Abstract
Diarrhoeagenic enterohaemorrhagic Escherichia coli and enteropathogenic E. coli attach to human intestinal epithelium and efface brush-border microvilli, forming an A/E (attaching and effacing) lesion. These human pathogens are phenotypically similar to the mouse pathogen Citrobacter rodentium. Genetically, they all have a homologous set of virulent genes involved in the A/E lesion, and these genes are organized on a LEE (locus of enterocyte effacement), a pathogenicity island. This island comprises 41 specific open reading frames, of which most are organized at five operons, LEE1, LEE2, LEE3, LEE4 and tir (LEE5). The expression of the LEE genes is regulated in a complicated manner, and current knowledge is that there are at least two positive regulators, Ler (LEE-encoded regulator) and GrlA (global regulator of LEE activator), and one negative regulator, called GrlR (global regulator of LEE repressor). In enterohaemorrhagic E. coli, GrlA is encoded by l0043, whereas GrlR is encoded by l0044. Here we report a fourth regulatory gene located in LEE3, namely l0036. Its expression is tightly controlled. When overexpressed, this factor, named Mpc (multiple point controller), interacts with Ler and suppresses the expression of the LEE proteins. When the translation is not initiated or terminated before maturation, the type III secretion of effectors is completely abolished. Therefore, together with the fact that several cis elements reside in the region that l0036 spans, l0036 appeared to have multiple functions in the regulation of LEE expression.
Keywords: enterohaemorrhagic Escherichia coli (EHEC), enteropathogenic Escherichia coli (EPEC), l0036, pathogenesis, regulation, type III secretion
Abbreviations: A/E, attaching and effacing; EHEC, enterohaemorrahgic Escherichia coli; EPEC, enteropathogenic E. coli; GrlA, global regulator of LEE activator; GrlR, global regulator of LEE repressor; GST, glutathione S-transferase; H-NS, histone-like, nucleoid structuring protein; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; LEE, locus of enterocyte effacement; Ler, LEE-encoded regulator; Mpc, multiple point controller; ORF, open reading frame; TTS, type III secretion; TTSS, type III secretion system
INTRODUCTION
Escherichia coli O157:H7 is associated with enterohaemorrhagic diarrhoea of humans in developed countries. Infection with this pathogen results in the formation of A/E (attaching and effacing) lesions of the host intestinal epithelial cells [1]. The property of forming an A/E lesion is shared by infection with EPEC (enteropathogenic E. coli) and of Citrobacter rodentium. The virulence factors involved are encoded by a chromosomal pathogenicity island of the bacteria, known as the LEE (locus of enterocyte effacement) [2]. The LEE island contains 41 ORFs (open reading frames), and a majority of them are organized into five operons (LEE1–LEE5) [3]. The functions of the products encoded can be categorized into transcriptional regulators, components for the TTSS (type III secretion system), translocators, effectors and specific chaperones. Whereas some of these ORFs have been well characterized, others remain poorly defined. The latter is exemplified by l0036 of enterohaemorrhagic E. coli (EHEC) that is reported here.
The expression of the LEE genes is under tight control, and current knowledge indicates that there are multiple regulatory molecules involved in this process. A quorum-sensing system regulates virulence gene expression in EHEC and EPEC [4]. An IHF (integration host factor) that is a global regulator also plays an essential role in the expression of several genes of LEE [5], among which is the ler gene located in LEE1. Ler (LEE-encoded regulator), the product of ler, is a DNA-binding protein that counteracts the repression of H-NS (histone-like, nucleoid structuring protein), another DNA-binding protein that negatively modulates the gene expression [6]. Many studies have revealed that LEE2, LEE3, LEE4 and tir (LEE5) are activated directly by Ler [3,7–9]. On the other hand, two newly reported regulators, GrlA (global regulator of LEE activator) and GrlR (global regulator of LEE repressor) function indirectly in regulation, probably through modulating the Ler effect [10,11]. LEE expression could be regulated in a number of additional ways. One of the speculated targets is l0036 in EHEC (corresponding to orf12 in EPEC and C. rodentium). The rationale is based upon the fact that this region contains binding sites for both H-NS and Ler [8,12]. Moreover, this DNA sequence is predicted to contain an attenuator regulatory sequence that may mediate the expression of LEE2 [6].
ORF l0036 is the first coding sequence in LEE3 and theoretically encodes a product with a calculated molecular mass of 12.7 kDa. In the previous studies [10,13], L0036 has been implicated as a component of TTSS, and a mutant of this gene phenotypically behaves in a similar manner to a deletion of other ORFs in the same operon or in LEE2. Here, we show that L0036 is not directly required for TTS (type III secretion), but its translation is pivotal for the whole TTSS. By overexpression of the gene product, we also found that L0036 may antagonize Ler's action by a direct interaction. Multiple roles for l0036 have thus been observed for the first time.
EXPERIMENTAL
Bacterial strains, primers and growth conditions
E. coli strain JM110 [F′ traD36 lacIqΔ(lacZ)M15 proA+B+/rpsL-(Strr) thr leu thi lacY galK galT ara fhuA dam dcm glnV44 Δ(lac-proAB); New England Biolabs] served as the host for plasmid preparation. E. coli strain BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dcm (DE3); Novagen] was used in the T7 promoter-driven transcription–translation assay. E. coli XL1-Blue MRF′ Kan (Stratagene) was used for the bacterial two-hybrid assay described below and has a genotype (mcrA)183 (mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′::Tn 10 proAB lacIqZ.M15 Tn5 (Kanr)]. EHEC strain 43888 (A.T.C.C.), which does not produce either Shiga-like toxin I or II and lacks stx (Shiga toxin) genes, was used as the parental strain for creating specific LEE gene deletions. The primers used in the present study are listed in Table 1. Bacteria were grown in LB (Luria–Bertani) broth (Difco) unless specified, and the medium was supplemented with appropriate antibiotics whenever necessary.
Table 1. Oligonucleotides used in the present study.
| Name | Sequence (5′ to 3′) |
|---|---|
| PUC(−)-FB | ACT AGT GGA TCC AGA ATT |
| PUC(−)-RK | ATG GTA CCG CCG CCA CTC AT |
| L0036-F | GCC TGC AGA TGA ATC TTT TAG |
| L0036-R | TCA TGA TGT CAT CCT GCG AA |
| L0054-F | GCA TGC GGA GAT TAT TTA TTA TG |
| L0054-R | CAT GTT AAA TAT TTT TCA GCG GTA T |
| SEPZ-R | GGT TAG GCA TAT TTC ATC GC |
| SEPQ-R | ATT CCT GAT TAA TCA CAT AC |
| ESCV-F-NcoI | CGC CAT GGG ATG ACA TCA TGA ATA AAC TC |
| P-SP6 | TAT TTA GGT GAC ACT ATA G |
| ESCV-R-SpeI | GGA CTA GTT GCT CTG AAA TCA TTT ACC G |
| 15850F-KpnI | GGG GTA CCA ATA CGC TTT TTC AAG C |
| ESCN-R-BamHII | CGG GAT CCG GCA ACC ACT TTG AAT AGG |
| 16440F-KpnI | GGG GTA CCA ATC CTC TTG AAC TAA CTT C |
| 16122F | AGC GTA CGT CAT GAG TGA TTG AAC ATA TTT TTG TC |
| 16122R | GAC AAA AAT ATG TTC AAT CAC TCA TGA CGT ACG CT |
| L0036-1F | TTA TTG CAT CGA AAC TAA TTT TGT A |
| L0036-1R-XbaI | TCT AGA ATT CAT CTG CAG GCT CTG AAG |
| L0036-2F-XbaI | TCT AGA GTT CGC AGG ATG ACA TCA TG |
| L0036-2R-SalI | ACG CGT CGA CGA CAA ATC AGC TCC CAT A |
| ESCV-R-SalI | ACG CGT CGA CTG CTC TGA AAT CAT TTA CCG |
| 15960F | GAT TTT AAT GGG TTT TCT TTT TTT ATT G |
Plasmid construction
To express the product of l0036, the l0036 sequence was PCR-amplified from EHEC genomic DNA using the primer pair L0036-F/L0036-R, and the PCR product was cloned into the vector pUC-T (MDBio, Inc.) to create pUC-L36. The l0036 sequence was then amplified again from pUC-L36 with primers PUC(−)-FB and PUC(−)-RK. After digestion with endonucleases BamHI/KpnI, the l0036-containing fragment was cloned into pQE30 (Qiagen) to yield pQL36, in which L0036 is expressed with an N-terminal RGS-His6 tag. The same DNA fragment was cloned into pQE31 (Qiagen) to create pQL36N, in which L0036 was out of frame and a stop codon was generated immediately after the N-terminal RGS-His6 tag. As a result, no L0036 was expressed. To create alternative expression of L0036 from plasmids with different origins of replication and copy numbers, the fragment of l0036 along with the T5 promoter in pQL36 was obtained after digestion with XhoI/NheI and cloned into the XhoI/NheI-treated pACYC177 (New England Biolabs), to give pACYL36. pACYL36 was then cleaved with HindIII, and the fragment containing T5::L0036 was inserted into the HindIII site of pCC1BAC (Epicentre) to give pCCL36.
Similarly, the Ler expression plasmid pQ-Ler was constructed by inserting pQE32 (Qiagen) with a product of ler that was obtained from PCR amplification of the EHEC DNA using primers L0054-F and L0054-R. Plasmids derived from the pQE30 series are ampicillin-resistant. To construct pACYLer that was chloramphenicol-resistant, the ler-containing DNA fragment was digested with XhoI/PvuII and ligated with SalI/EcoRV-cleaved pACYC184. An alternative plasmid, pACYLerK, which is kanamycin-resistant, was constructed by digesting pQ-Ler with AatII/PvuII, isolating the ler-containing fragment and subsequently ligating this fragment with AatII/ScaI-digested pACYC177.
To create pACY3731, a DNA fragment from sepZ to sepQ was PCR amplified from EHEC genomic DNA with the primer pair SEPZ-R/SEPQ-R and cloned into pGEM-T Easy vector (Promega) to yield pGE3731. pGE3731 was then digested with NcoI/SalI, and the sepZ to sepQ fragment was cloned into the BspHI/SalI-digested pACYC184 to yield pACY3731. To create pACY3531, a DNA fragment was PCR-amplified from pGE3731 with the primers ESCV-F-NcoI and P-SP6, digested with NcoI/SpeI, and cloned into NcoI/NheI-digested pQE60 to yield pQE3531. pQE3531 was then digested with XhoI/PvuII, and the l0036-escV-containing fragment was ligated with SalI/EcoRV-digested pACYC184 to yield pACY3531. To create pACY35, escV was PCR-amplified from pACY3531 with primers ESCV-F-NcoI and ESCV-R-SpeI, digested with NcoI/SpeI and cloned into NcoI/NheI-treated pQE60 to yield pQL35. pQL35 was then digested with XhoI/PvuII, and the escV-containing fragment was cloned into SalI/EcoRV-digested pACYC184 to yield pACY35.
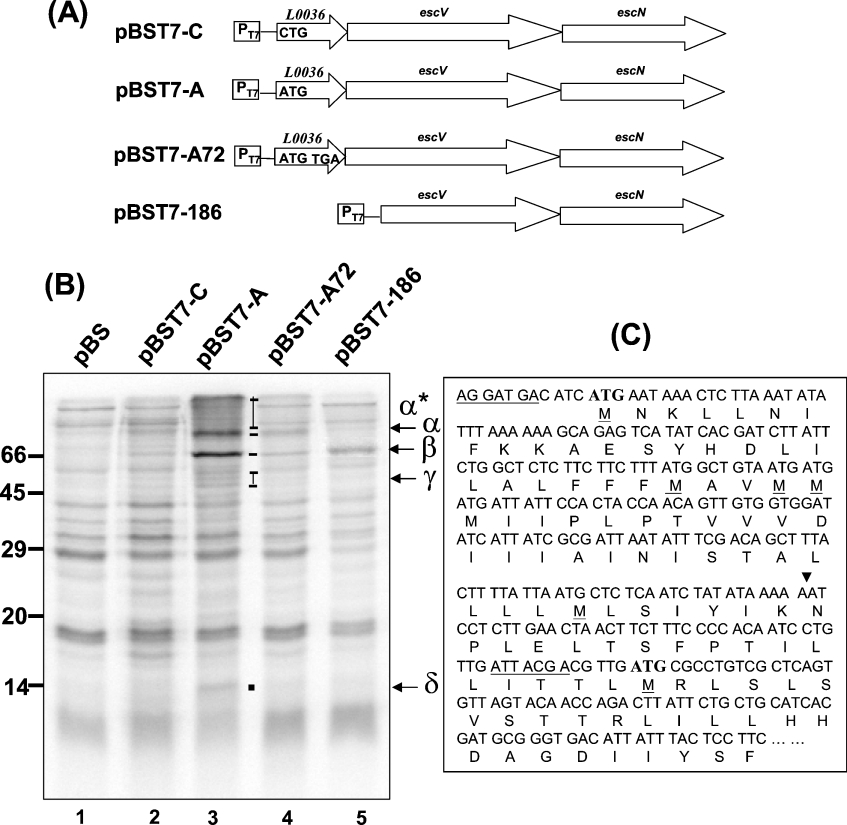
To create pBST7-A and pBST7-C, DNA fragments were amplified separately from the genomic DNAs of EHEC and AC36 (see below) with the primer pair 15850F-KpnI/ESCN-R-BamHI, digested with KpnI/BamHI and cloned into pBluescript II SK(+). To create pBST7-A72, the two DNA fragments covering the N-terminal portion and the C-terminal portion of l0036 were first PCR-amplified from pBST7-A with primer pairs 15850F-KpnI/16122R and 16122F/ESCN-R-BamHI respectively. Primers 16122R and 16122F were partially complementary and contained a TGA stop codon designed at codon 72 of l0036. As a result, when the above two PCR products were mixed, denatured and renatured, the partially end-overlapping sequences annealed. Taq DNA polymerase was then used to extend the mutually primed strands, and, finally, PCR was carried out to amplify a full-length product of l0036. The PCR product was then digested with KpnI/BamHI and cloned into pBluescript II SK(+). To create pBST7-186, the DNA fragment containing the alternative escV RBS (ribosome-binding site) and the start codon shown in Figure 3(C) (below) was amplified from pBST7-A with primers 16440F-KpnI and ESCN-R-BamHI. The product was digested with KpnI/BamHI and cloned into KpnI/BamHI-digested pBluescript II SK(+).
Figure 3. Analysis of the role of l0036 in bacterial transcription and translation.
(A) Illustration of the transcripts driven by T7 promoter in various plasmids. pBST7-A has the insert copied from the parental sequence; pBST7-C differs from pBST7-A by replacing the initiation codon of l0036 with CTG; pBST7-A72 differs by placing TGA at the 72nd codon; pBST7-186 has the 5′ LEE3 nucleotides deleted down to the 183rd base of the escV ORF. (B) Protein expression profiles derived from the transcripts in (A). Proteins synthesized in the plasmid-transformed bacteria were labelled with [35S]methionine in the presence of both IPTG and rifampicin. Bands labelled were those presumably derived from ORFs of the transcript (see the text for the product assignment). (C) Analysis of the 5′ nucleotides and the N-terminal amino acid residues encoded by escV. All methionine residues in this region are highlighted, and the possible ribosome-binding sites are underlined. The arrowhead indicates the 5′ nucleotide of the truncated escV, to which the T7 promoter was directly linked in pBST7-186.
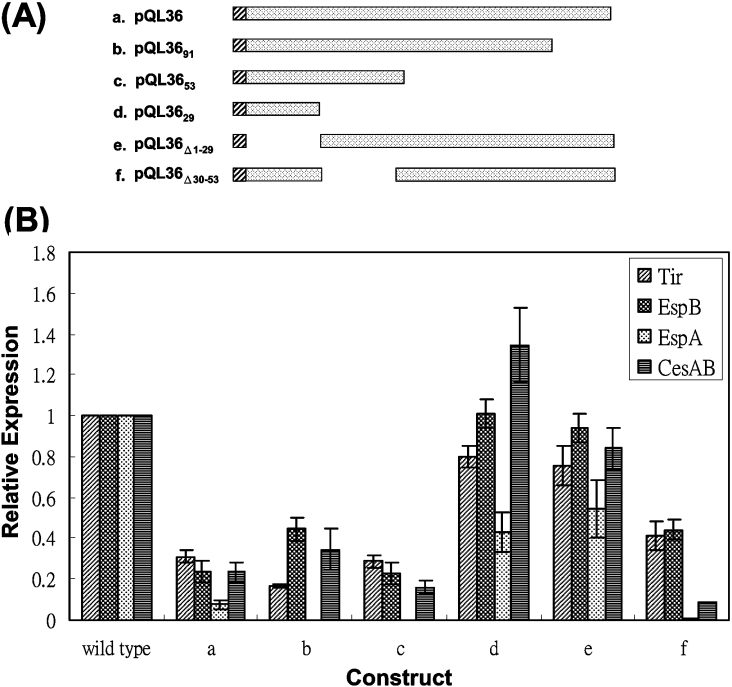
To express truncated L0036, pQL36 was used as the template for PCR inverse amplification with different primer pairs. Basically the forward and reverse primer pairs were end-phosphorylated with T4 polynucleotide kinase, and the products obtained by PCR were self-ligated to yield pQL36 derivatives that covered areas of L0036 as illustrated in Figure 7(A) (below).
Figure 7. Fine mapping the region of L0036 involved in the suppression of the LEE expression.
(A) Schematic illustration of the truncated L0036 expressed from individual plasmids. His6-tag fused to the N-terminus of L0036 is represented by hatched bars. (B) Bacterial lysates from the wild-type strain 43888 harbouring additional plasmids shown in (A) were analysed for the LEE proteins by Western blotting as described in legend to Figure 1. Each band on the blot was quantified by ImageQuant version 1.2 (Molecular Dynamics), and the digitalized intensity was compared with that derived solely from the wild-type bacteria. Data were summarized from three different experiments.
To prepare plasmids for the bacterial two-hybrid system, pQL36 was digested with BamHI and SalI. The DNA fragment containing l0036 was cloned into BamHI/XhoI-treated pBT and pTRG (Stratagene) to yield pBT-L36 and pTRG-L36 respectively. By the same strategy, ler was cloned into pBT and pTRG in similar, but reciprocal, experiments.
Generation of mutants on LEE
To delete l0036 in the EHEC chromosome, the method previously described [14] was followed. First, the 5′- and 3′-flanking regions of l0036 were PCR-amplified from EHEC genomic DNA with primer pairs L0036-1F/L0036-1R-XbaI and L0036-2F-XbaI/L0036-2R-SalI respectively. The PCR products were then cloned into pGEM-T Easy and confirmed by sequencing. These inserted fragments were then cleaved from plasmids separately with NotI/XbaI and XbaI/SalI. The fragments were then isolated and three-way ligated with NotI/SalI-restricted pKO3 [14], yielding pKO3-ΔL0036. pKO3-ΔL0036 was electroporated into a wild-type EHEC strain 43888. Gene deletion mediated by homologous recombination was then stepwise carried out by selection in chloramphenicol and then sucrose. The mutant obtained, named strain 43888 ΔL0036, was confirmed as a chromosomal deletion of l0036 by PCR amplification of the region spanning the target site and by sequencing the PCR product.
An alternative approach to construct the l0036-deleted mutant was to silence the expression of l0036 by altering the start codon of l0036 and leaving the rest of the sequence untouched. To accomplish this mutagenic step, a DNA fragment containing the upstream sequence of l0036 was amplified with primers L0036-1F/15960R so that the mutated start codon of l0036 was included in the 3′ end of the fragment. The second fragment covering l0036 and its downstream sequence was PCR-amplified with primer pairs 15960F/ESCV-R-SalI in which primer 15960F incorporated the altered l0036 start codon and was partially complementary to the primer 15960R. As a result, the above mentioned two fragments overlapped at the sequence flanking the altered start codon of l0036. After mixing the two fragments, followed by denaturation and renaturation, PCR was carried out in a manner similar to that described above for the construction of pBST7-A72. The product was then cleaved with SalI/BssHII and ligated with SalI/BssHII-digested pKO3-ΔL0036, yielding pKO3-AC36, which was in turn used to generate the l0036-silenced mutant EHEC, AC36, by stepwise selection as described above.
EHEC 43888 ΔLer has been described previously and was created by using the same strategy [11].
Protein preparation and immunoblotting
Bacterial proteins were prepared as previously described [15]. In brief, bacteria were cultured overnight in LB broth and diluted 1:100 in M9 minimal medium [11]. After 6 h cultivation at 37 °C in 5% CO2, the bacterial cultures were separated by centrifugation into pellets and supernatants, and it was from these that the cell lysates and secreted proteins were prepared. Proteins in the samples were then resolved by SDS/PAGE. Immunoblotting analyses of various antigens were performed as previously described [15,16]. Anti-GST [anti-(glutathione S-transferase)], anti-EspD, anti-EspA, anti-CesAB and anti-OmpA were prepared by immunizing mice with GST [17] and His6-tagged recombinant proteins, respectively [18]. Anti-Tir and anti-EspB have been described previously [15,19] (Tir is an effector from LEE5; EspA, EspD and EspB are translocators from LEE4; CesAB is a cytosolic chaperone from LEE1; OmpA is a constitutively expressed outer membrane porin of E. coli). Anti-RGS-His6 (Clontech) and anti-O157 (Difco) were purchased commercially.
Bacterial transcription/translation-coupled assay
The assay was performed as described previously [18], except for a slight modification. In brief, E. coli BL21 (DE3) harbouring a specified plasmid was cultured overnight in LB broth. Cell pellets from 100 μl cultures were resuspended in 1 ml of methionine-deficient assay medium (Difco) for 2 h in 37 °C. IPTG (isopropyl β-D-thiogalactoside) was added to a final concentration of 1 mM for 1 h to induce the synthesis of T7 polymerase. After induction, rifampicin was added to the cultures to a final concentration of 200 μg/ml which were incubated for a further 1 h, to inhibit the function of E. coli RNA polymerase. To label proteins mainly synthesized from the T7 promoter-transcribed RNA, 1 μl of [35S]methionine (Amersham) at 50 μCi/ml was then added to the cultures for 10 min. Cells were then pelleted and lysed in 50 μl of SDS sample buffer, and proteins were resolved by SDS/8%- or 15%-(w/v)-PAGE. The gels were dried and exposed for autoradiography, and the results were analysed using a Phosphorimager (Molecular Dynamics).
Bacterial two-hybrid assay
The two-hybrid system was performed according to the manufacturer's (Stratagene) instructions. In brief, derivatives of pBT (encoding the λcI protein that is used to fuse with the bait) and pTRG (coding for an N-terminal domain of RNA polymerase α subunit that is used to fuse with the target protein) were cotransformed into E. coli XL1-Blue MRF′ (Stratagene) and plated on LB-agar plates containing carbenicillin in addition to the plasmid-selecting antibiotics. E. coli XL1-Blue MRF′ contains the reporter genes ampr and lacZ that are turned on by a positive interaction of the bait and the target gene products. The bacterial transformants were scored for the numbers of colonies formed after 24 h cultivation at 30 °C and bacterial β-galactosidase activity [20] from three representative colonies was measured.
GST pull-down assay
Transformants of E. coli JM110 harbouring pGEX-2T, pGEX-L0036 or pQ-Ler were cultured overnight in LB broth. After bacterial dilution (1:100) into fresh LB broth and 2 h cultivation at 30 °C, IPTG (1 mM) was added to the culture. After an additional incubation for 2 h, the bacteria were pelleted and homogenized using glass beads. Next, the insoluble materials were removed by centrifugation and the supernatants were mixed with glutathione–Sepharose 4B (Pharmacia) beads and rocked at 4 °C for 2 h. After washing twice with PBS, the beads were mixed with clarified bacterial lysate containing His6-tagged Ler for another 2 h at 4 °C. After centrifugation and washing twice with PBS, the proteins bound on to the beads were dissolved in the SDS sample buffer, resolved by SDS/PAGE and analysed by immunoblotting as described above.
RESULTS
Aborted expression of l0036 abolished the secretion, but not the synthesis, of the type III secretion proteins
Examination of the proteins secreted revealed that TTS was totally abolished in our mutant strain 43888 ΔL0036 (results not shown), and these findings are consistent with those reported previously [10]. However, within the DNA sequence of l0036 there are several regulation factor-binding sites [8,12]. Truncation of l0036 may result in unpredicted effects caused by the deletion of these critical DNA elements. Therefore, to preserve the DNA integrity while knocking out the protein expression, an alternative approach was used. The parental bacteria were mutated only at the start codon of l0036, which was changed from ATG to CTG, and the resulted mutant so generated was named AC36.
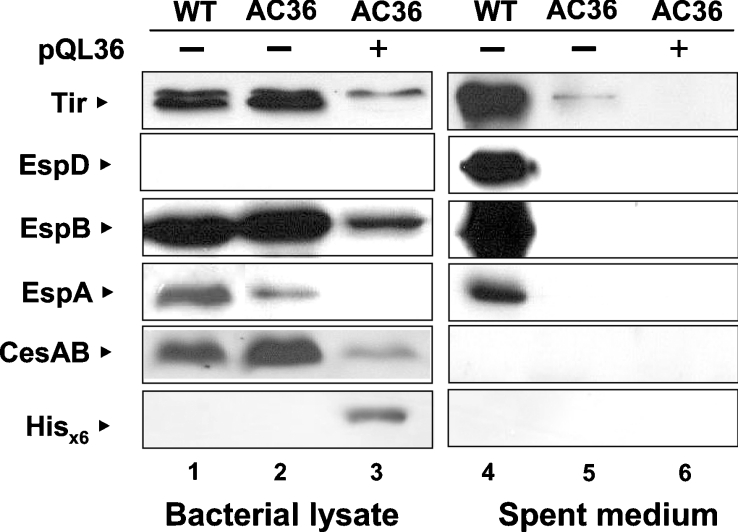
AC36 was analysed for the presence of five representative proteins in both the bacterial lysate and the spent medium (Figure 1) by Western blotting using specific antibodies. In the bacterial lysates, Tir and EspB were detected equally well in both strains of the wild-type and AC36, whereas the intensities of EspA and CesAB varied only slightly (lanes 1 and 2). On the other hand, a big difference was observed for the proteins secreted into the medium. Tir, EspD, EspB, and EspA were found abundantly in the spent medium of the wild-type EHEC (lane 4), but not in that of AC36 (lane 5). Since CesAB is a chaperone for EspA and EspB and is retained in the bacterial cells, it was not surprising that CesAB was not found in the spent medium (compare lanes 1 and 4, also lanes 2 and 5). On the other hand, EspD was hardly detected in the bacterial lysates of the wild-type strain (lane 1) unless more protein was loaded into the gels (results not shown), an observation reflecting the fact that EspD inherits the specific property of, perhaps, having a high efficiency of secretion. Furthermore, in the secretion-deficient AC36, whereas Tir, EspB and EspA (Figure 1, lane 2) were found in the bacterial lysates, EspD remained undetected in both bacterial cell preparations and medium. It is then reasonable to hypothesize that the expression of EspD differs from that of the others by having an additional layer of stringent control that is probably coupled to secretion. At any rate, mutating the putative start codon of l0036 apparently abolished TTS, but did not affect most of the syntheses of these proteins inside the bacteria.
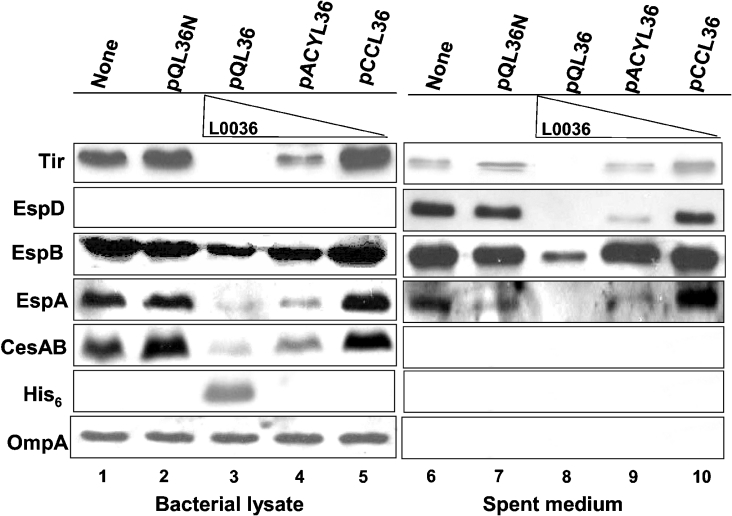
Figure 1. Effects on the expression and secretion of representative LEE proteins when the initiation codon of l0036 was altered.
Bacteria transformed with (+) or without (−) pQL36 were cultivated in M9 medium for 6 h. Bacterial lysates and proteins from spent medium were then prepared and analysed by Western blotting using antibodies specific to the antigen indicated. Abbreviations: WT, the wild-type strain 43888; AC36, a mutant strain 43888 with initiation codon of l0036 altered from ATG to CTG.
To confirm that the above effect is really mediated by the loss of a putative protein, a complementation experiment was performed by transforming AC36 with an l0036-expressing plasmid (i.e. pQL36). Plasmid pQL36 expresses a L0036 protein with a His6-tag at its N-terminus and is expected to be 16 kDa in size. Consistent with this expectation, Figure 1 (lane 3) shows that a protein detected by anti-(His6 tag) monoclonal antibody was seen in the bacterial lysate of the transformant but not in other preparations (lanes 1 and 2) or in the spent medium (lanes 4–6). This observation indicates that pQL36 did yield a cytosolic recombinant L0036. Intriguingly, the secretion of the LEE proteins by AC36 was not restored to the levels seen with the wild-type EHEC (comparing lane 4 with lane 6). Therefore the loss of TTS could not simply be explained by the loss of the protein L0036. Furthermore, and surprisingly, the synthesis of all the representative LEE proteins was decreased in the presence, in trans, of L0036 (compare lanes 1 and 3).
Complementation of AC36 requires gene products downstream of l0036
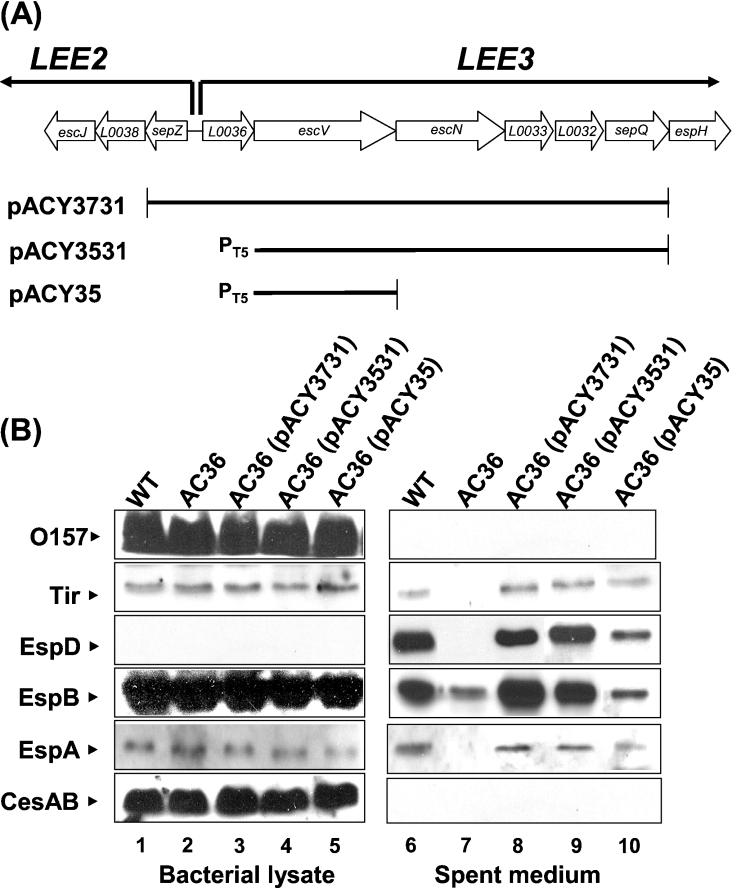
To clarify why AC36 after complementation with pQL36-expressed L0036 did not restore TTS, a polar effect on the downstream genes was speculated. To test this hypothesis, plasmid pACY3731 covering the putative promoter and almost the entire LEE3 except for espH (Figure 2A) was transformed into AC36. The results shown in lane 8 of Figure 2(B) indicated that the secretion capacity of AC36 was fully recovered (compare lanes 6 and 8). Therefore it is likely that mutation of the start codon of l0036 might have affected the expression of downstream genes essential for TTSS. To substantiate this notion, plasmid pACY3531 was constructed by replacing the DNA upstream to escV in pACY3731, including L0036 and the putative LEE3 promoter, with an independent T5 promoter (Figure 2A). Complementing AC36 with pACY3531 also gave full recovery of secretion capacity (Figure 2B, lane 9). On the other hand, complementation with pACY35 that covered solely escV only partially restored EHEC secretion activity (Figure 2B, lane 10). Therefore, these observations together suggest that the start codon of l0036 is critical for the expression of downstream genes in the same LEE3 operon. Possibly, the expressions of EscN and other downstream LEE3 proteins, together with EscV, contribute to the full recovery of secretion activity. Again, no effect was observed on the synthesis of the LEE proteins, at least Tir from LEE5, EspB and EspA from LEE4, and CesAB from LEE1, in AC36 regardless of whether the bacteria were or were not complemented by the plasmids discussed above (lanes 1–5 in Figure 2B). Therefore, the protein of l0036 is not, per se, critical for the synthesis of the LEE proteins, nor is important for TTS. It is most likely that the translation of L0036 is a necessary event for subsequent EscV and EscN translation.
Figure 2. LEE3 genes downstream of l0036 required for AC36 to complement the defect in the type III secretion.
(A) Schematic of gene organization (not to scale) flanking l0036 and illustration of the DNA inserts in the complementation plasmids. (B) Proteins from bacterial lysates and spent medium were analysed as described above in the legend to Figure 1. O157, the antigen detected by anti-O157 antibodies from Difco.
Translation of L0036 regulates the synthesis of downstream gene products in LEE3
To confirm that the synthesis of EscV is coupled with the translation of L0036, the translational products have to be visualized. Since the amounts of LEE3 products are too low to be detected by Western blotting in EHEC (results not shown), we constructed plasmids to express the genes and labelled the products with [35S]methionine in vivo [18]. In the first three plasmids of Figure 3(A), the inserts were driven by T7 promoter and all three contained the same length of LEE DNA that covers l0036, escV and escN, but differs in one nucleotide in l0036 (Figure 3A). pBST7-C differed from pBST7-A by a variation of CTG versus ATG in the start codon of l0036; pBST7-A72 differed from PBST7-A by replacing the 72nd codon of l0036 with a TGA stop codon. In PBST7-C, the translation of l0036 should be abolished at the beginning, whereas that in pBST7-A72 would prematurely terminate after reading through the 71st codon.
The results shown in lane 1 of Figure 3(B) revealed the background of proteins labelled when bacteria carrying the control vector pBST7 were cultured in the medium containing [35S]methionine and rifampicin. When the bacteria harbouring pBST7-A were similarly treated, several labelled proteins with different sizes were apparent (Figure 3B, lane 3). These extra proteins must be derived from the LEE DNA insert carried in pBST7-A. By comparing this pattern with the protein pattern displayed by the bacteria carrying pBST7-C, in which the start codon of l0036 was eliminated, at least five distinct protein bands could be identified (lane 2 versus lane 3). The δ protein had a size of 14 kDa, which matches the expected size of L0036 with 117 amino acid residues. EscV has 675 amino acid residues with an expected size of 75 kDa and has been reported to form oligomers previously [23]. Therefore, the α band may represent the product of escV and a series of bands (labelled ‘α*’) above the α band may have product(s) representing the oligomers of EscV. Downstream of escV is escN, which encodes a 446-amino-acid-residue protein. There are several possible bands of about 50 kDa in size, and these are labelled as the γ bands in lane 3 and may represent EscN. The β band, heavily isotope-labelled in lane 3, may be derived from the degradation of EscV or result from an alternative initiation within escV (see below). All together, these translation products supported the hypothesis that the synthesis of EscV, and also EscN, requires a prior translation of l0036, and no synthesis of L0036 gives little in the way of EscV and EscN products.
To substantiate the above hypothesis, the protein pattern displayed by bacteria carrying pBST7-A72 was examined (Figure 3B, lane 4). First, the 14 kDa product should disappear, owing to an early termination at the 72nd codon of l0036. Consistent with this, the δ band seen in lane 3 is no longer observed in lane 4. By following the same reasoning, when the translation of L0036 stops before maturation, the genes on the same LEE3 mRNA and downstream to l0036 should be poorly translated. Lane 4 in Figure 3 obviously shows that the intensities of α*, α, β, and γ bands all decreased, down to a background level. This fact strongly supports the central role of L0036 in controlling the translation of the LEE3 genes.
To distinguish whether the β band in lane 3 resulted from the degradation of EscV or is derived from an alternative initiation, the escV ORF was carefully examined (Figure 3C). Within the first 100 amino acids of EscV there are seven methionine residues, including the putative initiation methionine residue. Two DNA stretches may possibly act as the ribosome-binding site: one is located upstream and adjacent to the first methionine residue, whereas the second is proximal to the seventh methionine residue (Figure 3C). If translation is initiated at the seventh methionine residue, the new product would have the observed size of 66 kDa. To test this possibility, pBST7-186 was generated. In this construct, l0036 and the first 183 nucleotides of escV were deleted, and translation would be expected to be initiated at the first ATG codon in the newly established reading frame. Lane 5 in Figure 3(B) shows the result with a major product labelled at the size of 66 kDa (band β) and disappearance of band δ (the product of l0036), bands α* and α (products presumably derived from EscV). These observations are consistent with the suggestion that the translation of escV starts at two alternative initiation sites to give products with sizes of 75 and 66 kDa. They both have an identical C-terminus, but vary in their N-terminal regions. The former one has additional N-terminal segment and may form oligomers; this hypothesis agrees with the finding that this N-terminal segment contains a stretch of hydrophobic amino acids (Figure 3C). Whether there is a biological significance for these alternative initiations of escV remains to be explored.
L0036 has a down-regulation effect on the synthesis of the LEE proteins
Since an aborted initiation in translation of l0036 resulted in a failure of TTS, the translation of l0036 must be indispensable, and there must be a product translated from l0036. To study the possible function of l0036 in a detectable amount, we expressed l0036 in the context of a wild-type background. Driven by its authentic promoter and followed by a sequence coding for the His6-tag, l0036 was incorporated into plasmids with different copy numbers. These plasmids were then transformed into the wild-type strain 43888, and the representative LEE proteins were examined by Western blotting in both bacterial lysates and the spent medium (Figure 4). Since the copy numbers of the plasmids in the bacteria decrease as pQL36 was replaced by pACYL36 and then by pCCL36, the amounts of L0036 expressed were expected to decrease accordingly. Consistent with this expectation, L0036 expressed at the highest level was detected in the lysate of bacteria harbouring pQL36 (lane 3) by anti-His6 tag. No signal related to L0036 was seen in the lysates with either pACYL36 (lane 4) or pCCL36 (lane 5), perhaps because the expression levels were too low to be detected by the antibody. The fact that OmpA was detected in equal amounts in each preparation (lanes 1 to 5) excludes the possibility of unequal sample loading.
Figure 4. Dosage effect of L0036 on the expression and secretion of the LEE proteins.
The amount of L0036 expressed in bacteria was controlled by plasmids carrying the same expression cassette but with different copy numbers. pQL36N is identical with pQL36, except that L0036 was out of frame and a stop codon was generated immediately after the N-terminal RGS-His6 tag. Bacteria used were the wild-type strain 43888, and the protein analyses were carried out as described in the legend to Figure 1.
In the bacterial lysate, high-level L0036 expression by pQL36 appeared to severely suppress the expressions of Tir, EspB, EspA and CesAB. As the expression level of L0036 decreased, the suppression effect on these proteins was increasingly alleviated (comparing lane 3 with lane 5). The suppression effect was obviously due to the expression of l0036 per se, since pQL36N that was identical with pQL36, but differed by having a non-expressible l0036, gave no suppression at all (lane 2).
In the medium, secretion of the representative proteins was also suppressed by overexpressing L0036. Similar to what was observed with the lysates, decreasing the expression level of L0036 gradually restored the secretion capability to that seen for the wild-type (comparing lanes 8–10 with lane 6), and knocking out the expression of L0036 from the plasmid eliminated the suppression effect (lane 7).
Suppression effect of L0036 mediated by interaction with Ler
To identify how L0036 exerts its effect on suppressing the expression of the LEE proteins, a ler-deleted mutant (i.e. strain 43888 ΔLer) was used in the analysis (Figure 5A). Since Ler is a general positive regulator for LEE, it was expected that the expression of the LEE proteins in the ler-deleted strain (lane 2) would be lower than that in the wild-type strain 43888 (lane 1). Complementing the mutant with the Ler expression (i.e. pACY-Ler) recovered the levels of the LEE proteins synthesized (Figure 5A, lanes 1–3). However, when Ler and L0036 were co-expressed in strain 43888 ΔLer (lane 4), the positive regulatory effect of Ler seemed to be neutralized, and the transformant behaved as if it were the ler-deleted mutant. It is noteworthy that all syntheses of Tir, EspB, EspA and CesAB were down-regulated. This effect differed from that observed with the negative regulator GrlR [11], encoded by pAT-44 (Figure 5A), which suppressed only CesAB expression from LEE1 in the same situation (lanes 3 and 5).
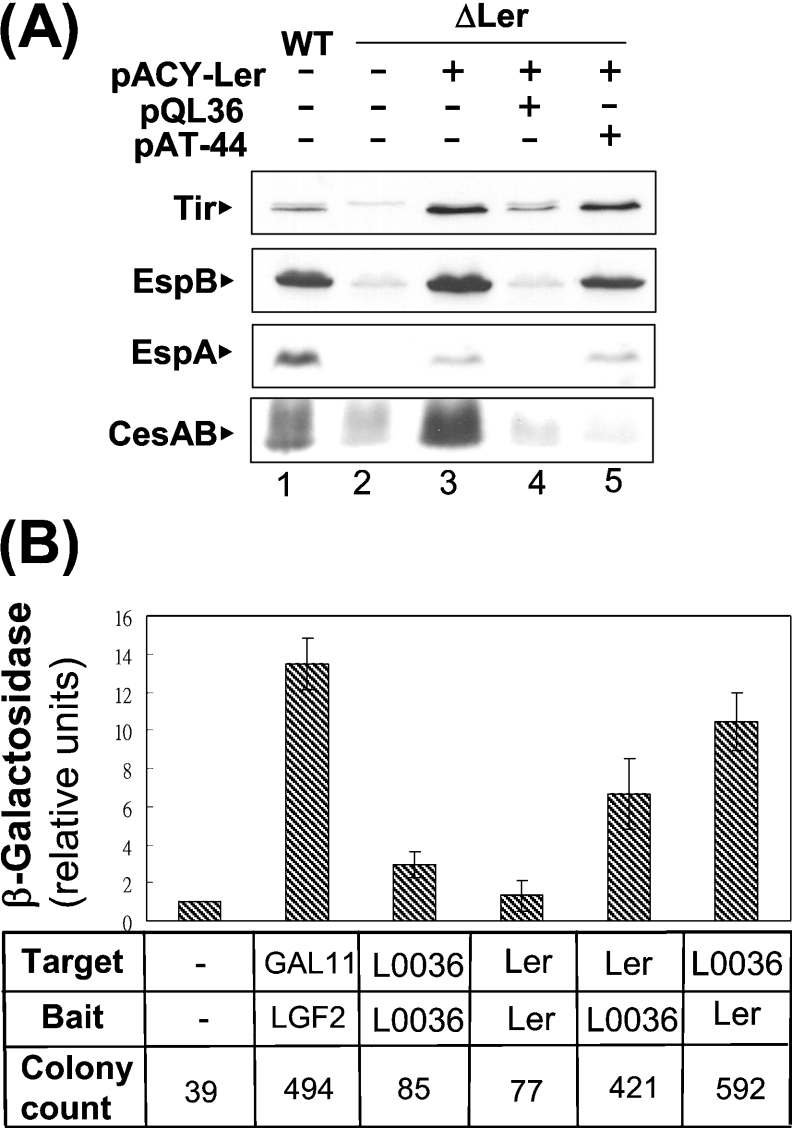
Figure 5. Ler is involved in the effects of L0036.
(A) Counteracting effect of L0036 on the activation of Ler on the LEE expression. Effects of expressing L0036 or L0044 in addition to Ler were examined by analysis of the LEE proteins in a Ler-deleted mutant, strain 43888 ΔLer. The mutant strain receiving various plasmids were analysed for the LEE proteins expressed in the bacterial lysates as described in the legend to Figure 1. (B) Interaction of Ler and L0036 as shown by a bacterial two-hybrid system. ORFs were cloned separately into expression plasmids to serve as target protein and bait. The interactions of the target and bait proteins in a reporter strain were revealed by an increase in colony number on a carbenicillin plate or by an increase in the bacterial β-galactosidase activity. Note: the known interaction between the dimerization domain of yeast transcriptional activator Gal4 (LGF2) and a domain derived from a mutant form of GAL11 (Stratagene) served as a positive control.
The above results suggest that L0036 suppressed the synthesis of the LEE proteins, perhaps by interaction with Ler, and therefore a bacterial two-hybrid system was used to test this possibility. In the system, interactions between the target protein and the bait were revealed by an increase in β-galactosidase activity as well in colony numbers when the bacteria were plated in the presence of carbenicillin (Figure 5B). Ler acted as the target, while L0036 acted as the bait, and gave a 6-fold increase in β-galactosidase activity as compared with the negative vector control. Consistently, the reciprocal experiment gave about a 10-fold increase in the activity. These results support the hypothesis that there is an interaction between L0036 and Ler, and this interaction seemed to be stronger than the self-interaction of Ler and that of L0036, since the latter two gave only 1.5-fold and 3-fold increases respectively. These β-galactosidase measurements were valid, since they agreed with the general tendency from the parallel scoring of the colony-formation units (Figure 5B).
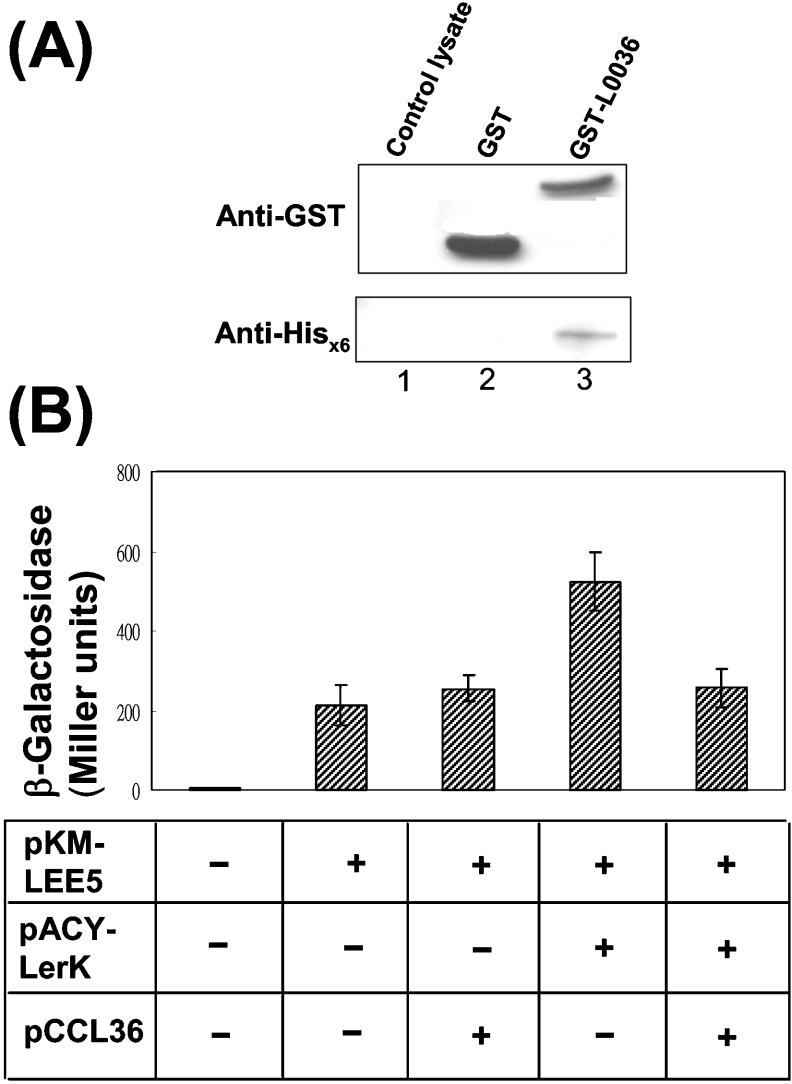
To further support the possible interaction between L0036 and Ler as suggested above by the two-hybrid system, a biochemical pull-down experiment was performed. Glutathione-conjugated agarose was used to retain the GST–L0036 fusion protein to which binding of Ler tagged with a His6 tag was addressed. Figure 6(A) shows that the His6-tagged Ler was bound to the agarose beads and effectively detected by Western blotting (lane 3). This binding was specific for GST-L0036 (lane 3), since Ler was not retained when GST (lane 2) or bacterial lysate (lane 1) was used as the control.
Figure 6. Interaction between L0036 and Ler.
(A) Interaction of L0036 with Ler shown by co-precipitation in a pull-down experiment. L0036 was fused to the C-terminus of GST, and this recombinant protein was bound to glutathione–Sepharose beads. Subsequent binding of His6-tagged Ler to the beads was examined by Western blotting using specific antibodies. (B) Co-expressing L0036 (from pCCL36) and Ler (from pACYLerK) decreased the activation effect of Ler on the LEE5 promoter. Bacteria (JM110) carrying a plasmid pKM-LEE5 that has lacZ driven by the LEE5 promoter were transformed by compatible plasmids. The extent of activation of the LEE5 promoter is reflected in the measured bacterial β-galactosidase activity. Measurements were taken from three separate colonies of the transformants.
As a result of interaction with L0036, the activation activity of Ler on LEE would be affected. To test this hypothesis, the LEE5 promoter that is regulated by Ler [9] was fused with lacZ, and the resulting pKM-LEE5 was used as a reporter plasmid. Figure 6(B) shows that E. coli JM 110, harbouring pKM-LEE5 alone, gave a low β-galactosidase activity similar to that shown by bacteria simultaneously carrying pKM-LEE5 and a low-copy L0036 expression plasmid, pCCL36. This observation suggests that L0036 exerts no direct function on the LEE5 promoter. In contrast, when the bacteria harboured pKM-LEE5 and pACYLerK, a pACY177-based Ler-expressing plasmid, the enzyme activity increased about 2-fold. This increase was as expected and agrees with the reported activation of Ler on the LEE5 promoter. In addition to pKM-LEE5, co-transformation of pCCL36 and pACYLerK gave β-galactosidase activity as low as the bacterium that harboured pKM-LEE5 alone. Apparently L0036 counteracted the effect of Ler. Therefore, the above observations consistently indicate that L0036 interacts with Ler directly, and this interaction could tune down the activation of Ler on LEE.
Mapping the suppression effect of L0036 to residues 1–29
To map the critical region of L0036 involved in the suppression of the LEE protein expression, a series of mutant plasmids were created with differently truncated l0036 (Figure 7A) in pQL36. These plasmids were then transformed into strain 43888, and their effects on the LEE expression were examined. Using the expression levels obtained with the wild-type bacteria as a reference, the relative amounts of LEE proteins expressed were estimated. Figure 7(B) shows the summarized results. An additional supply of the full-length L0036 to the bacteria suppressed the expression of Tir, EspB, EspA and CesAB by 70–90% relative to the reference (Figure 7A, construct a). Recombinant L0036 with increasing truncations from the C-terminus up to the residue 53 were stably produced in the bacterium, and these truncated products were all detectable by Western blotting using anti-His6 antibody (results not shown). These truncations did not effectively relieve the suppression effect (constructs b and c), a finding that suggests that the C-terminal portion of L0036 is not involved in such a function.
Construct d (i.e. pQL3629) contained the first 29 residues of L0036 and caused no obvious suppression of the expression of Tir, EspB, and CesAB and caused about 50% suppression of EspA expression. Since this construct was not detected in the bacterial lysate, no definite conclusion could be made. An alternative construct, pQL36Δ1-29 (e), was generated, in which the N-terminal first 29 residues were deleted. This construct was well expressed in the bacteria, and the bacteria gave LEE proteins in the lysate at relatively high levels (Figure 7B; construct e). When the truncation was shifted to residues 30–52 in pQL36Δ30-53 (construct f), LEE expression was suppressed again. Therefore, the critical residues of L0036 involved in suppressing LEE expression were concluded to be the N-terminal first 29 residues.
DISCUSSION
Defining the functions of ORFs in the LEE island is critical for a complete understanding of how EHEC and the related bacteria cause the A/E lesion. To reach this aim, a general strategy that has been used is to delete the specified ORFs and characterize the bacterial phenotypes and then link the observed phenotypes to the ORF's function [14]. Recently, all 41 genes of this island in C. rodentium have been systematically deleted. Parallel comparisons of the bacterial phenotypes have had a profound impact on the characterization of the undefined genes. One of the genes with properties so deduced is orf12, whose counterpart in EHEC is l0036. Thereby, l0036 of EHEC has been categorized by Deng et al. [10] into the group as part of the TTSS [10]. In contrast with their results, we have carried out detailed studies of l0036 by making mutants of this gene, complementing the protein encoded and characterizing how this gene and gene product affect expression of the other LEE proteins. The data provided all suggest that the function of l0036 and its gene product is to act on various aspects of LEE regulation.
Gene l0036 is located in LEE3, and many genes on LEE2 and LEE3 are part of the apparatus proteins responsible for the TTSS [22,23]. In the previously proposed TTSS, assembly of the secretion apparatus may be divided into stages and then proceed successively [21,24,25]. The secretion of the TTS proteins then depends on the integrity of the secretion apparatus. LEE4 and LEE5 encode proteins mainly for translocators and effectors to be secreted by TTSS. Thus, relative to LEE4 and LEE5, l0036, along with other genes responsible for the secretion apparatus on LEE2 and LEE3, may logically contribute to the assembly of the secretion apparatus. However, the fact that L0036 per se is not required for both secretion and synthesis of the LEE components (Figure 2) readily excludes the possibility that the function of this ORF is to encode a component of the apparatus [10]. Furthermore, escV and escN downstream of l0036 code for components of the secretion machinery, and their protein syntheses required the effective translation of l0036 (Figure 3B). Abolishing l0036 results in a loss of integrity of the TTSS and fully explains why a deletion of l0036 affects the secretion of the type III proteins, as observed previously [10]. Therefore the indispensable l0036 appears to function as a translational control that regulates the downstream genes of LEE3.
The fact that translation of l0036 has to occur has justified our attempts to determine what the function of the protein might be. Searching for homologous proteins by BLAST was unfruitful, given that the protein encoded by ORF12 of EPEC has been reported to have a low degree of homology with that of SsaM (a small protein encoded within Salmonella pathogenicity island 2) in Salmonella typhimurium [26]. We then used overexpression of this protein in the wild-type strain of EHEC and found a profound effect on the LEE protein expression. All the indicator proteins measured were severely down-regulated, and the degree of suppression was dosage-dependent upon the level of the L0036 protein expressed (Figure 4). Therefore, from the viewpoint of protein level, it is obvious that L0036 has to be maintained at a low level during the active stage of the LEE expression.
As to how L0036 mediates its suppression when its is overexpressed, our data in Figure 5(B) and Figure 6(A) have provided evidence, from both biochemical interaction and a genetic reporter system, to support the hypothesis that its action is mediated though Ler. Furthermore, in Figure 6(B), the activity of Ler was indeed proved to be down-regulated when L0036 was present.
In summary, l0036 has multiple functions and acts on the LEE expression. First, l0036 post-transcriptionally regulates the synthesis of the TTTS components that are encoded by the LEE3 operon; initiation of the L0036 translation is definitively required for the effective translation of the downstream genes. Secondly, the L0036 protein per se interacts with Ler and reduces its activation activity when overexpressed. In this regard, the increasing translation of L0036 seems to have a negative effect on the activation of LEE. Therefore, the required translation, but with the downsizing effect on the LEE activation of l0036, represents an intriguing property for this ORF. A third function of l0036 involved in the LEE regulation is attributed to the DNA sequence per se, which contains binding sites for the bacterial general regulatory protein H-NS as well as the specific LEE regulator Ler [8,12]. Therefore the DNA elements, the mRNA translation and the protein component from l0036 are all involved in the regulation of LEE expression, and lack of this gene adversely affects TTS, pedestal formation and virulence of the bacterium [10]. We therefore have named this important LEE gene mpc (multiple-point controller) and the encoded product Mpc.
The above mentioned functions of Mpc appear to be quite different from those observed with SsaM [26,27]. A deletion of ssaM does not have a polar effect on the downstream genes, although it does attenuate the bacterial virulence. In vitro, this ssaM-deleted mutant oversecretes SseJ and PipB, but fails to secrete proteins SseB, SseC and SseD. Furthermore, SsaM interacts with SpiC, another functionally similar protein originating from the same island, within the bacterial cell. Neither this peculiar interaction nor a secretion-switching effect of SaaM [27] have been seen with MpC. These functional differences between Mpc and SaaM could arise from the molecular difference rather than the homology, as a limited relatedness (17% identity and 41% similarity) exists between the sequence of SsaM and the first 93 amino acids encoded by ORF12. In addition, the interaction with SpiC has been attributed to the C-terminal region of SsaM [26], which is obviously distinct from that of Mpc.
Acknowledgments
We thank Dr R. Kirby (Department of Life Sciences, National Yang-Ming University, Taipei, Taiwan, Republic of China) for critical reading of the manuscript before its submission. We also thank Dr J. C. W. Lio and Dr Y. J. Wu (Institute of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan, Republic of China) for providing valuable plasmids. This work was supported in part by grant NSC 91-2320-B-010-091 MH from the National Science Council, Taiwan, Republic of China, and by grant 89-BFA22-2-4 from the Education Department, Taiwan, Republic of China.
References
- 1.Frankel G., Phillips A. D., Rosenshine I., Dougan G., Kaper J. B., Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: More subversive elements. Mol. Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 2.McDaniel T. K., Jarvis K. G., Donnenberg M. S., Kaper J. B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellies J. L., Elliott S. J., Sperandio V., Donnenberg M. S., Kaper J. B. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol. Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 4.Sperandio V., Mellies J. L., Nguyen W., Shin S., Kaper J. B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohaemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg D., Umanski T., Fang Y., Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 6.Bustamante V. H., Santana F. J., Calva E., Puente J. L. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 2001;39:664–678. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- 7.Elliott S. J., Sperandio V., Giron J. A., Shin S., Mellies J. L., Wainwright L., Hutcheson S. W., McDaniel T. K., Kaper J. B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohaemorrhagic Escherichia coli. Infect. Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperandio V., Mellies J. L., Delahay R. M., Frankel G., Crawford J. A., Nguyen W., Kaper J. B. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 2000;38:781–793. doi: 10.1046/j.1365-2958.2000.02168.x. [DOI] [PubMed] [Google Scholar]
- 9.Haack K. R., Robinson C. L., Miller K. J., Fowlkes J. W., Mellies J. L. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 2003;71:384–392. doi: 10.1128/IAI.71.1.384-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W., Puente J. L., Gruenheid S., Li Y., Vallance B. A., Vazquez A., Barba J., Ibarra J. A., O'Donnell P., Metalnikov P., Ashman K., et al. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lio J. C.-W., Syu W.-J. Identification of a negative regulator for the pathogenicity island of enterohaemorrhagic Escherichia coli O157:H7. J. Biomed. Sci. 2004;11:855–863. doi: 10.1007/BF02254371. [DOI] [PubMed] [Google Scholar]
- 12.Umanski T., Rosenshine I., Friedberg D. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology. 2002;148:2735–2744. doi: 10.1099/00221287-148-9-2735. [DOI] [PubMed] [Google Scholar]
- 13.Aizawa S. I. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 2001;202:157–164. doi: 10.1111/j.1574-6968.2001.tb10797.x. [DOI] [PubMed] [Google Scholar]
- 14.Link A. J., Phillips D., Church G. M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu H. J., Lin W. S., Syu W. J. Type III secretion of EspB in enterohaemorrhagic Escherichia coli O157:H7. Arch. Microbiol. 2003;180:218–226. doi: 10.1007/s00203-003-0579-7. [DOI] [PubMed] [Google Scholar]
- 16.Hsu S. C., Lin H. P., Wu J. C., Ko K. L., Sheen I. J., Yan B. S., Chou C. K., Syu W. J. Characterization of a strain-specific monoclonal antibody to hepatitis delta virus antigen. J. Virol. Methods. 2000;87:53–62. doi: 10.1016/s0166-0934(00)00147-6. [DOI] [PubMed] [Google Scholar]
- 17.Yan B. S., Lee K. M., Liu S. H., Syu W. J. Characterization of monoclonal antibodies to the 26-kDa glutathione S-transferase of Schistosoma japonicum. Hybridoma. 1996;15:429–433. doi: 10.1089/hyb.1996.15.429. [DOI] [PubMed] [Google Scholar]
- 18.Chang Y. Y., Yu S. L., Syu W. J. Organization of HIV-1 Pol is critical for pol polyprotein processing. J. Biomed. Sci. 1999;6:333–341. doi: 10.1007/BF02253522. [DOI] [PubMed] [Google Scholar]
- 19.Chuang C. H., Hsu S. C., Hsu C. L., Hsu T. C., Syu W. J. Construction of a tagging system for subcellular localization of proteins encoded by open reading frames. J. Biomed. Sci. 2001;8:170–175. doi: 10.1007/BF02256409. [DOI] [PubMed] [Google Scholar]
- 20.Miller J. H. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. Experiments in Molecular Genetics. [Google Scholar]
- 21.Gauthier A., Puente J. L., Finlay B. B. Secretion of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 2003;71:3310–3319. doi: 10.1128/IAI.71.6.3310-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis K. G., Giron J. A., Jerse A. E., McDaniel T. K., Donnenberg M. S., Kaper J. B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott S. J., Wainwright L. A., McDaniel T. K., Jarvis K. G., Deng Y. K., Lai L. C., McNamara B. P., Donnenberg M. S., Kaper J. B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli e2348/69. Mol. Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 24.Sukhan A., Kubori T., Galan J. E. Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 2003;185:3480–3483. doi: 10.1128/JB.185.11.3480-3483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimbrough T. G., Miller S. I. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X. J., Liu M., Holden D. W. SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators. Mol. Microbiol. 2004;54:604–619. doi: 10.1111/j.1365-2958.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- 27.Pallen M. J., Beatson S. A., Baile C. M. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 2005;5:9. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]