Abstract
The tumour suppressor ARF (alternative reading frame) is encoded by the INK4a (inhibitor of cyclin-dependent kinase 4)/ARF locus, which is frequently altered in human tumours. ARF binds MDM2 (murine double minute 2) and releases p53 from inhibition by MDM2, resulting in stabilization, accumulation and activation of p53. Recently, ARF has been found to associate with other proteins, but, to date, little is known about ARF-associated proteins that are implicated in post-translational regulation of ARF activity. Using a yeast two-hybrid screen, we have identified a novel protein, LZAP (LXXLL/leucine-zipper-containing ARF-binding protein), that interacts with endogenous ARF in mammalian cells. In the present study, we show that LZAP reversed the ability of ARF to inhibit HDM2's ubiquitin ligase activity towards p53, but simultaneously co-operated with ARF, maintaining p53 stability and increasing p53 transcriptional activity. Expression of LZAP, in addition to ARF, increased the percentage of cells in the G1 phase of the cell cycle. Expression of LZAP also caused activation of p53 and a p53-dependent G1 cell-cycle arrest in the absence of ARF. Taken together, our data suggest that LZAP can regulate ARF biochemical and biological activity. Additionally, LZAP has p53-dependent cell-cycle effects that are independent of ARF.
Keywords: alternative reading frame protein (ARF), C53, LXXLL/leucine-zipper-containing ARF-binding protein (LZAP), murine double minute 2 (MDM2), p53, ubiquitination
Abbreviations: ARF, alternative reading frame; CDK, cyclin-dependent kinase; DAPI, 4,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HA, haemagglutinin; HRP, horseradish peroxidase; INK4a, inhibitor of CDK4; IPTG, isopropyl β-D-thiogalactoside; KLH, keyhole-limpet haemocyanin; LZAP, LXXLL/leucine-zipper-containing ARF-binding protein; MDM2, murine double minute 2; HDM2, human homologue of MDM2; MEF, mouse embryonic fibroblast; pRb, retinoblastoma protein
INTRODUCTION
The tumour suppressor ARF (alternative reading frame) is a product of the INK4a [inhibitor of CDK (cyclin-dependent kinase) 4]/ARF locus, which encodes two unrelated cell-cycle inhibitors, p16INK4a and p14ARF (p19ARF in mice) [1]. These two proteins share nucleotide sequences in exons 2 and 3, but are specified by distinct first exons and are translated in different reading frames. The biochemical activities of the CDK inhibitor p16INK4a are well described. It binds cyclin-D-associated CDKs to prevent phosphorylation of the pRb (retinoblastoma protein), thereby maintaining pRb in its growth-suppressive state [2]. In contrast, ARF activates p53 through direct interaction with MDM2 (murine double minute 2) or HDM2 (human homologue of MDM2) in humans, the major negative regulator of p53 [3–5]. Activity of p53 is repressed by MDM2 through direct inhibition of p53 transcriptional activity [6] and through p53 degradation following HDM2-directed ubiquitination [7] and nuclear export [8]. ARF has been shown to antagonize all of these functions of MDM2 [9,10].
Acting as a sensor of hyperproliferative signals, ARF is induced by multiple oncogenes, including Myc [11], Ras [12], E1A [13], Abl [14] and E2F1 [15] (reviewed in [16]). Following oncogenic stimulation, increased levels of ARF induce nuclear accumulation and activation of p53, resulting in cell-cycle arrest and/or apoptosis [16]. ARF has also been reported to respond to other signals, such as DNA damage, microtubule disruption, morphological changes and short-lived perturbations in the cell cycle and in nucleolar function [17,18]. Furthermore, it has been reported that, in several human haemopoietic tumour cell lines that express abundant amounts of ARF mRNA, ARF protein cannot be detected, indicating that post-transcriptional regulation of ARF may play a role in certain tumour types [19]. A recent report also suggests that binding of Tat-binding protein 1 regulates ARF protein stability [20]. ARF stability has been found to be regulated by proteasomal degradation after N-terminal ubiquitination [21,22]. Although the transcriptional regulation of ARF by oncogenes and transcriptional factors has been demonstrated, little is known about the post-transcriptional regulation of ARF activity.
Several proteins, other than MDM2, have been reported to interact with ARF, including E2F1 [23,24], neurabin II [25], HIF-1α (hypoxia-inducible factor 1α) [26], Pex19P [27], CARF (collaborator of ARF) [28] and B23/nucleophosmin [29–32]. Some of these proteins alter ARF localization or ARF-dependent activation of p53 [25,27,28]. To identify ARF-interacting proteins that may regulate ARF activity, we screened for ARF-binding proteins using a yeast two-hybrid system. A novel human protein that bound ARF was identified in yeast and confirmed in mammalian cells. This protein is highly similar to a rat protein, C53, which has been identified previously using a yeast two-hybrid assay as interacting with p35, the precursor of CDK5 activator [33]. We show here that this LZAP (LXXLL/leucine-zipper-containing ARF-binding protein) directly and specifically binds to the N-terminal region of human ARF. Our data suggest that, upon direct binding to ARF, LZAP reverses ARF inhibition of HDM2's p53 ubiquitination activity. Remarkably, despite its ability to restore HDM2 ubiquitination of p53 in the presence of ARF, expression of LZAP did not lead to p53 degradation or decreased p53 transcriptional activity. Additionally, ectopic expression of LZAP resulted in a G1 cell-cycle arrest that was dependent on p53, but independent of ARF.
MATERIALS AND METHODS
Plasmid constructs
EST (expressed sequence tag) clone (clone ID CS0DI052YB01, GenBank® Accession number AL573636) containing LZAP cDNA was purchased from Invitrogen. The entire coding sequence of LZAP was amplified by PCR using the forward primer, 5′-CGGGGTACCCATGGAGGACCATCAGCACGTGCCC-3′, and the reverse primer, 5′-CCGTCTAGATCACAGAGAGGTTCCCATCAGGTTCAC-3′. The PCR product was digested with KpnI and XbaI and subcloned into the corresponding sites in the pcDNA3 and pcDNA3-FLAG expression vectors. All constructs derived from PCR products were verified by direct DNA sequencing. The pGL3-p21-luc p53-responsive luciferase reporter was a gift from Dr James J. Manfredi (Mount Sinai School of Medicine, New York, NY, U.S.A.). The p53 mutant pCEP4-p53V143A was kindly provided by Dr Jennifer A. Pietenpol (Vanderbilt University, Nashville, TN, U.S.A.). The yeast two-hybrid plasmid containing LZAP cDNA, pACT2-LZAP, was obtained from screening the human brain cDNA library using ARF as bait.
Yeast two-hybrid assay
The procedure of yeast two-hybrid has been described previously [3]. Briefly, yeast HF7C cells were co-transformed with pACT2 and pGBT8 constructs. Yeast transformed by both constructs were verified by growth on medium lacking leucine and tryptophan (−LW), and interaction between two fusion proteins was identified by yeast growth on medium lacking leucine, tryptophan and histidine (−LWH), and confirmed using a β-galactosidase activity assay.
Cell culture and transfection
Mammalian tumour cell lines were maintained at 37 °C under 5% CO2, in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) foetal bovine serum. All cell culture media and supplements were purchased from Invitrogen. Cells were plated 24 h before transfection, transfected at 30–50% confluence for cell-cycle analysis, and at 50–70% confluence for other experiments using FuGene6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. The total amount of transfected DNA in any single experiment was kept constant by adding empty vector (pcDNA3) as necessary.
Antibodies and immunochemistry procedures
Rabbit anti-human LZAP serum, 3486, was produced in male New Zealand white rabbits against the KLH (keyhole-limpet haemocyanin)-conjugated synthetic peptide CDISKRYSGRPVNLMGTSL corresponding to amino acid residues 489–506 of human LZAP. The first N-terminal cysteine residue was added to facilitate covalent KLH conjugation. This anti-LZAP antibody was affinity-purified using a Sulfolink kit (Pierce) following the manufacturer's instructions, which was used in immunofluorescence; and also was conjugated with HRP (horseradish peroxidase) for immunoblotting by the Vanderbilt Molecular Recognition Shared Resource. Mouse monoclonal antibodies specific to FLAG tag, M2 and HRP-conjugated M2 were purchased from Sigma; mouse anti-ARF antibody 14P02, mouse anti-Ku antibody Ab-2 and anti-actin antibody pan Ab-5 were purchased from Neomarker; flurochrome-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories; and other antibodies were from Santa Cruz Biotechnology.
Procedures for immunoprecipitation and immunoblotting have been described previously [3]. Briefly, cells cultured in 100-mm-diameter dishes were lysed in 0.5% (v/v) Nonidet P40 lysis buffer supplemented with protease inhibitor cocktail (Roche). Cell lysates were immunoprecipitated by incubating with specific antibodies followed by Protein A- or Protein G-conjugated agarose beads (Invitrogen) at 4 °C. Agarose beads were then washed three times with lysis buffer and resuspended in Laemmli buffer (4% SDS, 0.02% Bromophenol Blue and 100 mM Tris/HCl, pH 6.8) for gel electrophoresis followed by immunoblotting.
The procedure of indirect immunofluorescence was previously described [10]. Briefly, cells growing on six-well culture dishes were fixed in 1 ml of buffered 3% (w/v) paraformaldehyde for 10 min at room temperature (22 °C), permeabilized in 0.2% (v/v) Triton X-100 at 4 °C for 5 min, and blocked with 0.5% (v/v) BSA. Target proteins were visualized by immunostaining with specific primary antibodies for 1 h at room temperature followed by appropriate FITC- or rhodamine-conjugated secondary antibodies for 30 min.
Sequential immunoprecipitation
LZAP was immunoprecipitated from 250 μg of cell lysate twice using 5 μl of rabbit anti-LZAP serum (3486), and cleared with Protein A-conjugated agarose beads for a total of three times. Lysate after LZAP depletion was then similarly depleted of HDM2 by using mouse anti-HDM2 antibody (SMP14). Finally, ARF was immunoprecipitated using 3 μl of rabbit anti-ARF serum (10437). Repeated immunoprecipitates using identical antibodies were combined, separated by electrophoresis and immunoblotted as described previously.
Ubiquitin ligase assays
The procedure for ubiquitin ligase activity assay was essentially the same as has been previously described [34]. For the in vivo ubiquitin ligase assay, U2OS (human osteosarcoma) cells in a 100-mm-diameter dish were transfected with plasmids including pSK-HA-Ub (0.5 μg), pCMV-p53 (0.5 μg), pCMV-HDM2 (0.5 μg), pCI-neo-ARF (1 μg) and pcDNA3-LZAP (2 μg). The total amount of plasmid DNA in each transfection was adjusted to 4.5 μg with an empty pcDNA3 vector when needed. At 2 days after transfection, cells were treated with 25 μM proteasome inhibitor MG132 for 4 h. Cells were then lysed in 100 μl of SDS lysis buffer (50 mM Tris/HCl, pH 7.5, 0.5 mM EDTA, 1% SDS and 1 mM dithiothreitol) by boiling for 10 min and were diluted in 1 ml of 0.5% (v/v) Nonidet P40 buffer. p53 was immunoprecipitated from cell lysates with 0.6 μg of goat polyclonal anti-p53 antibody (FL393), resolved by SDS/15% PAGE followed by immunoblotting with mouse monoclonal anti-HA (haemagglutinin) antibody (F7).
For in vitro ubiquitin ligase assays, U2OS cells were transfected as in the in vivo assay excluding pSK-HA-Ub. At 2 days after transfection, cells were lysed, and protein complexes were immunoprecipitated with 0.6 μg of mouse anti-HDM2 antibody (SMP14) or 2 μl of rabbit anti-ARF serum (10437). Immunoprecipitates immobilized on agarose beads were washed, and 30 μl of ubiquitination reaction mixture containing 10 nM okadaic acid, 2 mM ATP, 0.75 μg of ubiquitin, 60 ng of E1 and 300 ng of E2 within the reaction buffer (50 mM Tris/HCl, pH 7.4, 5 mM MgCl2, 2 mM NaF and 0.6 mM dithiothreitol) was added. Reaction mixtures were incubated at 37 °C for 1 h with gentle shaking at 100 rev./min. The reaction then was terminated by adding 30 μl of 2× Laemmli buffer and boiling for 5 min. p53 was resolved by SDS/15% PAGE and visualized by immunoblotting with goat polyclonal anti-p53 antibody (FL393).
Luciferase reporter assay
U2OS cells cultured in six-well dishes were transfected with 500 ng of pCI-neo-ARF and/or 500 ng of pcDNA3-LZAP together with 100 ng of the pGL3-p21-luc reporter and 10 ng of pRL-TK (Promega) as the internal control, which expresses luciferase from Renilla reniformis under the regulation of HSV-TK (herpes simplex virus thymidine kinase) promoter. Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega), and light emission was quantified using a Monolight 2010 luminometer (BD Biosciences). To control for transfection efficiency, firefly luciferase results after normalization to Renilla luciferase are presented, but similar effects were observed before normalization to this control.
Stability of p53
Cycloheximide-stability assay
RKO cells cultured in six-well dishes were transfected with 1 μg of total DNA constructs including 0.1 μg of pCMV-p53, 0.2 μg of pCMV-HDM2, 0.2 μg of pCI-neo-ARF and/or 0.5 μg of pcDNA3-LZAP. At 2 days after transfection, 60 μg/ml cycloheximide was added to inhibit further protein synthesis. At the indicated time points after cycloheximide treatment, cells were trypsinized and lysed in 1% (v/v) SDS lysis buffer. The samples were subjected to SDS/15% PAGE and immunoblotting.
Pulse–chase assay
RKO cells in six-well plates were transfected with plasmids including 0.1 μg of pCMV-p53, 0.4 μg of pCMV-HDM2, 0.2 μg of pCI-neo-ARF and/or 0.5 μg of pcDNA3-LZAP. At 2 days after transfection, cells were incubated in 1 ml of pre-warmed medium without methionine and cysteine for 40 min and were pulselabelled for 4 h with 100 μCi of EasyTag EXPRESS35S protein labelling mix (PerkinElmer). After labelling, cells were washed with pre-warmed PBS, incubated in complete medium for various times as indicated. p53 was immunoprecipitated using 0.6 μg of goat anti-p53 antibody (FL393), separated by electrophoresis and visualized with a PhosphoImager (FLA-5000; FujiFilm). Band intensity was quantified after subtraction of background using ImageGauge software (version 4.1; FujiFilm).
Detection of ubiquitinated p53 in subcellular fractions
U2OS cells were transfected with plasmids as described in the in vitro ubiquitin ligase assay. At 2 days after transfection, cells were treated with 25 μM proteasome inhibitor (MG132) for 4 h, trypsinized, resuspended and incubated on ice for 5 min in 1 ml of cytoplasmic extraction buffer containing 33 μg/ml digitonin, 20 mM Hepes, pH 7.4, 110 mM sodium acetate, 2 mM magnesium acetate and 0.5 mM EDTA. Cell nuclei were pelleted by centrifugation at 16100 g for 10 min, and the supernatant was collected as the cytoplasmic fraction. The nuclear pellet was washed twice with 1 ml of ice-cold PBS, lysed and diluted as described in the in vivo ubiquitin ligase assay, and designated as the nuclear fraction. From both cytoplasmic and nuclear fractions, p53 was immunoprecipitated with 0.6 μg of goat anti-p53 antibody (FL393), and immunoblotted with a mouse anti-p53 monoclonal antibody (DO1). Human cytoplasmic protein GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and nuclear protein Ku were immunoblotted as markers of cytoplasmic and nuclear fractions with rabbit anti-GAPDH antibody (FL335) and mouse anti-Ku antibody (Ab-2) respectively.
Heterokaryon assay
Nucleocytoplasmic shuttling was detected using a heterokaryon assay. U2OS cells cultured in p100 dishes were transfected with plasmids as described in the ‘Ubiquitin ligase assays’ Materials and methods section. At 5 h after transfection, cells were trypsinized, and 2×105 cells were seeded on a six-well tissue culture dish together with p53−/− MEF (mouse embryonic fibroblast) cells at a ratio of 1:1. After 16 h, mixed cells were treated with 60 μg/ml cycloheximide for 30 min, then fused by addition of 500 μl of pre-warmed 50% (v/v) poly(ethylene glycol) 1500 at 37 °C for 150 s. Cells then were washed with pre-warmed PBS three times and cultured further for 1 h in medium containing 60 μg/ml cycloheximide. Thereafter, cells were fixed, and ectopic proteins were visualized by indirect immunofluorescence and nuclei were stained with 5 ng/ml DAPI (4,6-diamidino-2-phenylindole; Sigma).
Flow cytometry and clonogenic growth analysis
Cells cultured in 100-mm-diameter plates were transfected at low density (30–50%) with pCI-neo-ARF (1.5 μg) and/or pcDNA3-LZAP (1.5 μg) as well as 0.3 μg of plasmid directing expression of GFP (green fluorescent protein)–spectrin. NARF2 and NARF2-E6 cells were treated with 1 mM IPTG (isopropyl β-D-thiogalactoside) 24 h before fixation. At 2 days after transfection, cells were trypsinized and fixed in 70% ethanol, stained with 10 μg/ml propidium iodide, and the cell-cycle distribution of at least 5000 GFP-positive cells was analysed by flow cytometry as described previously [35].
For clonogenic growth analyses, cells were infected with control retrovirus (pHIT-puro) or retroviruses directing expression of ARF or LZAP (pHIT-puro-ARF or pHIT-puro-LZAP). At 1 day after infection, 1 μg/ml puromycin was added to the medium. On day 3 after infection, aliquots of cells were lysed for SDS/15% PAGE and immunoblotting, and equal numbers of cells were split on to 150-mm-diameter dishes and were selected for a total of 2 weeks before counting colonies.
RESULTS
LZAP binds ARF
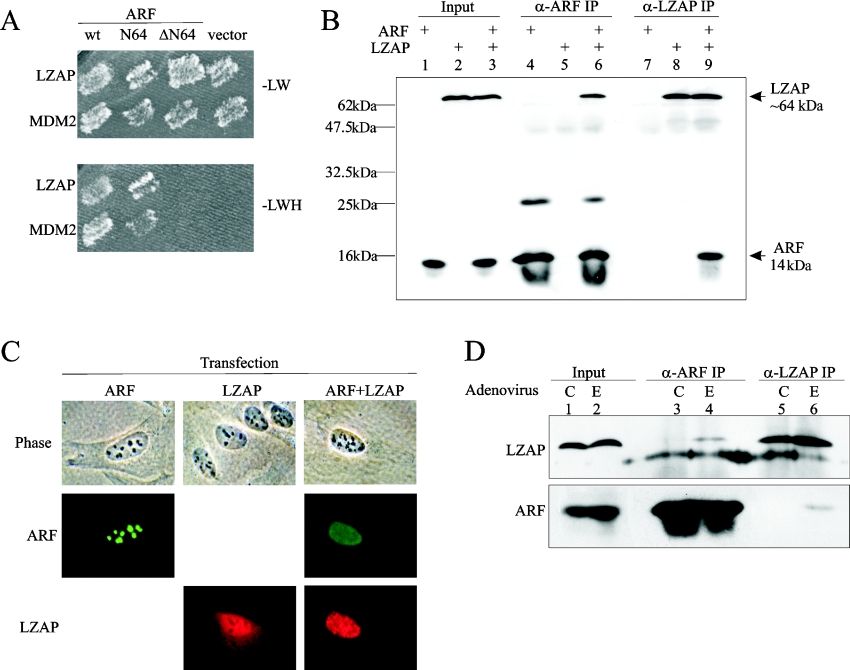
To identify proteins that are capable of interacting with human ARF, we screened a human brain cDNA library in a yeast two-hybrid system using full-length ARF as bait. ARF-interacting clones (232) were isolated and ARF binding of a novel protein, LZAP, was confirmed in yeast (Figure 1A). Rabbit polyclonal anti-LZAP antibody (3486) was made against a KLH-conjugated peptide containing the C-terminal 18 amino acids of LZAP that are conserved between human and mouse proteins. Binding between ARF and LZAP was confirmed in human cells, by expressing ARF and/or LZAP in U2OS cells followed by co-immunoprecipitation (Figure 1B). U2OS cells were chosen because they do not express ARF, but do express low levels of LZAP. Neither anti-ARF nor anti-LZAP antibodies precipitated the reciprocal protein in the absence of expressed target.
Figure 1. LZAP binds to ARF.
(A) Yeast HF7C cells were co-transformed with the indicated plasmids, and grown on medium lacking leucine and tryptophan (−LW) to verify the presence of both bait (pGBT8, Leu+) and prey (pACT2, Trp+) plasmids. Only interactions between bait and prey proteins allow growth on medium lacking leucine, tryptophan and histidine (−LWH). N64, amino acid residues 1–64; ΔN64, amino acid residues 65–132. (B) U2OS cells were transfected with pCI-neo-ARF and pcDNA3-LZAP as indicated. Immunoprecipitates (IP) were prepared using a rabbit anti-ARF (α-ARF) serum (10437) or a rabbit anti-LZAP (α-LZAP) serum (3486), separated by SDS/15% PAGE and immunoblotted using HRP-conjugated 3486 and mouse anti-ARF monoclonal antibodies (14P02). Molecular-mass sizes are given in kDa. (C) U2OS cells were transfected with pCI-neo-ARF and/or pcDNA3-LZAP, and fixed cells were immunostained with mouse anti-ARF antibody (14P02) and affinity-purified rabbit anti-LZAP antibody (3486) before visualization by indirect immunofluorescence (ARF–FITC, LZAP–Rhodamine). (D) H1299 cells were infected by control adenovirus (C) or adenovirus directing expression of E2F1 (E) at MOI (multiplicity of infection) of 30. At 30 h after infection, cells were lysed, and ARF–LZAP complexes were detected as described in (B). E2F1 induced ARF expression 2.5-fold in this representative experiment.
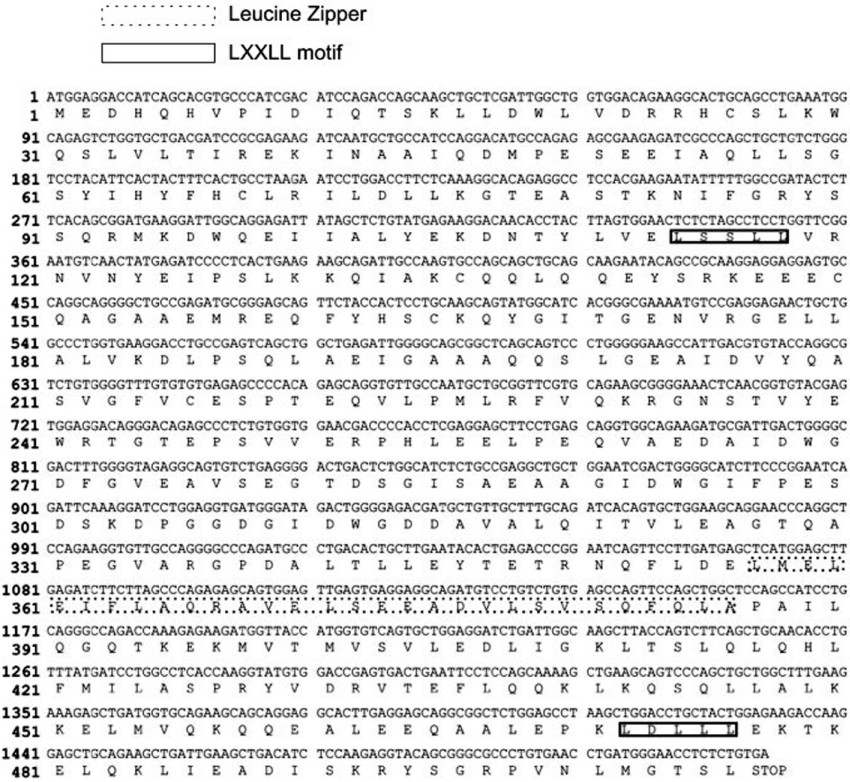
Database queries to identify homologues and conserved domains revealed that LZAP shared no significant amino acid homology with any known protein and had no conserved functional domains, except a putative leucine zipper (amino acids 357–385) and two LXXLL motifs (amino acids 114–118 and amino acids 472–476) (Figure 2). To identify endogenous expression of LZAP, eight cultured human cell lines from multiple tumour types and tissue origins, as well as one mouse cell line, were examined by immunoblotting. LZAP was detected in all cell lines examined, but expression levels varied (results not shown). In these cell lines, no obvious relationship was identified between LZAP protein levels and known ARF expression or p53 mutation status.
Figure 2. Nucleotide and corresponding amino acid sequences of LZAP.
The leucine zipper and the LXXLL motif of LZAP are indicated.
ARF and LZAP co-localize and alter subcellular localization of one another
Since endogenous LZAP was not detectable by immunofluorescence, subcellular localization of LZAP was determined in transfected U2OS cells, which lack endogenous ARF (endogenous LZAP is detectable by Western blotting with anti-LZAP, 3486). Proteins were visualized by fluorescence microscopy after indirect immunofluorescence staining (Figure 1C). ARF, when expressed alone, was primarily nucleolar. In contrast, in the absence of ARF, LZAP localized to both cytoplasm and nucleus, but was clearly excluded from the nucleolus in all cells examined (Figure 1C). Localization patterns of expressed LZAP were similar in H1299 (human large cell lung carcinoma) and Saos2 (human osteosarcoma) cells (results not shown). ARF localization was significantly altered from primarily nucleolar to both nuclear and nucleolar when expressed with LZAP. Likewise, LZAP localization was altered if ARF was co-expressed. Nucleolar LZAP, which was never detected in cells expressing LZAP without ARF, was found in the majority of ARF-expressing cells (88%). Co-expression of ARF also decreased cytoplasmic localization of LZAP (51% in cells expressing ARF and LZAP, 100% in cells expressing LZAP alone; Figure 1C, and results not shown). These data suggest that co-expression of ARF and LZAP results in alteration of ARF and LZAP subcellular localization with resultant co-localization of these two proteins. All results presented are derived from the examination of at least 200 cells from at least three independent experiments.
Oncogenic stimulation of endogenous ARF induces ARF–LZAP interaction
Our results suggest that exogenously expressed ARF and LZAP bind one another in both yeast and human cells and that subcellular localization patterns of LZAP and ARF are altered, resulting in co-localization. To determine whether endogenous ARF and LZAP bound to one another, we analysed H1299 cells, because these cells express both ARF and LZAP. An ARF–LZAP complex was not detected in untreated H1299 cells using either ARF- or LZAP-specific antibodies (Figure 1D, lanes 3 and 5). Lack of ARF–LZAP interaction may be based on compartmentalization of ARF to the nucleolus with LZAP excluded from the nucleolus, as suggested by our localization studies (Figure 1C). To determine whether increased endogenous ARF levels would bind to endogenous LZAP, we induced endogenous ARF by expression of E2F1. In E2F1 stimulated H1299 cells, binding of endogenous ARF and LZAP was detected using antibodies specific to either ARF or LZAP (Figure 1D, lanes 4 and 6). ARF levels were only moderately increased (2.5-fold) in response to E2F1, and LZAP levels were not significantly altered. Induction of the LZAP–ARF complex suggests that LZAP may play a role in the ARF-mediated response to the oncogenic stimulus of E2F1.
LZAP binds to the N-terminal region of ARF
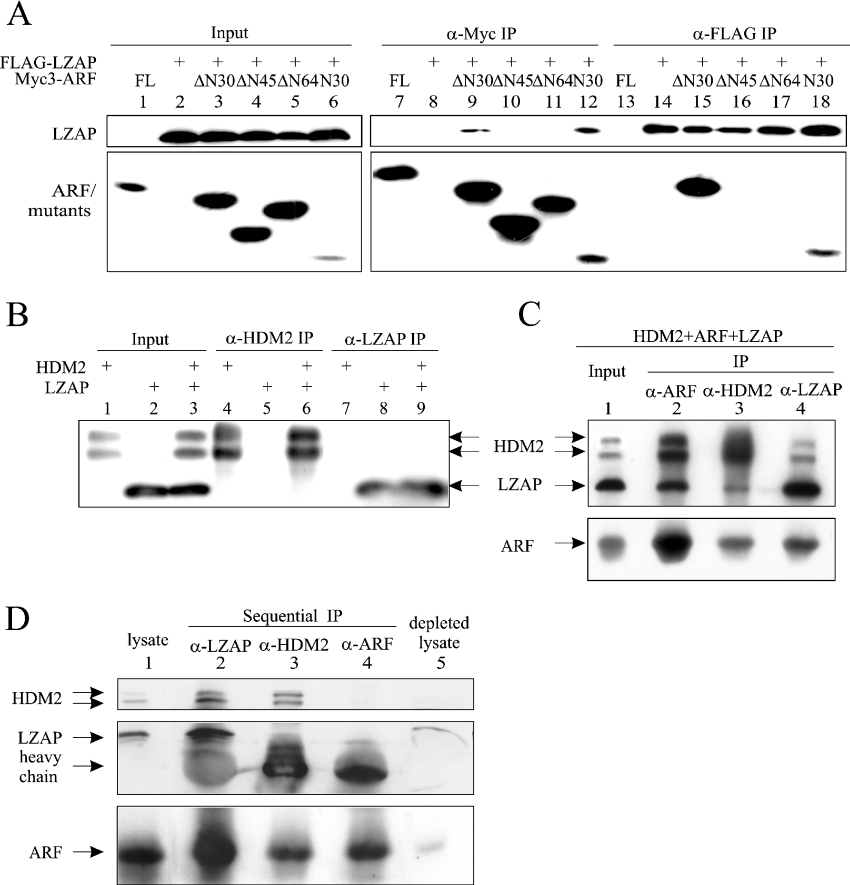
ARF-binding proteins described previously, including MDM2, interact with the first 64 amino acid residues of ARF. To initially identify the region of ARF that is important for LZAP binding, a yeast two-hybrid assay was performed. As expected, co-expression of HDM2 with the full-length or the N-terminal 64 amino acid residues of ARF (N64), but not the remainder of ARF (ΔN64), allowed HF7C yeast cells to grow on −LWH medium (Figure 1A). Likewise, full-length and N64 ARF, but not ΔN64 ARF, were able to interact with LZAP (Figure 1A). To define further the region of ARF that is critical for LZAP binding, FLAG-tagged full-length LZAP and truncation mutants of Myc-tagged ARF were co-expressed in U2OS cells, and then ARF or LZAP were immunoprecipitated from total cell lysates using either anti-FLAG or anti-Myc antibodies. Among the ARF mutants tested, LZAP reciprocally co-precipitated ARF peptides containing either the first 30 amino acid residues (N30) or the remainder of ARF (ΔN30), but not mutants lacking the N-terminal 45 (ΔN45, amino acids 46–132) or 64 (ΔN64, amino acids 65–132; note that ΔN64 has slower electrophoretic mobility than predicted) amino acid residues (Figure 3A). The ARF mutants N30 and ΔN30 do not share amino acid sequence, indicating that each contains at least one site sufficient to bind LZAP.
Figure 3. LZAP binds to the N-terminal region of ARF, and forms a ternary complex with ARF and HDM2.
(A) U2OS cells were transfected with pcDNA3-FLAG-LZAP and pcDNA3-Myc3-ARF (full-length or deletion mutants, see text for details) as indicated. Protein complexes were immunoprecipitated (IP) with anti-FLAG (α-FLAG) (M2) or anti-Myc (α-Myc) (9E10) antibodies, and proteins were detected by blotting with HRP-conjugated M2 or 9E10 antibodies. The Myc3-tagged ΔN64–ARF revealed an unusually slower mobility than the Myc3-tagged ΔN45–ARF, with relevant plasmids verified by direct sequencing. (B) U2OS cells were transfected with pcDNA3-LZAP and/or pCMV-HDM2. Immunoprecipitates (IP) were prepared by antibodies recognizing HDM2 (α-HDM2) (SMP14) or LZAP (α-LZAP) (3486) before SDS/15% PAGE and immunoblotting with the corresponding antibodies. (C) U2OS cells were transfected as indicated and immunoprecipitates (IP) were prepared by antibodies specifically recognizing HDM2 (α-HDM2), LZAP (α-LZAP) or ARF (α-ARF), separated by SDS/15% PAGE and immunoblotted with corresponding antibodies. (D) LZAP, HDM2 and ARF were sequentially depleted from 250 μg of cell lysate as described in the Materials and methods section. Immunoprecipitates (IP), as well as the original and depleted lysates, were subjected to SDS/15% PAGE and immunoblotting.
LZAP, ARF and HDM2 form a ternary complex
All known activities of ARF map to the exon 1β encoded N-terminal region, which is necessary and sufficient for binding of HDM2 [36,37]. Our data suggest that the LZAP-binding site of ARF at least partially overlaps the HDM2-binding site, raising two possibilities: (i) LZAP competes with HDM2 for ARF binding, and (ii) LZAP, ARF and HDM2 can exist in a ternary complex. To explore these possibilities, we first expressed LZAP and HDM2 singly or together in U2OS cells to determine whether they bound one another directly in the absence of ARF. No binding of HDM2 and LZAP was detected in U2OS cells that do not express ARF (Figure 3B). To determine whether binding of HDM2 and LZAP to ARF were mutually exclusive, LZAP, ARF and HDM2 were expressed in U2OS cells, and protein complexes were immunoprecipitated. As expected, ARF immunoprecipitates contained both LZAP and HDM2 (Figure 3C, lane 2). Even though LZAP and HDM2 binding sites of ARF at least partially overlap, immunocomplexes precipitated by antibodies specific to either LZAP or HDM2 contained all three proteins (Figure 3C, lanes 3 and 4), suggesting that LZAP, ARF and HDM2 can form a ternary complex bridged by ARF. However, these data do not rule out the possibility that complexes of ARF singly bound to either LZAP or HDM2 exist when all three proteins are simultaneously expressed.
To help to determine the proportion of HDM2 and ARF bound to LZAP, we performed sequential immunoprecipitation of U2OS cell lysate following expression of the three proteins (Figure 3D). LZAP was depleted from the lysate by two rounds of immunoprecipitation. Consistent with our previous observation, LZAP immunoprecipitates contained both ARF and HDM2 (Figure 3D, lane 2). To determine the portion of HDM2 bound to LZAP, HDM2 was then twice immunoprecipitated from the LZAP-depleted lysate. Comparison of HDM2-bound (Figure 3D, lane 2) and -unbound (Figure 3D, lane 3) to LZAP suggests that in these cells a significant portion of HDM2 was complexed with LZAP and ARF. Similarly, ARF immunoprecipitation from the HDM2- and LZAP- depleted lysate revealed that a substantial portion of cellular ARF was bound to LZAP (Figure 3D, compare lane 2 with lanes 3 and 4). Immunoblotting of the triply depleted lysate revealed that all proteins were efficiently immunoprecipitated with only a small amount of ARF being detectable.
LZAP reverses ARF's abrogation of HDM2-mediated ubiquitination of p53
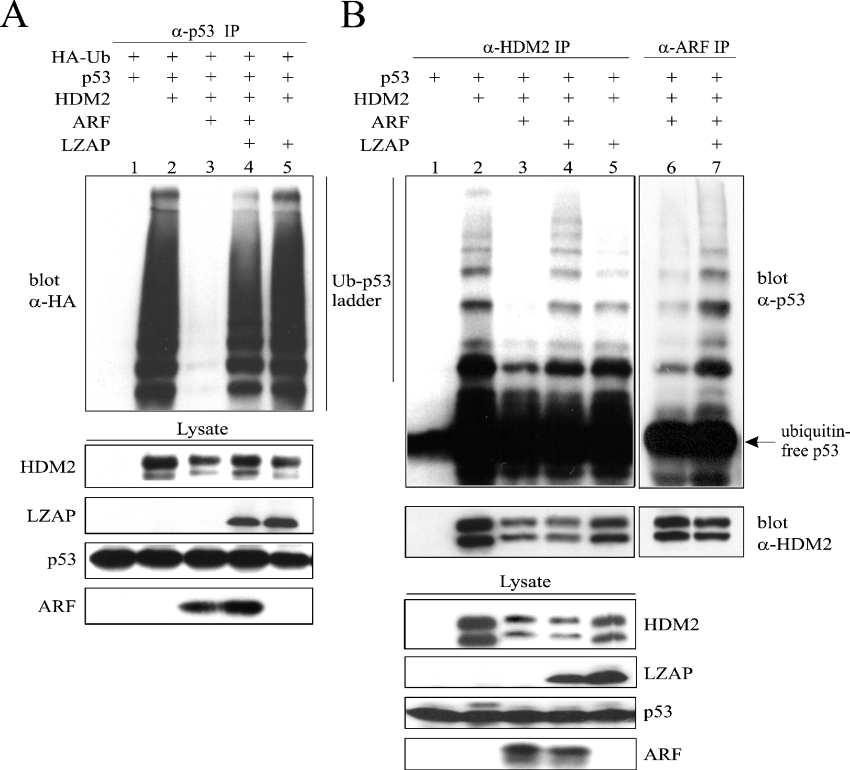
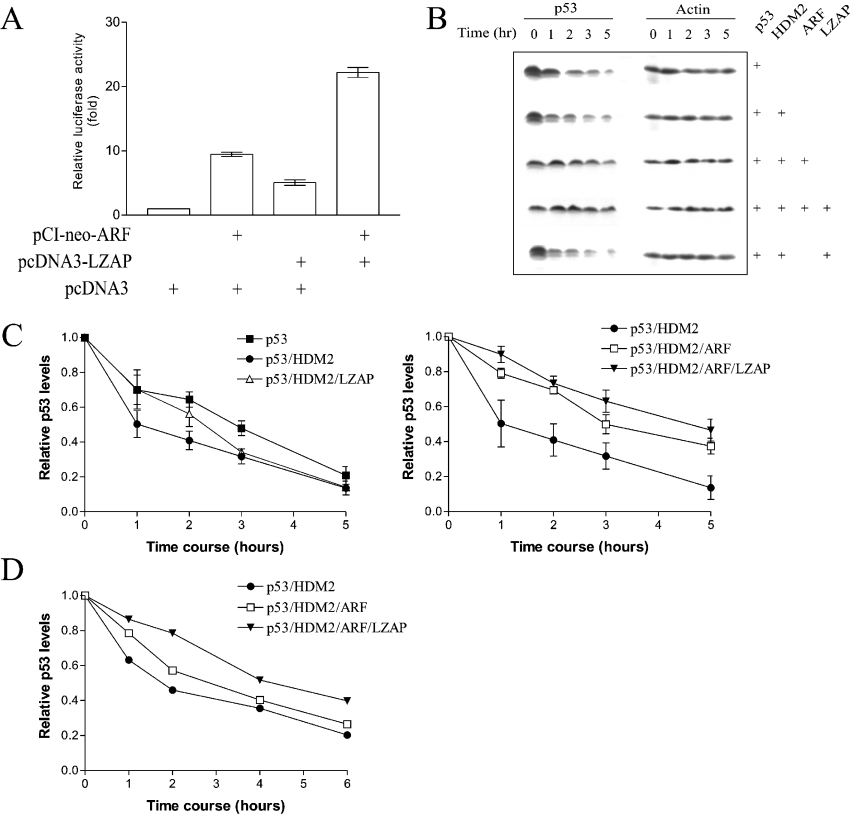
ARF inhibits HDM2 ubiquitin E3 ligase activity towards p53 in vitro and is thought to do so in vivo [38]. Our data suggest that LZAP binds ARF without disruption of the HDM2–ARF complex, raising the possibility that LZAP may regulate ARF inhibition of HDM2-directed p53 ubiquitination. To test this hypothesis, we employed both in vivo and in vitro E3 ubiquitin ligase assays. The in vivo ubiquitin ligase assay system takes advantage of endogenous E1 and E2. Transfection of U2OS with combinations of plasmids as indicated plus pSK-HA-Ub was followed by p53 immunoprecipitation and HA blotting to detect ubiquitinated p53 (Figure 4A). Before immunoprecipitation, the 26 S proteasome was inhibited with MG132 to allow accumulation of ubiquitinated proteins. As expected, co-expression of HDM2 and p53 produced a ubiquitinated p53 ladder, which was markedly decreased by expression of ARF (Figure 4A, lanes 2 and 3). Surprisingly, expression of LZAP in addition to p53, HDM2 and ARF restored the ubiquitinated p53 ladder (Figure 4A, lane 4), suggesting that LZAP could reverse ARF inhibition of HDM2-mediated p53 ubiquitination. This is remarkable, since ARF strongly inhibits HDM2's E3 ubiquitin ligase activity towards p53. In the absence of ARF, LZAP did not measurably alter p53 ubiquitination (Figure 4A, lane 5).
Figure 4. LZAP restores HDM2 ubiquitin E3 ligase activity towards p53 in the presence of ARF.
(A) U2OS cells were transfected with combinations of plasmids as indicated and, 4 h before lysis, treated with the proteasome inhibitor MG132. Lysates were immunoprecipitated with 0.6 μg of goat anti-p53 antibody (α-p53) (FL393), and HA–ubiquitin-modified p53 was visualized using a HRP-conjugated mouse anti-HA antibody (α-HA) (HRP-F7). (B) U2OS cells were transfected as indicated. Immunocomplexes immobilized on Protein G–beads were prepared using mouse anti-HDM2 antibody (α-HDM2) (SMP14, lanes 1–5) or anti-ARF serum (α-ARF) (10437, lanes 6 and 7), followed by an ubiquitination reaction. Identical lysates were immunoprecipitated (IP) in lanes 3 and 6, as well as lanes 4 and 7. Top panel: ubiquitinated p53 ladders were detected using goat anti-p53 antibody (α-p53) (FL393). Middle panel: the presence of HDM2 was confirmed by immunoblotting with SMP14. Bottom panels: for both in vivo and in vitro assays, lysates were separated by SDS/15% PAGE and immunoblotted with antibodies against the indicated proteins as a marker of protein expression (bottom panels). Ub, ubiquitin.
To confirm these results, an in vitro ubiquitination assay was performed. U2OS cells were transfected with combinations of plasmids as with the in vivo assay excluding HA–ubiquitin. HDM2 immunoprecipitates served as the source of both the E3 ubiquitin ligase and the substrate (p53). The ubiquitination reaction was performed following addition of purified E1, E2, ATP and ubiquitin to the immunocomplexes. As expected, HDM2 catalysed ubiquitination of p53 that was inhibited by ARF (Figure 4B, lanes 2 and 3). In the absence of ARF, HDM2 ubiquitin ligase activity towards p53 persisted in the presence of LZAP (Figure 4B, lane 5). Confirming the results of the in vivo ubiquitination assay, LZAP partially restored HDM2-directed p53 ubiquitination in the presence of ARF (Figure 4B, lane 4).
Despite our data suggesting that LZAP can bind to ARF without disrupting the ARF–HDM2 binary complex (Figures 3C and 3D), LZAP antagonism of ARF inhibition of HDM2 could occur through disrupting a portion of the ARF-HDM2 complexes, thus allowing free HDM2 to ubiquitinate p53. Since these ubiquitination assays measure enzymatic activity, even a small amount of active HDM2 could result in substantial p53 ubiquitination. Alternatively, LZAP may reverse ARF inhibition of HDM2 within an LZAP–ARF–HDM2 ternary complex. To elucidate further the mechanism by which LZAP abrogates ARF inhibition of HDM2, we repeated the in vitro ubiquitin ligase assay; however, instead of precipitating ubiquitin ligase complexes using anti-HDM2 antibody, we assessed the p53 ubiquitination capacity of HDM2 contained in anti-ARF immunoprecipitates. If LZAP inhibited ARF activity towards HDM2 through dissociation of a portion of ARF–HDM2 complexes, then, in the presence of LZAP, ARF immunoprecipitates should contain less HDM2 and have less p53 ubiquitin ligase activity. Notably, the immunoprecipitates prepared using anti-ARF antibody contained higher ubiquitin E3 ligase activity in the presence of LZAP (Figure 4B, lanes 6 and 7) despite having less or equivalent levels of HDM2 in the complex (Figure 4B, lanes 6 and 7, lower panel).
Although we have not ruled out the possibility that LZAP may recruit another p53-specific E3 ligase to the complex therefore negating the need for active HDM2, the most straightforward interpretation of the data including immunoprecipitation of HDM2 directly (Figure 4B, lanes 1–5) and of ARF-associated HDM2 (Figure 4B, lanes 6 and 7) is that the increased ubiquitin ligase activity seen in lanes 4 and 5 of Figure 4B is attributable to HDM2 that is in a complex with ARF and LZAP. Taken together, the ubiquitin ligase assays and the binding assays suggest that LZAP probably restores HDM2 ubiquitin E3 ligase activity towards p53 in the presence of ARF through formation of an LZAP–ARF–HDM2 ternary complex.
LZAP increases p53 transcriptional activity and does not destabilize p53
To decrease p53 protein levels and activity during normal cell growth, ubiquitinated p53 is targeted for proteasome-mediated degradation. In a cell line harbouring a temperature-sensitive mutant E1, the ubiquitin-activating enzyme, it was demonstrated that p53 degradation is largely ubiquitin-dependent [39]. However, recent evidence suggests that p53 ubiquitination alone is not sufficient to drive degradation [40]. Given our data that LZAP restores p53 ubiquitination in the presence of HDM2 and ARF, we wished to determine whether LZAP altered p53 transcriptional activity and stability. The transcriptional activity of p53 was determined using a Dual-Luciferase® Reporter Assay System with a p53-responsive firefly luciferase reporter (pGL3-p21-luc). Ectopic expression of ARF increased transcriptional activity of endogenous p53 (9.5-fold; Figure 5A). Surprisingly, expression of LZAP in addition to ARF resulted in a further increase in transcription of the p53-responsive reporter (22-fold; Figure 5A). In the absence of ARF, LZAP also increased p53 transcriptional activity (5-fold; Figure 5A). A similar pattern of p53-responsive luciferase reporter activation following expression of ARF and LZAP was observed in RKO cells (results not shown), which express p53 and HDM2, but lack ARF expression [41].
Figure 5. LZAP increases p53 transcriptional activity and does not destabilize p53.
(A) U2OS cells were transfected with the indicated plasmids in addition to 100 ng of pGL3-p21-luc (firefly luciferase) and 10 ng of pRL-TK (Renilla luciferase). Renilla luciferase activity was used as the internal control. Normalized firefly luciferase activity of cells transfected with vector only was assigned as 1. Results are means±S.E.M. for three independent experiments with three data points within each experiment. (B, C) RKO cells were transfected as indicated. Total cell lysates were prepared at 0, 1, 2, 3 and 5 h after cycloheximide treatment. Actin was blotted as the loading control. After cycloheximide treatment and immunoblotting, the amount of p53 protein at each time point was quantified (ImageQuant software version 5.0; Amersham Biosciences) and normalized based on actin expression. p53 levels at zero time was designated 1. Results in (C) are means±S.E.M. for three independent experiments. (D) RKO cells were transfected with the indicated plasmids and, after 2 days, were labelled with [35S]methionine/[35S]cysteine, followed by chase with unlabelled medium for the indicated times. p53 was immunoprecipitated from lysate and separated by SDS/15% PAGE. Bands corresponding to p53 were imaged and quantified using a PhosphoImager. Results are p53 band intensities after subtraction of background.
Our data suggest that expression of LZAP allowed p53 ubiquitination by HDM2 in the presence of ARF, yet increased p53 transcriptional activity. Ubiquitination of p53 need not inhibit transcriptional activity; however, ubiquitination-associated protein degradation is a major regulator of p53 activity. These seemingly conflicting data may be explained if LZAP in the presence of HDM2 and ARF: (i) represses degradation of ubiquitinated p53, (ii) increases p53 de novo synthesis, or (iii) increases ubiquitinated p53 transcriptional activity to compensate for increased degradation associated with ubiquitination of p53. To begin distinguishing between these possibilities, we determined p53 stability in the presence of HDM2, ARF and LZAP in RKO cells using cycloheximide to block protein synthesis (Figures 5B and 5C). At 2 days after transfection, protein synthesis was inhibited, and p53 protein levels were measured by immunoblotting and quantification of band intensity at the indicated time points (Figure 5C). As expected, ARF increased p53 stability in the presence of expressed HDM2. Notably, p53 stability following expression of LZAP in addition to HDM2 and ARF was not decreased relative to p53 stability following expression of ARF and HDM2 (Figure 5C). Similar results were observed in HeLa cells (results not shown). Also, LZAP in the absence of ARF did not result in destabilization of p53 (Figure 5C, p53/HDM2/LZAP). To rule out indirect effects of cycloheximide on p53 stability, a 35S pulse–chase experiment was performed in RKO cells (Figure 5D). Consistent with our cycloheximide data, p53 was not destabilized following expression of LZAP, ARF and HDM2 when compared with expression of ARF and HDM2. These data indicate that LZAP expression, despite increasing p53 ubiquitination in the presence of ARF and HDM2, does not result in destabilization of p53.
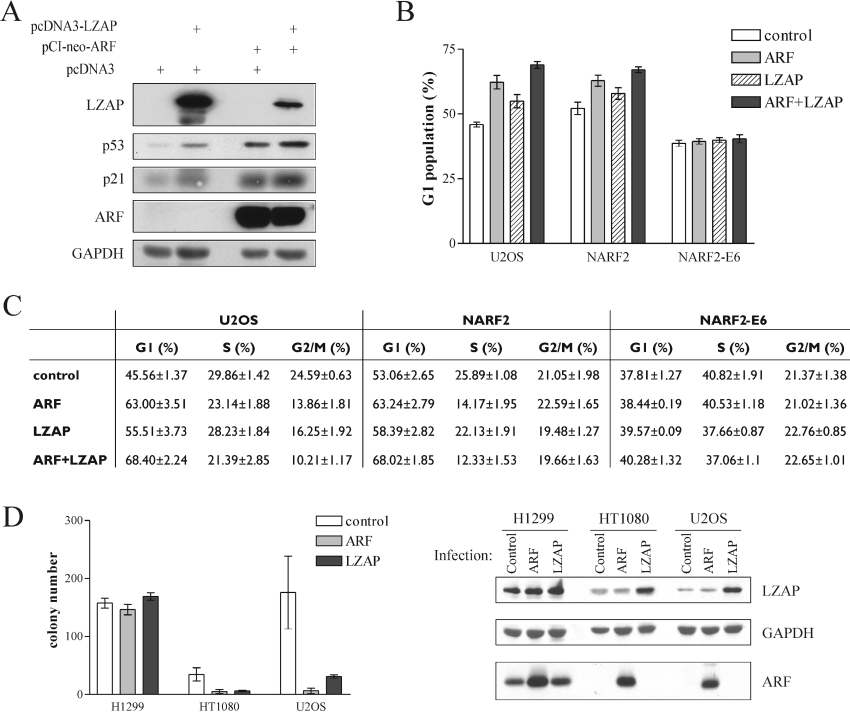
LZAP increases steady-state p53 levels and inhibits cellular proliferation
Since our data suggest that LZAP induces p53 transcriptional activity in U2OS cells, we wished to determine whether LZAP expression had biological consequences. A major downstream effector and transcriptional target of p53 is the CDK inhibitor p21. We determined p53 and p21 steady-state levels after transfection of U2OS cells with ARF sequences, LZAP sequences or the combination (Figure 6A). Expression of LZAP increased p21 levels less than 2-fold. Expression of ARF increased p21 levels 3.5-fold, and expression of LZAP and ARF resulted in the largest increase of p21 (3.9-fold). Increased p21 levels correlated with increased p53 levels following expression of LZAP or ARF. These results are in agreement with our luciferase reporter assays using a p21 promoter sequence (Figure 5A). To determine whether the increased p21 levels were associated with cell-cycle arrest, asynchronous U2OS cells were transfected with plasmids directing expression of ARF and/or LZAP as well as GFP–spectrin as a marker of transfected cells. After 2 days, the cell-cycle distribution of GFP-positive U2OS cells was analysed by flow cytometry (Figure 6C). On average, expression of ARF or LZAP singly, or the combination of ARF and LZAP, led to significant increases in the percentage of G1 cells (one-way ANOVA: P<0.0001, for the four groups) (Figure 6B). Combined LZAP and ARF expression resulted in G1 proliferative arrest that was significantly greater than cells expressing ARF (P<0.05) or LZAP (P<0.0001) singly.
Figure 6. LZAP increases p21 protein levels and inhibits cell proliferation.
(A) U2OS cells were transfected with the indicated plasmids and lysed 2 days after transfection. Protein levels were determined from cell lysates by immunoblotting with GAPDH serving as a loading control. (B, C) Cells were transfected with the indicated DNA constructs in addition to a plasmid directing expression of GFP–spectrin. ARF expression was induced in NARF2 and NARF2-E6 cells by treatment with 1 mM IPTG. Cell-cycle distribution of GFP-positive cells was determined by flow cytometry on a FACSCalibur (BD Bioscience) and analysed using ModFit 2.0 (Verity Software House). Results are means±S.E.M. for at least three independent experiments. (D) Cells were infected with retroviruses as indicated, then selected for 2 weeks before analyses of colony formation. Expression of LZAP and ARF was confirmed in infected cells after 2 days of selection by SDS/15% PAGE and immunoblotting. Results are means±S.E.M. for three experiments.
Since LZAP increased p53 transcriptional activity and G1 cell-cycle arrest in the absence of ARF (Figures 5A and 6B), we used isogenic U2OS-derived cell lines (NARF2 and NARF2-E6) to determine whether the ARF-independent cell-cycle arrest observed with LZAP expression was p53-dependent. Both NARF2 and NARF2-E6 cells inducibly express ARF, but p53 activity is blocked in NARF2-E6 cells by constitutive expression of human papillomavirus oncoprotein E6 [5]. As observed with U2OS cells, expression of LZAP with or without IPTG-induced ARF in NARF2 cells resulted in a significant increase of G1 cell population (one-way ANOVA: P=0.0002) (Figure 6B). In contrast, expression of LZAP and induction of ARF, either alone or in combination, did not cause a G1 arrest in NARF2-E6 cells (one-way ANOVA: P=0.3226) (Figure 6B). The G1 cell-cycle arrest caused by ectopic expression of LZAP in U2OS cells was also abolished by co-expression of p53V143A, a dominant-negative p53 mutant that is deficient in DNA binding (results not shown). Consistent with these observations, ectopic expression of ARF, LZAP or the combination of ARF and LZAP did not cause a cell-cycle arrest in two p53-deficient cell lines, H1299 and Saos2 cells (results not shown). Likewise, colony-formation assays were performed in p53-negative H1299 cells and p53-positive U2OS and HT1080 following infection with retroviruses directing expression of ARF or LZAP (Figure 6D). Expression of LZAP and ARF inhibited colony formation in both p53-positive cell lines, but not in the p53-negative H1299 cells.
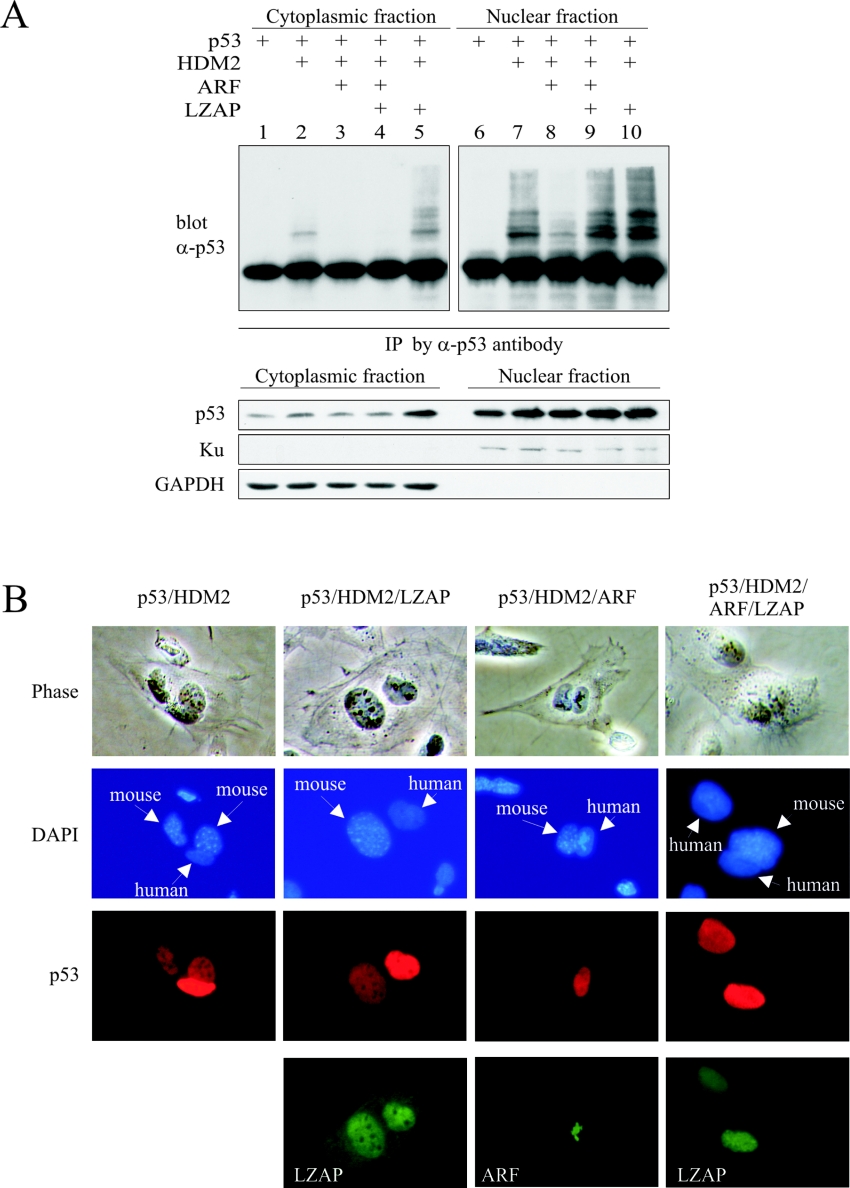
LZAP increases nuclear ubiquitinated p53
When expressed in the presence of HDM2 and ARF, LZAP restored p53 ubiquitination (Figure 4), but did not result in p53 degradation or decreased p53 transcriptional activity (Figure 5). To confirm these data and to explore potential mechanisms, we inhibited the 26 S proteasome and determined levels of ubiquitinated p53 in nuclear and cytoplasmic fractions of U2OS cells following expression of p53, HDM2, ARF and LZAP (Figure 7A). p53 was immunoprecipitated from nuclear and cytoplasmic lysates, and ubiquitinated p53 ladders were identified by p53 immunoblotting. As expected, ubiquitinated p53 was detected in cytoplasmic fractions of cells expressing HDM2 and p53 (Figure 7A, lane 2). Also, cytoplasmic ubiquitinated p53 was decreased in cells expressing ARF in addition to HDM2 and p53 regardless of the presence of LZAP (Figure 7A, lanes 3 and 4). Ubiquitinated p53 species were more abundant in the nuclear fractions (Figure 7A, lanes 6–10). As expected, HDM2 expression increased ubiquitinated p53 species, and co-expression of ARF decreased accumulation of nuclear ubiquitinated p53 (Figure 7A, lanes 6–8). In the presence of ARF and HDM2, LZAP restored nuclear ubiquitinated p53 species (Figure 7A, lane 9). Expression of LZAP in the absence of ARF did not alter ubiquitination patterns of p53 in either cytoplasmic or nuclear fractions. Human Ku and GAPDH immunoblotting confirmed nuclear and cytoplasmic fractionation. HDM2, LZAP and ARF expression was confirmed by immunoblotting (results not shown).
Figure 7. p53 is ubiquitinated and retained in nuclei in the presence of HDM2, ARF and LZAP.
(A) U2OS cells were transfected with the indicated plasmids. At 2 days after transfection, the proteasome was inhibited, and cytoplasmic and nuclear fractions of cell lysates were prepared. p53 was immunoprecipitated (IP) using a goat anti-(full-length p53) antibody (α-p53) (FL393), and ubiquitinated p53 ladders were detected by immunoblotting with mouse anti-p53 antibody (DO1). Human cytoplasmic protein GAPDH and nuclear protein Ku were blotted as the markers of cytoplasmic and nuclear fractions respectively. (B) Heterokaryon assay was performed by transfection of U2OS cells with the indicated plasmids before fusion with p53−/− MEFs. Nucleocytoplasmic shuttling of p53 was determined by indirect immunofluorescence. LZAP and ARF expression was verified by indirect immunofluorescence. Representative fused cells are shown from more than 30 examined for each experiment. Mouse nuclei were distinguished by punctate staining with DAPI.
Our data suggest that, in the presence of ARF and HDM2, LZAP restores HDM2-mediated p53 ubiquitination, but under these circumstances, ubiquitinated p53 is not destabilized and is primarily localized to the nucleus. To determine whether LZAP alters p53 nucleocytoplasmic shuttling, we performed a heterokaryon assay in which human U2OS cells expressing p53, HDM2, ARF and LZAP were fused with p53−/− MEFs (Figure 7B). If p53 shuttles from the human nucleus to the fused cellular cytoplasm, random re-import of p53 into either the mouse or human nucleus is possible. Shuttling is indicated by the presence of p53 in the p53−/− mouse cell nucleus. Localization of p53 was determined by indirect immunofluorescence, and mouse cell nuclei were identified by DAPI staining. As expected, p53 shuttling was observed in cells that expressed p53 and HDM2, as indicated by the presence of p53 in mouse nuclei (Figure 7B, p53/HDM2 column). p53 shuttling was not altered by the expression of LZAP in the absence of ARF (Figure 7B, p53/HDM2/LZAP column). As expected, expression of ARF in addition to p53 and HDM2 inhibited p53 shuttling (Figure 7B, p53/HDM2/ARF column). Likewise, no p53 shuttling was observed following expression of LZAP in the presence of ARF and HDM2 (Figure 7B, p53/HDM2/ARF/LZAP column). For all shuttling experiments, more than 30 heterokaryons were observed, and p53 shuttling was consistent in all scored cells.
These findings are consistent with our ubiquitination assays, as well as p53 stability and transcriptional activity data (Figures 4 and 5), and together suggest that LZAP in the presence of ARF and HDM2 allows for nuclear localization of ubiquitinated p53 forms. Taken together, our data suggest that ubiquitinated p53 in the presence of ARF and LZAP does not shuttle, and is localized to the nucleus where it is transcriptionally active.
DISCUSSION
p53 activity within the cell is tightly controlled, primarily through regulation of p53 stability. In embryonic development, stabilization of p53 as a result of homozygous loss of MDM2 results in embryonic lethality that can be rescued by simultaneous loss of p53, suggesting that, at least in development, MDM2/HDM2 is the major controller of p53 [42,43]. Because of its central role in regulation of p53, HDM2 activity is also regulated at multiple levels. ARF binds HDM2 and inhibits its E3 ubiquitin ligase activity towards p53, while simultaneously reversing HDM2's inhibition of p53 transactivation [36]. ARF expression is induced by multiple different oncogenic stimuli resulting in p53-dependent cell-cycle arrest and/or apoptosis [16,44]. In this capacity, ARF seems to direct a major cellular defence, protecting the organism from cells with excessive proliferative signalling and, as such, many human tumour types frequently lack expression of ARF. In this study, we identified and characterized a novel protein, LZAP, which regulates ARF activity towards HDM2. Our data suggest that LZAP also has ARF-independent cell-cycle regulatory activities that depend on p53.
At least two mechanistic models of ARF regulation of HDM2 exist: (i) ARF dissociates HDM2 from p53 and sequesters MDM2 in the nucleolus [9,45], and (ii) ARF binds to HDM2–p53 in the nucleus, forms a ternary complex and within the complex inhibits HDM2's ubiquitination and transcriptional inhibition of p53 [10,36]. The nucleolar sequestration model suggests that ARF must dissociate p53 from MDM2 and that sequestration of HDM2 in the nucleolus is vital for ARF activity. Direct experimental evidence has shown that ARF mutants excluded from nucleoli are capable of stabilizing and activating p53 equally as well as wild-type ARF [37], and recent data suggest that nucleolar ARF may be inactive towards p53 and that B23/nucleophosmin may play a role in antagonizing ARF activity by sequestering ARF in the nucleolus [32]. The role of nucleolar ARF in the regulation of p53 remains controversial.
Our data favours the ternary p53–HDM2–ARF complex model. In addition to the binding data (Figure 3) which suggest that HDM2, ARF and LZAP are capable of forming a complex, ubiquitination and p53 stability data support the existence of quaternary p53–HDM2–ARF–LZAP complexes. ARF complexes precipitated from in vitro ubiquitination assays (Figure 4B, lanes 6 and 7) not only contained p53 protein, but also contained p53 ubiquitination activity that was increased in the presence of LZAP. Had LZAP disrupted the p53–HDM2–ARF complex by competing for ARF, then ARF immunoprecipitates would contain LZAP, but no p53 and no HDM2 ubiquitin ligase activity towards p53. Likewise, if LZAP competed with HDM2 for ARF, the addition of LZAP to p53–HDM2–ARF complexes should decrease p53 stability and increase nuclear cytoplasmic shuttling. Since we observed no decrease in p53 stability and no nuclear cytoplasmic shuttling of p53 following the addition of LZAP in the presence of HDM2 and ARF (Figures 5 and 7), our data suggest that LZAP alters ARF activity within a complex containing p53–HDM2–ARF.
ARF inhibits HDM2-mediated p53 degradation while simultaneously inhibiting HDM2 ubiquitination of p53; however, p53 ubiquitination and degradation activities of HDM2 can be separated, since mutants of MDM2 have been described that can ubiquitinate p53, but that cannot direct p53 degradation [46]. Also, ARF inhibition of HDM2 ubiquitin ligase activity is not absolute since HDM2 bound to ARF still has ligase activity, just not towards p53 [47]. The fate of ubiquitinated p53, and the sequence and subcellular localization of p53 ubiquitination has been the subject of intense investigation and debate. Some studies suggest that p53 degradation requires nuclear export [8,48,49]. Other studies suggest that nuclear ubiquitinated p53 can be degraded without export [50,51]. Our data suggest that LZAP reverses ARF inhibition of HDM2 and allows p53 ubiquitination in the nucleus. Although ubiquitinated p53 exists in the presence of HDM2, ARF and LZAP, it is not exported from the nucleus (Figure 7). Assuming that both nuclear and cytoplasmic degradation of ubiquitinated p53 contribute to its degradation, inhibition of nuclear export may partially explain LZAP restoration of HDM2-directed p53 ubiquitination without subsequent destabilization. Our data does not exclude the possibilities that LZAP in the presence of ARF may restore HDM2 mono-ubiquitination, but not polyubiquitination of p53, that LZAP may allow only low levels of p53 ubiquitination that would require nuclear export for degradation, or that LZAP may inhibit targeting of ubiquitinated p53 to nuclear proteasomes as additional mechanisms of stabilizing ubiquitinated p53.
Our data suggest that LZAP allows nuclear accumulation of ubiquitinated and transcriptionally active p53 in the presence of ARF and HDM2. Given that p53 is tightly regulated by HDM2 and ARF, why would additional regulation of ARF be necessary? Mechanisms that ensure a rapid increase in p53 activity, such as post-translational modification, are necessary to prevent growth of damaged cells, but perhaps as important, mechanisms to ensure rapid inhibition of p53 activity must be in place to allow cellular survival and division under appropriate conditions. Ubiquitinated, but active, p53 in the nucleus may be very susceptible to rapid nuclear export and degradation following loss or inhibition of ARF. Although regulators that increase ARF expression are characterized, and loss of ARF in tumour cells is very common through deletion or transcriptional silencing of the ARF gene, mechanisms of ARF inactivation under normal cellular growth conditions are not well characterized.
We report identification of an ARF-binding protein that abrogates ARF inhibition of HDM2 ubiquitin ligase activity. Despite increasing ubiquitination of p53 in the presence of HDM2 and ARF, LZAP expression did not result in loss of stabilization of p53 protein and enhanced p53 transcriptional activity further. These characteristics, as well as induction of endogenous ARF–LZAP interaction following E2F1 oncogene stimulation, imply a possible role of LZAP in ARF-mediated tumorigenesis surveillance. Regulators of LZAP are presently unknown, but, given its biochemical activity, LZAP could serve as a means of prolonging ARF-directed p53 responses that would otherwise be dampened by increased HDM2 expression as part of a p53-directed negative-feedback loop. Since our data suggest that LZAP allows ubiquitination of p53 with maintenance of p53 activity, ARF down-regulation under these circumstances may result in a rapid decrease in p53 protein levels and p53 activity. Our data also suggest that LZAP can activate p53 transcription and cause a p53-dependent cell-cycle arrest in the absence of ARF. Further studies are needed to determine the mechanism of ARF-independent p53 activation by LZAP.
Acknowledgments
We thank James Manfredi and Jennifer Pietenpol for generously providing expression plasmids, and Gordon Peters for providing NARF2 and NARF2-E6 cells. Thanks to Yanping Zhang and to Jennifer Pietenpol for reading and helpful discussion of the paper. We especially thank Yue Xiong for providing extensive support with materials and for insightful discussion. Support for this research was provided by grant number (2 R01-DE013173-05) from the National Institute of Dental and Craniofacial Research to W.G.Y. and from the Barry Baker Endowment at Vanderbilt University.
References
- 1.Quelle D. E., Zindy F., Ashmun R. A., Sherr C. J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 2.Serrano M. The tumor suppressor protein p16INK4a. Exp. Cell Res. 1997;237:7–13. doi: 10.1006/excr.1997.3824. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Xiong Y., Yarbrough W. G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 4.Pomerantz J., Schreiber-Agus N., Liegeois N. J., Silverman A., Alland L., Chin L., Potes J., Chen K., Orlow I., Lee H. W., et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 5.Stott F. J., Bates S., James M. C., McConnell B. B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K. H., Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 7.Honda R., Tanaka H., Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 8.Freedman D. A., Levine A. J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 11.Zindy F., Eischen C. M., Randle D. H., Kamijo T., Cleveland J. L., Sherr C. J., Roussel M. F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmero I., Pantoja C., Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature (London) 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 13.de Stanchina E., McCurrach M. E., Zindy F., Shieh S. Y., Ferbeyre G., Samuelson A. V., Prives C., Roussel M. F., Sherr C. J., Lowe S. W. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong F., Zou X., Hinrichs K., Calame K., Goff S. P. Inhibition of v-Abl transformation by p53 and p19ARF. Oncogene. 1999;18:7731–7739. doi: 10.1038/sj.onc.1203290. [DOI] [PubMed] [Google Scholar]
- 15.Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., Vousden K. H. p14ARF links the tumour suppressors RB and p53. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 16.Sherr C. J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 17.Khan S. H., Moritsugu J., Wahl G. M. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3266–3271. doi: 10.1073/pnas.050560997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David-Pfeuty T., Nouvian-Dooghe Y. Human p14Arf: an exquisite sensor of morphological changes and of short-lived perturbations in cell cycle and in nucleolar function. Oncogene. 2002;21:6779–6790. doi: 10.1038/sj.onc.1205871. [DOI] [PubMed] [Google Scholar]
- 19.Della Valle V., Duro D., Bernard O., Larsen C. J. The human protein p19ARF is not detected in hemopoietic human cell lines that abundantly express the alternative β transcript of the p16INK4a/MTS1 gene. Oncogene. 1997;15:2475–2481. doi: 10.1038/sj.onc.1201417. [DOI] [PubMed] [Google Scholar]
- 20.Pollice A., Nasti V., Ronca R., Vivo M., Lo Iacono M., Calogero R., Calabro V., La Mantia G. Functional and physical interaction of the human ARF tumor suppressor with Tat-binding protein-1. J. Biol. Chem. 2004;279:6345–6353. doi: 10.1074/jbc.M310957200. [DOI] [PubMed] [Google Scholar]
- 21.Rodway H., Llanos S., Rowe J., Peters G. Stability of nucleolar versus non-nucleolar forms of human p14ARF. Oncogene. 2004;23:6186–6192. doi: 10.1038/sj.onc.1207854. [DOI] [PubMed] [Google Scholar]
- 22.Kuo M. L., den Besten W., Bertwistle D., Roussel M. F., Sherr C. J. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eymin B., Karayan L., Seite P., Brambilla C., Brambilla E., Larsen C. J., Gazzeri S. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene. 2001;20:1033–1041. doi: 10.1038/sj.onc.1204220. [DOI] [PubMed] [Google Scholar]
- 24.Martelli F., Hamilton T., Silver D. P., Sharpless N. E., Bardeesy N., Rokas M., DePinho R. A., Livingston D. M., Grossman S. R. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4455–4460. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vivo M., Calogero R. A., Sansone F., Calabro V., Parisi T., Borrelli L., Saviozzi S., La Mantia G. The human tumor suppressor arf interacts with spinophilin/neurabin II, a type 1 protein-phosphatase-binding protein. J. Biol. Chem. 2001;276:14161–14169. doi: 10.1074/jbc.M006845200. [DOI] [PubMed] [Google Scholar]
- 26.Fatyol K., Szalay A. A. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1α (HIF-1α) and inhibits HIF-1-mediated transcription. J. Biol. Chem. 2001;276:28421–28429. doi: 10.1074/jbc.M102847200. [DOI] [PubMed] [Google Scholar]
- 27.Sugihara T., Kaul S. C., Kato J., Reddel R. R., Nomura H., Wadhwa R. Pex19p dampens the p19ARF–p53–p21WAF1 tumor suppressor pathway. J. Biol. Chem. 2001;276:18649–18652. doi: 10.1074/jbc.C100011200. [DOI] [PubMed] [Google Scholar]
- 28.Hasan M. K., Yaguchi T., Sugihara T., Kumar P. K., Taira K., Reddel R. R., Kaul S. C., Wadhwa R. CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53. J. Biol. Chem. 2002;277:37765–37770. doi: 10.1074/jbc.M204177200. [DOI] [PubMed] [Google Scholar]
- 29.Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–64. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 30.Bertwistle D., Sugimoto M., Sherr C. J. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady S. N., Yu Y., Maggi L. B., Jr, Weber J. D. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol. Cell. Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ching Y. P., Qi Z., Wang J. H. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene. 2000;242:285–294. doi: 10.1016/s0378-1119(99)00499-0. [DOI] [PubMed] [Google Scholar]
- 34.Furukawa M., Zhang Y., McCarville J., Ohta T., Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 2000;20:8185–8197. [Google Scholar]
- 35.Yarbrough W. G., Aprelikova O., Pei H., Olshan A. F., Liu E. T. Familial tumor syndrome associated with a germline nonfunctional p16INK4a allele. J. Natl. Cancer Inst. 1996;88:1489–1491. doi: 10.1093/jnci/88.20.1489. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12:175–186. [PubMed] [Google Scholar]
- 37.Midgley C. A., Desterro J. M., Saville M. K., Howard S., Sparks A., Hay R. T., Lane D. P. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 38.Honda R., Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdary D. R., Dermody J. J., Jha K. K., Ozer H. L. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., Yao T. P. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards S. J., Dix B. R., Myers C. J., Dobson-Le D., Huschtscha L., Hibma M., Royds J., Braithwaite A. W. Evidence that replication of the antitumor adenovirus ONYX-015 is not controlled by the p53 and p14ARF tumor suppressor genes. J. Virol. 2002;76:12483–12490. doi: 10.1128/JVI.76.24.12483-12490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S. N., Roe A. E., Donehower L. A., Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 43.Montes de Oca Luna R., Wagner D. S., Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 44.Sherr C. J., Weber J. D. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 45.Tao W., Levine A. J. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Q., Yao J., Wani G., Wani M. A., Wani A. A. Mdm2 mutant defective in binding p300 promotes ubiquitination but not degradation of p53: evidence for the role of p300 in integrating ubiquitination and proteolysis. J. Biol. Chem. 2001;276:29695–29701. doi: 10.1074/jbc.M102634200. [DOI] [PubMed] [Google Scholar]
- 47.Pan Y., Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Keefe K., Li H., Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 2003;23:6396–6405. doi: 10.1128/MCB.23.18.6396-6405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth J., Dobbelstein M., Freedman D. A., Shenk T., Levine A. J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xirodimas D. P., Stephen C. W., Lane D. P. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 51.Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]