutant doubled every 27.7 h (95% CI, 25.7 to 30.2 h) (Fig. 3A). Cell viability in HIV-1 D64N mutant-infected cells averaged 98.2%. As was observed in cultures infected with wild type HIV-1, the number of 2-LTR circles/μg of DNA decreased with a slope that was very close to the inverse slope of the doubling time of the cells (Fig. 3B). In the experiment shown, the t1/2 was 28.3 h (95% CI, 26.1 to 30.9 h), which was statistically indistinguishable (P = 0.692) from the doubling time of the infected culture. In contrast, the decay of total HIV-1 DNA (t1/2, 21 h; 95% CI, 17.8 to 25.6 h) differed significantly, reflecting the fact that the D64N mutant virus is incapable of integration (P = 0.0426). This more rapid decay likely reflects the contribution of the rapid decay of linear unintegrated molecules in the absence of stable integrated forms (T. Pierson and R. F. Siliciano, unpublished results). Analysis of the data combined from both wild-type and D64N mutant infections indicated no additional decay of 2-LTR circles relative to the rate of cell division (P = 0.400).
FIG. 3.
Decay of 2-LTR circles following infection with an integrase mutant HIV-1. MT-2 cells were infected as described with a virus carrying a single point mutation in integrase (D64N) that renders it incapable of multiple rounds of replication. (A) At the indicated times, cells were counted to determine the rate of proliferation, and a sample was harvested for quantification of 2-LTR circles and total HIV-1 DNA, as described in the legend to Fig. 1. (B) Observed decay of 2-LTR circles and total HIV-1 DNA measured by phosphorimager analysis.
In addition to determining the t1/2s of 2-LTR circles, we determined the residual decay in 2-LTR circles after taking the doubling time of the cells into account (Table 1). In some experiments, there was no residual decay after accounting for the dilutional effects of cells division, precluding the calculation of a t1/2. In other experiments, very slight residual decay was detected. The precision of the measurement of residual decay is indicated by the 95% confidence intervals about the residual decay t1/2s (Table 1). Even when the lower 95% confidence interval for the residual decay slope is used to estimate the most rapid decay consistent with the data, very slow decay is predicted (t1/2 = 112.8, 291.5, and 367.5 h in the experiments shown).
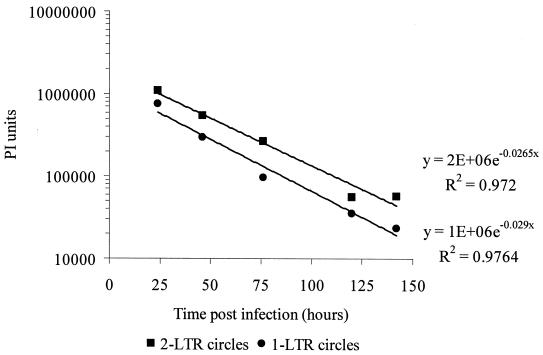
To determine whether 1-LTR circles are also stable, we monitored their decay in the same culture system. As was the case for 2-LTR circles, the decrease in the number of 1-LTR circles/μg of DNA could not be distinguished from the decrease expected from dilution (P = 0.1859). The stability of both 1- and 2-LTR circles was observed following infections of MT-2 cells with either wild-type virus (data not shown) or the D64N mutant HIV-1. Additionally, the rates of decay of 1- and 2-LTR circles were indistinguishable (P = 0.5225) (Fig. 4). Taken together, these data suggest that both circular forms of HIV-1 are highly stable.
FIG. 4.
Both 1- and 2-LTR circles decay at similar rates. The decay of circular forms of the viral genome was measured as described following the infection of MT-2 cells with D64N mutant HIV-1 IIIb. The observed decay rates of 2-LTR circles and 1-LTR circles were measured by phosphorimager analysis.
The results presented here demonstrate that the circular DNA forms of the HIV-1 genome are intrinsically stable in a CD4+ T-cell line. The decrease in 2-LTR circles observed in our in vitro experiments can be completely accounted for by dilution through cell division (Fig. 2). We suggest that the reported lability of the circles in vivo needs to be reevaluated before this assay can be used to measure recent HIV-1 infection of susceptible cells in infected individuals.
Extensive use of circular forms of HIV-1 in the study of viral dynamics has been reported (9, 21, 29). Interpreting these studies depends on understanding the stability of 1- and 2-LTR circles in HIV-1-infected individuals. The factors that regulate decay of the circles in T-cell populations in vivo are complex and may involve more than the intrinsic stability of these circular pieces of DNA. Studies of HIV-1 dynamics in response to antiretroviral therapy have revealed that the life span of productively infected CD4+ T lymphoblasts in vivo is relatively short (t1/2 of 1 to 2 days). The relatively rapid clearance of productively infected cells in vivo may be a function of the immune response and/or the cytopathic effects of the virus (reviewed in reference 26). This complex relationship between viral and cellular dynamics must be considered when evaluating the rate at which 2-LTR circles decay in vivo in response to HAART. Decreases in the number of circles detected in a cell population will reflect not only the intrinsic stability of the 2-LTR circles, but also the life span of the relevant population of infected cells and its distribution between the blood and other compartments. A recent study by Sharkey et al. (29) measured the decay of 2-LTR circles in vitro and in four HIV-1 infected individuals. The authors suggest that the circles are short-lived and are a reasonable measure of recent HIV-1 replication, but there are other possible interpretations. While a t1/2 for the decay of 2-LTR circles was not reported in this study, analysis of the fall in the number of 2-LTR circles in response to HAART suggests that 2-LTR circles in these patients decay with a t1/2 of 0.4 to 7 weeks. It is possible that these values reflect the decay or redistribution of cells bearing circles rather than the actual molecular decay of 2-LTR circles themselves. In the study by Sharkey et al., decay of the circles following in vitro infection was suggested, but the effects of cell proliferation and death were not described.
The present study took a reductionist approach to the analysis of circular forms of the viral genome. The intrinsic stability of circular forms at the molecular level was evaluated in vitro in a system in which results were not complicated by cell death or redistribution. There is no evidence that circular forms of HIV DNA allow high levels of transcription, and therefore cells carrying circles are not expected to die from viral cytopathic effects, especially since a vpr− virus (HIV-1 IIIb) was used. In fact, when infections were carried out at low MOIs, minimal cell death was evident in our culture system, and cells maintained a logarithmic state of growth. It should be noted that the death of cells bearing circles would produce a more rapid decay than expected from dilution, and therefore cell death cannot be an explanation for the stability observed here. Thus, having accounted for the effects of cell division and death, we demonstrate that circular forms of the viral genome are highly stable.
It is interesting that several other episomal DNA circles within the nucleus, both viral and cellular, have been shown to be highly stable. Circular intermediates of hepadnavirus infection have been shown to persist in infected liver cultures, decreasing in number only as a function of cell death (35). The long-term stability of T cell receptor excision circles has allowed their use in several studies as a marker of emergence of T lymphocytes from the thymus (4). Thus, it is not surprising that circular forms of HIV-1 DNA would be highly stable as well.
The ability of suppressive HAART regimens to reduce virus levels in plasma to a level below the limit of detection makes it imperative that assays capable of studying viral dynamics in individuals who have responded well to HAART be developed. In patients with measurable viremia, the extremely rapid rate of clearance of free virus from the plasma allows real-time measurements of viral replication in vivo (reviewed in reference 26). While the formation of 2-LTR circles occurs relatively soon after infection, our data suggest that the intrinsic stability of these circles may allow them to persist indefinitely should the infected cell survive and remain in the compartment being sampled. The in vitro stability of 2-LTR circles has recently been confirmed by the work of Butler et al. (1). Therefore, although measurement of 2-LTR circles in vivo offers some indication of previous viral replication, it may not provide the same type of real-time measure of viral replication that plasma HIV-1 RNA assays provide. In conclusion, our results raise the possibility that the use of 2-LTR circles as a surrogate marker of ongoing HIV-1 replication may be limited by the stability of these forms of HIV-1 DNA.
Acknowledgments
T.P. and T.L.K. contributed equally to this work.
We thank Stuart Ray, Daphne Monie, Rick Bushman, and members of the Siliciano laboratory for helpful discussions.
This work was supported by NIH grant AI 43222 to R.F.S.
REFERENCES
- 1.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism studied by fluorescence-monitored PCR: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 2.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, L. Livornese, Jr., M. J. Ingerman, J. Witek, R. J. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282:1627-1632. [DOI] [PubMed] [Google Scholar]
- 4.Douek, D. C., R. D. McFarland, P. H. Keiser, E. A. Gage, J. M. Massey, B. F. Haynes, M. A. Polis, A. T. Haase, M. B. Feinberg, J. L. Sullivan, B. D. Jamieson, J. A. Zack, L. J. Picker, and R. A. Koup. 1998. Changes in thymic function with age and during the treatment of HIV infection. Nature 396:690-695. [DOI] [PubMed] [Google Scholar]
- 5.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 8.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 9.Furtaldo, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 10.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 11.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, M. A. Fischl, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 12.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 13.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed] [Google Scholar]
- 14.Hockett, R. D., J. M. Kilby, C. A. Derdeyn, M. S. Saag, M. Sillers, K. Squires, S. Chiz, M. A. Nowak, G. M. Shaw, and R. P. Bucy. 1999. Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA. J. Exp. Med. 189:1545-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, T., K. Drlica, A. Pinter, and E. Murphy. 1991. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J. Virol. 65:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkosky, J., R. A. Katz, and A. M. Skalka. 1990. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J. Acquir. Immune Defic. Syndr. 3:852-858. [PubMed] [Google Scholar]
- 18.Lazdins, J. K., J. Mestan, G. Goutte, M. R. Walker, G. Bold, H. G. Capraro, and T. Klimkait. 1997. In vitro effect of alpha1-acid glycoprotein on the anti-human immunodeficiency virus (HIV) activity of the protease inhibitor CGP 61755: a comparative study with other relevant HIV protease inhibitors. J. Infect. Dis. 175:1063-1070. [DOI] [PubMed] [Google Scholar]
- 19.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panther, L. A., R. W. Coombs, S. A. Aung, C. dela Rosa, D. Gretch, and L. Corey. 1999. Unintegrated HIV-1 circular 2-LTR proviral DNA as a marker of recently infected cells: relative effect of recombinant CD4, zidovudine, and saquinavir in vitro. J. Med. Virol. 58:165-173. [DOI] [PubMed] [Google Scholar]
- 22.Panther, L. A., R. W. Coombs, J. E. Zeh, A. C. Collier, and L. Corey. 1998. Unintegrated circular HIV-1 DNA in the peripheral mononuclear cells of HIV-1-infected subjects: association with high levels of plasma HIV-1 RNA, rapid decline in CD4 count, and clinical progression to AIDS. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 17:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Pauza, C. D., P. Trivedi, T. S. McKechnie, D. D. Richman, and F. M. Graziano. 1994. 2-LTR circular viral DNA as a marker for human immunodeficiency virus type 1 infection in vivo. Virology 205:470-478. [DOI] [PubMed] [Google Scholar]
- 24.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 25.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 27.Shank, P. R., S. H. Hughes, H. J. Kung, J. E. Majors, N. Quintrell, R. V. Guntaka, J. M. Bishop, and H. E. Varmus. 1978. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 15:1383-1395. [DOI] [PubMed] [Google Scholar]
- 28.Shank, P. R., and H. E. Varmus. 1978. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J. Virol. 25:104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison, I. I. I., B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, J. S., S. Y. Kim, and M. J. Roth. 1990. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 64:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 64:4903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 33.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 34.Zazzi, M., L. Romano, M. Catucci, G. Venturi, A. De Milito, P. Almi, A. Gonnelli, M. Rubino, U. Occhini, and P. E. Valensin. 1997. Evaluation of the presence of 2-LTR HIV-1 unintegrated DNA as a simple molecular predictor of disease progression. J. Med. Virol. 52:20-25. [DOI] [PubMed] [Google Scholar]
- 35.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]