Abstract
Ectodomain shedding is a proteolytic mechanism by which transmembrane molecules are converted into a soluble form. Cleavage is mediated by metalloproteases and proceeds in a constitutive or inducible fashion. Although believed to be a cell-surface event, there is increasing evidence that cleavage can take place in intracellular compartments. However, it is unknown how cleaved soluble molecules get access to the extracellular space. By analysing L1 (CD171) and CD44 in ovarian carcinoma cells, we show in the present paper that the cleavage induced by ionomycin, APMA (4-aminophenylmercuric acetate) or MCD (methyl-β-cyclodextrin) is initiated in an endosomal compartment that is subsequently released in the form of exosomes. Calcium influx augmented the release of exosomes containing functionally active forms of ADAM10 (a disintegrin and metalloprotease 10) and ADAM17 [TACE (tumour necrosis factor α-converting enzyme)] as well as CD44 and L1 cytoplasmic cleavage fragments. Cleavage could also proceed in released exosomes, but only depletion of ADAM10 by small interfering RNA blocked cleavage under constitutive and induced conditions. In contrast, cleavage of L1 in response to PMA occurred at the cell surface and was mediated by ADAM17. We conclude that different ADAMs are involved in distinct cellular compartments and that ADAM10 is responsible for shedding in vesicles. Our findings open up the possibility that exosomes serve as a platform for ectodomain shedding and as a vehicle for the cellular export of soluble molecules.
Keywords: ADAM (a disintegrin and metalloproteinase), CD44, ectodomain shedding, exosome, L1, ovarian carcinoma
Abbreviations: ADAM, a disintegrin and metalloprotease; APMA, 4-aminophenylmercuric acetate; APP, amyloid precursor protein; BOG, β-octylglucopyranoside; CHO, Chinese-hamster ovary; IL-6, interleukin-6; mAb, monoclonal antibody; MCD, methyl-β-cyclodextrin; MVB, multivesicular body; NHS, N-hydroxysuccinimido; PE, phycoerythrin; siRNA, small interfering RNA; TNFα, tumour necrosis factor α; TACE, TNFα-converting enzyme; TNFR1, TNF receptor 1
INTRODUCTION
Ectodomain shedding is a proteolytic mechanism by which transmembrane molecules are converted into a soluble form [1–4]. This process plays an essential role in many cellular processes and allows the cell to rapidly adapt the surface phenotype and generate soluble mediators that can act on other cells. Shed proteins are diverse in structure and function and comprise molecules such as TNFα (tumour necrosis factor α), Fas ligand, IL-6 (interleukin-6) receptor, L-selectin, TGFα (transforming growth factor α), APP (amyloid precursor protein) and CD44. The ADAM (a disintegrin and metalloprotease) family of metalloproteases plays a pivotal role in ectodomain shedding [1–4]. Cleavage proceeds in a constitutive fashion but can be augmented by a variety of stimuli such as PMA [5–7], pervanadate [8], APMA (4-aminophenylmercuric acetate) [9–11], MCD (methyl-β-cyclodextrin) [12–15], calmodulin inhibitors [16–18] and substances that induce calcium influx [11,17,19]. It is unclear whether such diverse stimuli activate common or distinct pathways of shedding. A recent study on CD44 cleavage has shown that PMA-induced cleavage acts via ADAM17 [TACE (TNFα-converting enzyme)], whereas calcium influx-induced cleavage is mediated by ADAM10 [17]. Although initially believed to be a cell-surface event, there is increasing evidence that ectodomain cleavage also can take place in intracellular compartments [13,20]. However, the mechanism by which cleaved/soluble forms of the molecules are released into the extracellular space is unclear.
Exosomes are membrane vesicles that are released from a variety of different cell types including tumour cells, red blood cells, platelets, lymphocytes and dendritic cells [21,22]. Exosomes are formed by invagination and budding from the limiting membrane of late endosomes [23]. They accumulate in cytosolic MVBs (multivesicular bodies) from where they are released by fusion with the plasma membrane [23]. The process of vesicle release is particularly active in proliferating cells, such as cancer cells, where the release can occur continuously [24]. Exosomes can recruit various cytosolic and plasma membrane proteins including MHC molecules, tetraspanins, adhesion molecules and metalloproteases [21,22]. It has been proposed that exosomes released from tumour cells, due to their enrichment in proteolytic and adhesive activity, could promote cellular invasion and migration during metastasis [24,25].
The adhesion molecule L1 plays a crucial role in axon guidance and cell migration in the nervous system [26–28] and promotes motility and growth of human carcinoma cells [29–31]. Although a type I transmembrane molecule, L1 can undergo ectodomain shedding to release a soluble form into the extracellular space [8,13,29]. Soluble L1 stimulates cell migration [29] and is significantly increased in the serum and ascites of ovarian carcinoma patients [32,33]. Elevated levels of other shed molecules such as ErbB2 (HER2 or Neu) [34,35] and CD44 [36] have also been detected in ascites. For L1, we have reported that constitutive shedding involves the metalloprotease ADAM10 [29,33] and that cleavage occurs at the cell surface or in membrane vesicles released from tumour cells [13,33]. Membrane vesicles containing L1 cleavage fragments and ADAM10 could be isolated from media conditioned by carcinoma cell lines and ascites from ovarian carcinoma patients [33]. However, a direct link between ADAM10 cleavage of L1 in exosomes and the cleavage of other molecules is yet to be established.
In the present paper, we have more closely analysed the role of exosomes in ectodomain shedding. We demonstrate that ADAM10 and ADAM17 are present in exosomes from ovarian carcinoma cells and ascites fluid and are functionally active. We show that in addition to L1, CD44 is also cleaved in exosomes. We observed that stimuli including ionomycin, APMA and MCD, but not PMA, activated the release of exosomes and the cleavage of L1 and CD44 by ADAM10. Our findings open up the possibility that exosomes serve as a major platform for ectodomain shedding and as a vehicle for the export of soluble molecules.
EXPERIMENTAL
Cells
The ovarian carcinoma cell lines OVMz and SKOV3ip have been described previously [29,33]. Cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) foetal bovine serum, 100 units/ml penicillin/100 μg/ml streptomycin and 10 mM glutamine at 37 °C, 5% CO2 and 100% humidity.
Chemicals and antibodies
Antibodies specific for the ectodomain [L1-11A, subclone of mAb (monoclonal antibody) UJ 127.11] or cytoplasmic domain (pcytL1) of human L1 have been described [29]. mAb IM-7 to the ectodomain of CD44 was a gift from Dr Margot Zoeller of the German Cancer Research Center. The antibody to the cytoplasmic region of CD44 (pcytCD44 or CT1) was a gift from Dr Claire Isacke (Institute of Cancer Research, London, U.K.). The antibody to the C-terminus of ADAM10 (serum no. 71) was prepared in our laboratory [13] and the antibody to the C-terminus of ADAM17 was obtained from Chemikon (Hofheim, Germany). The mAb to the ADAM10 ectodomain (mAb 11G2) will be described in a separate publication (E. Rubinstein, unpublished work). This mAb reacts in Western blot with a human ADAM10–Fc fusion protein but does not recognize a human ADAM17–Fc (Y. Issa and P. Altevogt, unpublished work). The mAb to the ectodomain of ADAM17 (mAb 34/4D) was a gift from Dr Martin Humphries (Faculty of Life Sciences, Manchester, U.K.). A second mAb to the ectodomain of ADAM17 (mAb 9301) was from R&D Systems (Wiesbaden, Germany). The mAb to annexin I was from BD Transduction (Heidelberg, Germany) and the mAb to CD9 was described in [33]. APMA, PMA and EGTA were obtained from Sigma (Taufkirchen, Germany). Ionomycin-calcium salt was from Calbiochem (Bad Soden, Germany) and the metalloprotease inhibitor TAPI-0 was from Peptides International (Louisville, KY, U.S.A.). Triton X-100 and BOG (β-octylglucopyranoside) were from Gerbu (Gaiberg, Germany) and MCD was from Fluka (Buchs, Switzerland). Membrane-impermeable, thiol-cleavable EZ-link-NHS-SS-biotin (where NHS stands for N-hydroxysuccinimido) was obtained from Pierce (Perbio Science, Germany).
siRNA (small interfering RNA) transfection
ADAM10 (5′-AGACAUUAUGAAGGAUUAUTT-3′) and ADAM17 (5′-GAGAAGCUUGAUUCUUGCTT-3′) siRNAs were synthesized by MWG-Biotech (Ebersberg, Germany). Cells were transfected with annealed siRNAs using Oligofectamine (Life Technologies) and analysed 72 h later.
Isolation of membrane vesicles
Cells were cultivated overnight in a serum-free medium and then treated with or without ionomycin (1 μM), APMA (50 μM), MCD (10 mM) or PMA (50 ng/ml) for the indicated length of time. Tissue culture supernatants were collected and centrifuged for 10 min at 300 g and for 20 min at 10000 g to remove cellular debris. Membrane vesicles were collected by centrifugation at 100000 g for 2 h at 4 °C using a Beckman SW40 rotor. Vesicles were directly dissolved in SDS-sample buffer [30% sucrose, 80 mM Tris/HCl (pH 8.8), 3% SDS and 0.01 mg/ml Bromphenol Blue] or processed further for gradient centrifugation (see below).
Sucrose-density-gradient fractionation
Vesicles resuspended in 0.25 M sucrose were loaded on to the top of a step gradient comprising layers of 2, 1.3, 1.16, 0.8, 0.5 and 0.25 M sucrose as described in [13,33]. The gradient was previously calibrated for the floating of marker proteins for distinct cellular membranes [13,33]. The gradients were centrifuged at 100000 g for 2.5 h using a Beckman SW40 rotor. Twelve 1 ml fractions were collected from the top of the gradient and precipitated with chloroform/methanol (1:4, v/v). Samples were analysed by SDS/PAGE and Western blotting as described below.
Cell lysis and immunoprecipitation
Cells were detached from tissue flask ware by treatment with PBS/5 mM EDTA. Cell pellets or isolated vesicles were lysed in lysis buffer (20 mM Tris/HCl, pH 8.0, containing 50 mM BOG, 10 mM NaF, 10 mM orthovanadate, 1 mM PMSF and 1 μg/ml each of leupeptin, aprotinin and pepstatin). Lysates were cleared by centrifugation and either mixed with reducing SDS-sample buffer or used for immunoprecipitation. For immunoprecipitation, antibodies were preincubated with Protein G–Sepharose (Amersham Biosciences, Freiburg, Germany) for 60 min followed by washing with PBS. The precoated beads were then incubated with lysates for 60 min, washed three times with the lysis buffer and boiled with SDS-sample buffer for direct gel analysis. Alternatively, beads were suspended in cleavage buffer [50 mM Tris/HCl, pH 7.4, containing 2 mM CaCl2 and 0.1% (w/v) Triton X-100] and used in peptide cleavage assays. Immunoprecipitation of soluble L1 from conditioned medium was performed using L1-11A coupled with Sepharose as described previously [13].
Peptide cleavage assay
ADAM10 and ADAM17 immunoprecipitates in 90 μl were incubated with 10 μl (final concentration 50 mM) of the fluorescent quenching peptide Abz-Leu-Ala-Gln-Ala-Val-Arg-Ser-Ser-Ser-Arg-Dap(Dnp)-NH2 [where Abz is 2-aminobenzoyl and Dnp is (2,4-dinitrophenyl)-L-2,3-diaminopropionyl]; Peptide International, Louisville, KY, U.S.A.). Cleavage was followed in a fluorescent plate reader (Fluoroskan Ascent FL Thermo-Electron Corp.) using filters for excitation at 320 nm and emission at 405 nm. Where indicated, the metalloprotease inhibitor TAPI-0 was used at a final concentration of 10 μM.
FACS analysis
The staining of cells with mAbs and PE (phycoerythrin)-conjugated secondary antibodies has been described in [13,29]. Stained cells were analysed using a FACScan instrument and Cellquest software (Becton and Dickinson, Heidelberg, Germany). FACS analysis of vesicles was performed after adsorbtion of isolated vesicles on 4 μm (surfactant-free) aldehyde–sulphate latex beads (Interfacial Dynamics Corp., Portland, OR, U.S.A.) as described in [37].
Biochemical analysis
Cell-surface biotinylation was performed by the method of Warner et al. [38]. SDS/PAGE under reducing conditions and transfer of proteins on to Immobilon membranes using semi-dry blotting has been described previously [8,13]. After blocking with 5% (w/v) non-fat milk in TBS (Tris-buffered saline), blots were developed with the respective primary antibody followed by peroxidase-conjugated secondary antibody and ECL® detection. Densitometric analysis of band intensities was performed using the TINA software (Raytest, Straubenhardt, Germany).
RESULTS
Characterization of carcinoma-released microvesicles
We examined the expression of CD44, L1 and the metalloproteases ADAM10 and ADAM17 (TACE) in the ovarian carcinoma cell lines OVMz and SKOV3ip. All four molecules were expressed at the cell surface (Figure 1A). Both cell lines also released membrane vesicles into the medium that could be isolated by ultracentrifugation.
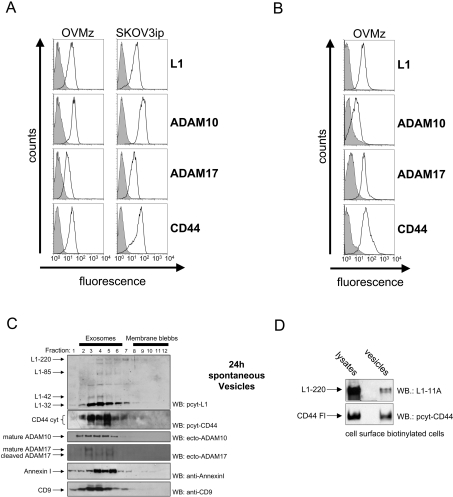
Figure 1. Characterization of tumour cell-derived exosomes.
(A) The indicated ovarian carcinoma cell lines were analysed by cytofluorographic analysis using mAbs to the indicated antigens followed by PE-conjugated secondary antibody to mouse IgG. (B) Membrane vesicles isolated from a medium conditioned by OVMz cells (cultivated under serum-free conditions) were adsorbed on latex beads and stained as described above. (C) Sucrose-density-gradient centrifugation of membrane vesicles. Gradient fractions were boiled with SDS-sample buffer and analysed by Western blotting (WB) with antibodies to exosomal marker proteins CD9 and annexin I, the metalloproteases ADAM10 and ADAM17, and L1 (pcytL1) and CD44 (pcytCD44) cleavage fragments. Separation of exosomes or membrane blebs based on sucrose density flotation was performed as described in [33]. Results are from one representative experiment of three performed. Note that L1-220 is often weakly detectable due to rapid cleavage. Molecular masses of each molecule are as follows: CD44cyt: multiple bands at 15–25 kDa; full-length CD44: approx. 97 kDa; mature ADAM10: 66 kDa; mature ADAM17: 95 kDa; cleaved ADAM17: 80 kDa; annexin I: 38 kDa; CD9: 20 kDa. (D) EZ-link-NHS-SS-biotin cell surface labelled OVMz cells were cultivated for 90 min in complete medium at 37 °C. Cells were then treated with glutathione at 4 °C to remove the non-internalized biotin label and were further cultivated overnight. Exosome release was induced by treatment with ionomycin-calcium salt (1 μM; for 1 h at 37 °C) and exosomes were collected by ultracentrifugation. Cells and exosomes were lysed in BOG lysis buffer and the lysates was adsorbed on streptavidin–agarose to isolate biotinylated proteins. After washing, the bound material was eluted by boiling with SDS-sample buffer. Blots were probed with the indicated antibodies to detect biotinylated L1 and CD44.
We next investigated whether these isolated vesicles were exosomes. Exosomes are released from cells by fusion of MVBs with the plasma membrane and have an orientation of antigens facing outside [21–23]. To determine the orientation of antigens, we immobilized vesicles from OVMz cells on to latex beads and carried out FACS analysis. Consistent with the profile of exosomes, the isolated vesicles were readily stained with antibodies to the ectodomains of L1, CD44, ADAM10 and ADAM17 (Figure 1B).
We carried out sucrose density centrifugation, in combination with Western-blot analysis for exosomal markers, to further characterize the released membrane vesicles. Fractions in the middle of the gradient were positive for the exosome markers CD9 and annexin I (Figure 1C) and the cytoskeletal proteins moesin and Hsp70 (heat-shock protein 70; results not shown) [21]. Western-blot analysis using antibodies to ADAM10 and ADAM17 demonstrated that both proteases co-localized in these exosomal fractions (Figure 1C). ADAM10 was present exclusively in the mature form. ADAM17 was detected in the mature form but smaller fragments most probably generated by proteolysis were also detected. Previous studies have shown that ADAM17 can undergo proteolysis after stimulation of cells [5] or under cell-lysis conditions [39,40].
Antibodies to the cytoplasmic domains of L1 and CD44 detected the full-length form and the cytoplasmic cleavage fragments (Figure 1C). In the case of CD44, multiple cytoplasmic cleavage fragments were detectable consistent with previous reports [17,41]. Western-blot analysis with pcytL1 detected the full-length L1-220 form and the major cleavage fragment (L1-32), all within the exosomal fractions (Figure 1C). These results indicate that full-length and ectodomain-processed forms of both CD44 and L1 and the relevant proteases implicated in ectodomain shedding are present in exosomes.
We assessed whether L1 and CD44 in exosomes were derived from the cell surface. Cell-surface proteins were coupled with the membrane-impermeable covalent linker EZ-link-NHS-SS-biotin and then returned to complete tissue culture medium to induce internalization. After 90 min at 37 °C, the remaining biotin label was cleaved from the proteins by the reducing agent, glutathione, at 4 °C as described in [38]. The cells were then cultivated overnight and the released exosomes were analysed for the content of biotinylated L1 and CD44. As shown in Figure 1(D), biotinylated forms of both molecules were indeed detected in exosomes. These findings suggest that the exosomal content is, at least in part, derived from the cell surface after endocytosis.
Ionomycin-mediated cleavage and exosome release
A recent study showed that the release of exosomes in K562 cells is augmented by substances that increase intracellular calcium levels [42]. Importantly, it has also been shown that ionophore-induced calcium influx can enhance the ADAM10-mediated ectodomain shedding of CD44 as well as other molecules [11,17]. Given the role of calcium influx in exosome release, we reasoned that ionomycin might activate vesicle-based proteases and augment the release of exosomes.
To test this, we stimulated SKOV3ip cells with ionomycin for various lengths of time in the presence or absence of the Ca2+ chelator EGTA. The onset of cleavage was analysed by sucrose-density-gradient centrifugation of cell homogenates. This fractionation allows the analysis of cleavage in distinct subcellular membranes [13]. L1 can be cleaved in the ectodomain by several proteases and the cleavage fragments generated have been described previously [13,29]. As shown in Figure 2(A), ionomycin led to a clear induction of the L1-32 cleavage fragment. Cleavage was mostly confined to the endosome/Golgi fractions of the gradient and could be blocked by EGTA. Although fulllength L1 was present in the plasma membrane fractions of the gradient, there was little L1-32 cleavage induced. In contrast, L1-85 cleavage that can be generated by plasmin and the proprotein convertase PC5A [18] was significantly enhanced. This observation suggested that L1-32 cleavage in ovarian carcinoma cells occurred mostly within the cells and not at the plasma membrane (see below).
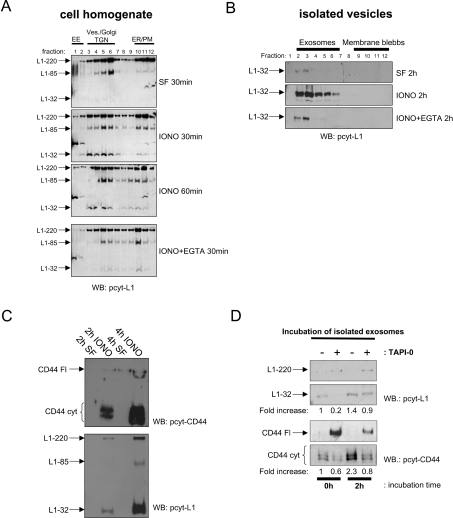
Figure 2. Effect of ionomycin on cleavage and exosomal release.
(A) Effect of ionomycin on L1 cleavage in SKOV3ip cells. Equal numbers of SKOV3ip cells in serum-free medium (SF) were treated with ionomycin-calcium salt (1 μM; IONO) for the indicated length of time in the presence or absence of EGTA (10 mM) at 37 °C. Cells and medium were separated and cell homogenates were layered on a stepwise sucrose gradient to separate intracellular and plasma membrane compartments according to buoyant density. Gradient fractions were analysed by Western blotting (WB) using the pcytL1 antibody. EE, early endosome; ves, exosomes; TGN, trans-Golgi network; ER, endoplasmic reticulum; PM, plasma membrane. The positions of the organelle marker proteins EEA-1 (endosome) and gm130 (Golgi) are shown. (B) Analysis of exosomes released from ionomycin-treated cells using sucrose-density-gradient centrifugation. For each gradient, 4×107 cells were treated. Fractions were analysed by Western blotting using pcytL1. (C) Timecourse analysis of exosomal release. Exosomes from 4×107 cells were examined, after 2 or 4 h of ionomycin treatment (1 μM), by Western blotting with the indicated primary antibodies. (D) Batches of 2×107 cells were treated with ionomycin-calcium salt for 2 h in the presence or absence of TAPI-0 (10 μM). Exosomes were isolated, split in half and further incubated at 37 °C for the indicated length of time and analysed for CD44 or L1 cleavage by Western blotting as described above. Note the relationship between cleaved and uncleaved forms of L1 and CD44 due to the blocking effect of TAPI-0. Results are from one representative experiment of three performed. Refer to Figure 1 for molecular masses.
We noticed a loss of L1-32 signal intensity upon prolonged ionomycin treatment (≥60 min; see Figure 2A). This suggested that cleavage fragments may be released from the cells. Therefore we examined the supernatant of treated cells for levels of exosomes using sucrose gradients as described above. Indeed, following ionomycin exposure, medium conditioned by SKOV3ip cells had increased amounts of exosomes containing the L1-32 cleavage fragment (Figure 2B). The buoyant density of vesicles was similar to the endosome/Golgi-containing fractions of the cell homogenate gradients (Figure 2B). Importantly, the effect of ionomycin on exosome release and L1 cleavage was largely prevented in the presence of the Ca2+ chelator EGTA, suggesting a crucial role for calcium influx (Figure 2B).
Cleavage of CD44 and L1 in exosomes proceeds after release
To further investigate whether cleavage was initiated in an intracellular compartment that subsequently was released from cells in exosomes, we carried out a time-course analysis. Prolonged treatment showed an increase in levels of CD44 and L1 cleavage fragments in exosomes (Figure 2C).
We also examined whether CD44 and L1 cleavage could continue after exosome release. Exosomes were isolated from supernatants of ionomycin-treated cells that were cultivated in the absence or presence of the metalloprotease inhibitor TAPI-0. Exosomes were further incubated at 37 °C and analysed at various time points for cleavage fragments. As indicated by the progressive appearance of L1-32 and CD44cyt, cleavage could proceed in isolated exosomes in a time-dependent fashion (Figure 2D).
Effects of other shedding inducers on exosome release
We tested various other compounds known as inducers of ectodomain shedding such as the protein kinase C activator PMA, the cholesterol-extracting agent MCD and the metalloprotease activator APMA. As shown in Figure 3(A), sucrose-density-gradient centrifugation of cell homogenates revealed induction of L1 cleavage by all compounds as measured by the appearance of the L1-32 cleavage fragment. Again, cleavage was mostly confined to the endosome/Golgi fractions of the gradient.
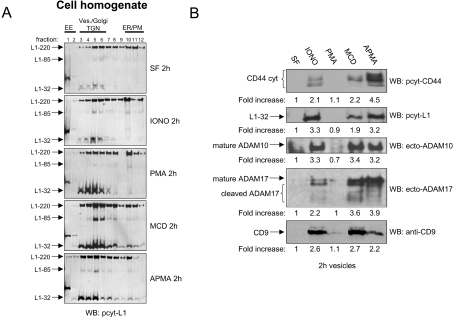
Figure 3. Effects of other shedding inducers on cleavage and exosomal release.
(A) Batches of 107 SKOV3ip cells in serum-free medium (SF) were treated with ionomycin-calcium salt (IONO 1 μM), PMA (50 ng/ml), APMA (50 μM) or MCD (10 mM) for 2 h at 37 °C. Cells and medium were separated and cell homogenates were layered on a stepwise sucrose gradient to separate intracellular organelles and plasma membranes. Gradient fractions were analysed by Western blotting (WB) using pcytL1. (B) Analysis of exosomes by Western blotting with the indicated antibodies. Results are from one representative experiment of three performed.
We examined the vesicle content of conditioned medium for the presence of L1 and CD44 cleavage fragments. Cleavage fragments of L1 and CD44 were clearly enhanced in released vesicles after ionomycin, MCD and APMA treatment (Figure 3B). Importantly, the vesicles released in response to ionomycin, APMA and MCD were identified as exosomes, as indicated by the presence of CD9 (Figure 3B). PMA treatment did not result in the release of cleavage fragments in exosomes although induction of cleavage in intracellular compartments was observed (see Figure 3A). PMA treatment was also without effect on the rate of exosome release from cells as indicated by Western-blot analysis of CD9 levels (Figure 3B). These findings suggested that PMA-induced ectodomain shedding of L1 and CD44 is distinct from that activated in response to ionomycin, APMA and MCD (see below).
ADAM10 and ADAM17 in exosomes are proteolytically active
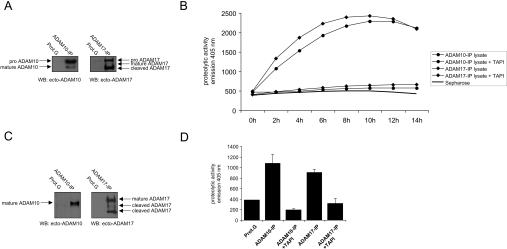
In order to more closely define the protease involved in exosomal cleavage of L1 and CD44, we analysed whether ADAM10 and ADAM17 were active. As both metalloproteases have been reported to cleave a TNFα peptide [43], we established a new peptide cleavage assay using immunoprecipitated enzymes. As shown in Figures 4(A) and 4(B), ADAM10 and ADAM17 were efficiently purified from cell lysates of SKOV3ip and OVMz cells and were able to cleave the TNFα peptide in a time-dependent fashion. Cleavage was inhibited in the presence of TAPI-0. A similar analysis was carried out for ADAM10 and ADAM17 from exosomes. As shown in Figures 4(C) and 4(D), immunaffinity-purified ADAM10 and ADAM17 isolated from BOG-solubilized exosomes were also functional in peptide cleavage assays and were again inhibited by TAPI-0.
Figure 4. Proteolytic activity of immunoaffinity-isolated ADAM10 and ADAM17.
Batches of 2×107 SKOV3ip cells were lysed in BOG lysis buffer and immunoprecipitated (IP) with ADAM10 (mAb 11G2) and ADAM17 (mAb 9301) specific mAbs. (A) Purity of immunoprecipitated ADAM10 and ADAM17 from lysates. An aliquot of each immunoprecipitated protease was subjected to Western-blot (WB) analysis using the indicated antibodies. (B) Measurement of proteolytic activity of lysate-derived metalloproteases using a fluorescent TNFα peptide cleavage assay. The time-course analysis was carried out in triplicate and means±S.D. were calculated. Only mean values are given. S.D. was less than 5% and is not shown. (C) Purity of immunoprecipitated ADAM10 and ADAM17 from exosomes. Isolated exosomes were lysed in BOG lysis buffer and used for immunoprecipitation. An aliquot of each immunoprecipitate was subjected to Western-blot analysis. (D) Measurement of proteolytic activity of exosome-derived proteases using a fluorescent TNFα peptide cleavage assay. The analysis was carried out in triplicate for 12 h. Results are presented as means±S.D. Results are from one representative experiment of at least five performed. Refer to Figure 1 for molecular masses. Additional bands are as follows: proADAM10, 85 kDa; proADAM17, 120 kDa. An additional cleavage fragment of ADAM17 with a mass of 60 kDa is visible in (C).
ADAM10-mediated cleavage of L1 and CD44 in exosomes
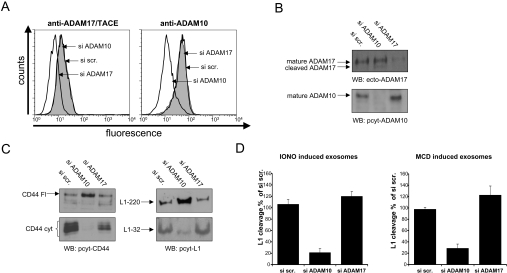
We used siRNA-mediated depletion of ADAM10 and ADAM17 to further investigate the role of each metalloprotease in exosomal cleavage of L1 and CD44. Transfection of OVMz cells with siRNAs led to down-regulation of the respective protease at the cell surface as detected by FACS analysis (Figure 5A) and in released exosomes as detected by Western blotting (Figure 5B). Cleavage of CD44 was completely blocked in ADAM10-depleted cells and an approx. 60% reduction was seen with ADAM17 depletion (Figure 5C). L1 cleavage was only inhibited following ADAM10 depletion.
Figure 5. siRNA-mediated depletion of ADAMs: effect on cleavage in exosomes.
Depletion of ADAM10 and ADAM17 in OVMz cells after transfection with specific siRNAs. A scrambled siRNA (si scr.) was used as the control. (A) Depletion at the cell surface. Cells were analysed, 48 h after transfection, by FACS using antibodies to ADAM10 (mAb 11G2) and ADAM17 (mAb 9301) followed by PE-conjugated goat anti-mouse IgG. (B) Depletion in exosomes. Exosomes were isolated from cells 48 h after transfection and analysed by Western blotting (WB). (C) Inhibition of constitutive CD44 and L1 cleavage in exosomes in ADAM10 siRNA-transfected cells. Note that inhibition of cleavage is associated with enhanced presence of full-length CD44 and L1. (D) Inhibition of ionomycin- and MCD-induced cleavage in exosomes by ADAM10-specific siRNA. Results are given as relative means±S.D. from duplicate samples and are based on densitometric analysis of L1-32 as depicted in (C). Results are from one representative experiment of at least three performed.
We also addressed the role of ADAM10 and ADAM17 in the induced ectodomain cleavage in exosomes. siRNA-transfected cells were treated with MCD or ionomcyin for 2 h and the released exosomes were analysed. Densitometric analysis of L1-32 band intensities in exosomes showed that ADAM10 was the dominant protease responsible for L1 and CD44 cleavage following MCD or ionomycin treatment (Figure 5D). Together, these results suggest that ADAM10 is the primary mediator of ectodomain cleavage in exosomes.
L1 cleavage at the cell surface
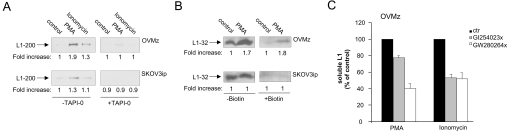
Our results so far suggested that PMA- and ionomycin-induced shedding might work via different mechanisms. We compared the amounts of soluble L1 released in response to PMA or ionomycin treatment. As shown in Figure 6(A), PMA induced the release of significant amounts of soluble L1 from OVMz cells, whereas SKOV3ip cells showed less induction of release. All cells responded to ionomycin although the amount of soluble L1 released was lower. As expected, the cleavage induced by PMA and ionomycin was blocked by TAPI-0.
Figure 6. L1 shedding at the cell surface.
(A) Analysis of soluble L1-200. Batches of 3×106 cells were stimulated with PMA (50 ng/ml) or ionomycin (1 μM) in the absence or presence of TAPI-0 (10 μM) for 1 h at 37 °C and supernatants were collected for analysis. Soluble L1 was immunoprecipitated with L1-11A coupled with Sepharose and analysed by Western-blot analysis. (B) The indicated cells were cell surface-biotinylated after PMA treatment (50 ng/ml) for 1 h at 37 °C or without treatment. Cells were lysed and then half of the lysate was adsorbed on streptavidin–agarose to isolate biotinylated proteins. The second half served as positive control. After washing, the bound material was eluted by boiling with SDS-sample buffer. Blots were probed with pcytL1 to detect the biotinylated L1-32 cleavage product. (C) Effect of specific metalloprotease inhibitors GW280264X (equally specific for ADAM17 and ADAM10) and GI254023X (100-fold more specific for ADAM10) on the cleavage of L1 in OVMz cells using PMA and ionomycin stimulation. Conditioned medium was analysed for soluble L1 using an L1-specific ELISA as previously described [29]. Results are from one representative experiment of at least three performed. ctr, control.
It has been reported that ADAM17 is responsible for the PMA-induced cleavage of CD44 at the cell surface [17]. Since FACS staining results (see Figure 1A) and cell-surface biotinylation experiments (results not shown) detected cell-surface localization of ADAM10 and ADAM17, we examined whether cleavage was indeed taking place at the plasma membrane. We took advantage of the fact that the membrane-retained L1-32 cleavage fragment can be biotinylated at the cell surface. As shown in Figure 6(B), biotinylation experiments detected L1-32 at the surface of OVMz but not SKOV3ip cells following treatment with PMA for 2 h. Non-biotinylated L1-32 was clearly detected in all cells.
To analyse whether PMA or ionomycin used different ADAMs for cleavage, we used specific inhibitors of ADAM10 and ADAM17. The compound GW280264X blocks both ADAM10 and ADAM17 with comparable IC50 values, whereas compound GI254023X has 100-fold higher specificity towards ADAM10 [44]. As shown in Figure 6(C), we observed significantly higher inhibition with GW280264X upon PMA treatment of OVMz cells. In contrast, both inhibitors worked about equally well under ionomycin induction of L1 cleavage. These results suggest that PMA-induced L1 shedding in OVMz cells is ADAM17-dependent. The shedding process becomes ADAM10-dependent and occurs in exosomes when cells are treated with ionomycin.
DISCUSSION
An important role for exosomes in normal and malignant cell–cell communication is increasingly recognized. Soluble forms of a variety of molecules such as TNFR1 (TNF receptor 1), IL-1β and FGF-2 (fibroblast growth factor-2) can be released from cells within vesicles [45–47]. In the case of TNFR1, the full-length transmembrane molecule is released in exosome-like vesicles and this serves as a mechanism for the generation of a soluble form of this cytokine receptor [45]. In addition, the ability of exosomes to mediate MHC class I- and class II-restricted T-cell stimulatory activity suggests that they may have a potential application in immunotherapy [22,48]. In the present paper, we demonstrate that exosomes represent a novel platform for cellular export of cleaved L1 and CD44. This conclusion is based on the findings that (i) both L1 and CD44 are detectable in exosomes released from carcinoma cells, (ii) the metalloproteases ADAM10 and ADAM17 implicated in shedding are present in an active form in exosomes, (iii) siRNA specific for ADAM10 but not ADAM17 blocked cleavage in exosomes and (iv) the cytoplasmic cleavage products of CD44 and L1 accumulate in exosomes in response to shedding inducers such as ionomycin, APMA and MCD. We suggest that the export of cleaved proteins via exosomes could be an additional pathway for the release of transmembrane molecules.
We observed that ionomycin, APMA and MCD initiated cleavage within an intracellular compartment with a profile consistent with endosome/Golgi-derived membranes. At the same time, these treatments augmented the release of vesicles. This suggested a link between ectodomain cleavage and exosome release. Pharmacological agents such as calcium ionophores (e.g. ionomycin and A23187), APMA and MCD have previously been shown to induce ectodomain shedding of a variety of transmembrane molecules and, consistent with our findings, in some cases this has been shown to involve ADAM10 [9–12,14,15]. However, a particular role for ectodomain shedding within vesicles has not been previously investigated. Exosomal proteins are believed to be derived from early endocytic vesicles [23]. Such vesicles can fuse with the outer membrane of MVBs, which requires interaction with several factors such as ESCRT-I (endosomal sorting complex required for transport I)–ESCRT-III, VPS4 (vacuolar protein sorting) and ALIX (ALG-2 interacting protein X) [23]. Calcium influx can activate the docking and fusion of vesicles in a Rab-11-dependent fashion [49]. Exosomes are then formed by inward budding from the limiting membrane of MVBs and are released into the extracellular space by fusion of MVBs with the plasma membrane [23]. Our previous studies have shown that in addition to lowering cellular cholesterol, MCD and the HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) reductase inhibitor lovastatin caused enhanced membrane vesicle release and L1 cleavage [13]. In the present study, we extend these findings to CD44 and demonstrate that ionomycin, APMA and MCD act via a common pathway involving cleavage and release of these proteins in exosomes.
Our studies identified functionally active ADAM10 and ADAM17 in exosomes from ovarian carcinoma cells (the present study) and importantly in vivo within ascites fluid ([33]; S. Keller, unpublished work). ADAM10 was the predominant protease responsible for L1 and CD44 cleavage in exosomes from OVMz and SKOV3ip carcinoma cells. This was not due to an absence of active ADAM17, as identified by the functional activity of this enzyme in peptide cleavage assays. It is also unlikely that the dominant activity of ADAM10 is due to substrate specificity for L1 and CD44, since CD44 can be cleaved by ADAM17 [17] and additional metalloproteases such as MT1-MMP (membrane type-1 matrix metalloprotease) [41], whilst L1 can be cleaved by ADAM17 in CHO-hL1 cells [CHO (Chinese-hamster ovary) cells stably transfected with human L1] (S. Riedle, unpublished work) and OVMz cells (see below). It is possible that the unique preference for ADAM10 over ADAM17 in exosomal cleavage is due to the ability of ADAM10 to become activated in response to stimuli such as ionomycin, APMA and MCD. A recent report indicated that ADAM10, but not ADAM17, can directly bind to calmodulin [17]. It was proposed that calcium influx or calmodulin inhibitors trigger the dissociation of calmodulin from pro-ADAM10 and subsequent maturation to active ADAM10 which can cleave CD44. In the case of MCD, activation of ADAM10- and ADAM17-dependent ectodomain shedding events is believed to involve a reduction in cellular cholesterol levels [14]. Our findings support the notion that CD44 cleavage in response to calcium flux is mediated by ADAM10 and extend this to other stimuli and a role for exosomes. Further studies are warranted to determine whether ADAM10–calmodulin interactions, as well as modification of ADAM10 function, could possibly take place within endosomes/MVBs.
PMA did not augment exosome release. We have shown previously that PMA treatment predominantly activates ectodomain shedding events at the plasma membrane [13]. Indeed, cell-surface cleavage of L1 in OVMz cells was up-regulated by PMA and involved ADAM17 as shown by inhibitor analysis. There was less PMA-induced cleavage seen in SKOV3ip cells. It appears that in some cell types the constitutive cleavage is intact but PMA-inducible cleavage is impaired. Indeed, defective PMA-induced cleavage has been observed previously. For example, ADAM17 knockout cells display functional constitutive ectodomain shedding of APP but are defective in PMA-induced cleavage in comparison with wild-type cells [49,50]. Meanwhile, cleavage-deficient CHO mutant cells (M2) display deficient PMA-induced cleavage of several transmembrane molecules [40,51]. The differences in shedding events between alternative cell types require further investigation.
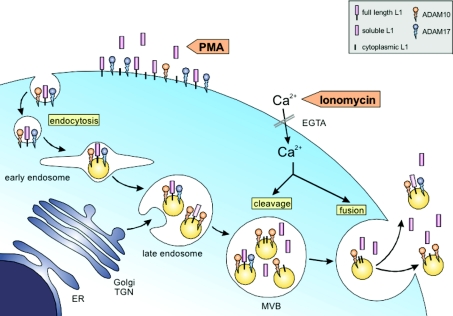
Our findings have identified two distinct pathways of cleavage for the transmembrane molecules L1 and CD44 that are depicted in the model shown in Figure 7. The first involves ADAM17-mediated cleavage at the cell surface, leading to shedding of soluble forms into the extracellular space. The second pathway involves ADAM10-mediated cleavage in endosomes that are subsequently released from the cell as exosomes where further ADAM10-dependent cleavage can also occur. In summary, our results underline the important role of ADAM10 and exosomes in the constitutive and stimulus-induced ectodomain shedding of L1 and CD44 from carcinoma cells. We propose that this release via exosomes is a novel pathway for ectodomain shedding of L1 and CD44 and indeed may be applicable to other established ADAM10 substrates.
Figure 7. Ectodomain cleavage in exosomes: a model.
Cleavage of L1 can be triggered by PMA and proceeds predominantly at the cell surface, leading to a rapid increase of soluble L1 in the extracellular space. This cleavage is mediated by ADAM17. A second pathway of cleavage is initiated in endosomes/MVBs and is triggered by calcium influx. This causes enhanced cleavage and release of L1 in the form of exosomes. After exosome release from the cell, further cleavage can also occur. Enhanced cleavage and release are also seen in response to MCD and APMA (not shown). Cleavage in exosomes is predominantly mediated by ADAM10. ER, endoplasmic reticulum, TGN, trans-Golgi network.
Acknowledgments
We thank Verena Gschwend and Sascha Hinterkopf for excellent technical assistance. This work was supported by a grant from the Deutsche Krebshilfe (Schwerpunktprogramm Migration and Invasion) and the European Community (EC-Strep Signalling & Traffic) to P. A.
References
- 1.Schlondorff J., Blobel C. P. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- 2.Arribas J., Borroto A. Protein ectodomain shedding. Chem. Rev. 2002;102:4627–4638. doi: 10.1021/cr010202t. [DOI] [PubMed] [Google Scholar]
- 3.Seals D. F., Courtneidge S. A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 4.Blobel C. P. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell. Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 5.Endres K., Anders A., Kojro E., Gilbert S., Fahrenholz F., Postina R. Tumor necrosis factor-alpha converting enzyme is processed by proprotein-convertases to its mature form which is degraded upon phorbol ester stimulation. Eur. J. Biochem. 2003;270:2386–2393. doi: 10.1046/j.1432-1033.2003.03606.x. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald M. L., Wang Z., Park P. W., Murphy G., Bernfield M. Shedding of syndecan-1 and -4 ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-3-sensitive metalloproteinase. J. Cell Biol. 2000;148:811–824. doi: 10.1083/jcb.148.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasbarri A., Del P. F., Girnita L., Martegani M. P., Natali P. G., Bartolazzi A. CD44s adhesive function spontaneous and PMA-inducible CD44 cleavage are regulated at post-translational level in cells of melanocytic lineage. Melanoma Res. 2003;13:325–337. doi: 10.1097/00008390-200308000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Gutwein P., Oleszewski M., Mechtersheimer S., Agmon L. N., Krauss K., Altevogt P. Role of Src kinases in the ADAM-mediated release of L1 adhesion molecule from human tumor cells. J. Biol. Chem. 2000;275:15490–15497. doi: 10.1074/jbc.275.20.15490. [DOI] [PubMed] [Google Scholar]
- 9.Merlos-Suarez A., Ruiz-Paz S., Baselga J., Arribas J. Metalloprotease-dependent protransforming growth factor-alpha ectodomain shedding in the absence of tumor necrosis factor-alpha-converting enzyme. J. Biol. Chem. 2001;276:48510–48517. doi: 10.1074/jbc.M103488200. [DOI] [PubMed] [Google Scholar]
- 10.Allinson T. M., Parkin E. T., Condon T. P., Schwager S. L., Sturrock E. D., Turner A. J., Hooper N. M. The role of ADAM10 and ADAM17 in the ectodomain shedding of angiotensin converting enzyme and the amyloid precursor protein. Eur. J. Biochem. 2004;271:2539–2547. doi: 10.1111/j.1432-1033.2004.04184.x. [DOI] [PubMed] [Google Scholar]
- 11.Sanderson M. P., Erickson S. N., Gough P. J., Garton K. J., Wille P. T., Raines E. W., Dunbar A. J., Dempsey P. J. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J. Biol. Chem. 2005;280:1826–1837. doi: 10.1074/jbc.M408804200. [DOI] [PubMed] [Google Scholar]
- 12.Kojro E., Gimpl G., Lammich S., Marz W., Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha-secretase ADAM 10. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Gast D., Joumaa S., Zentgraf H., Fogel M., Altevogt D. P. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003;17:292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 14.Matthews V., Schuster B., Schutze S., Bussmeyer I., Ludwig A., Hundhausen C., Sadowski T., Saftig P., Hartmann D., Kallen K. J., et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J. Biol. Chem. 2003;278:38829–38839. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 15.von Tresckow B., Kallen K. J., von Strandmann E. P., Borchmann P., Lange H., Engert A., Hansen H. P. Depletion of cellular cholesterol and lipid rafts increases shedding of CD30. J. Immunol. 2004;172:4324–4331. doi: 10.4049/jimmunol.172.7.4324. [DOI] [PubMed] [Google Scholar]
- 16.Kahn J., Walcheck B., Migaki G. I., Jutila M. A., Kishimoto T. K. Calmodulin regulates L-selectin adhesion molecule expression and function through a protease-dependent mechanism. Cell (Cambridge, Mass.) 1998;92:809–818. doi: 10.1016/s0092-8674(00)81408-7. [DOI] [PubMed] [Google Scholar]
- 17.Nagano O., Murakami D., Hartmann D., De S. B., Saftig P., Iwatsubo T., Nakajima M., Shinohara M., Saya H. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 2004;165:893–902. doi: 10.1083/jcb.200310024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalus I., Schnegelsberg B., Seidah N. G., Kleene R., Schachner M. The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J. Biol. Chem. 2003;278:10381–10388. doi: 10.1074/jbc.M208351200. [DOI] [PubMed] [Google Scholar]
- 19.Jones S. A., Horiuchi S., Novick D., Yamamoto N., Fuller G. M. Shedding of the soluble IL-6 receptor is triggered by Ca2+ mobilization, while basal release is predominantly the product of differential mRNA splicing in THP-1 cells. Eur. J. Immunol. 1998;8:3514–3522. doi: 10.1002/(SICI)1521-4141(199811)28:11<3514::AID-IMMU3514>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 20.Skovronsky D. M., Moore D. B., Milla M. E., Doms R. W., Lee V. M. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J. Biol. Chem. 2000;275:2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- 21.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 22.Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 23.Raiborg C., Rusten T. E., Stenmark H. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 24.Ginestra A., La Placa M. D., Saladino F., Cassara D., Nagase H., Vittorelli M. L. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998;18:3433–3437. [PubMed] [Google Scholar]
- 25.Dolo V., Ginestra A., Cassara D., Violini S., Lucania G., Torrisi M. R., Nagase H., Canevari S., Pavan A., Vittorelli M. L. Selective localization of matrix metalloproteinase 9, beta1 integrins, and human lymphocyte antigen class I molecules on membrane vesicles shed by 8701-BC breast carcinoma cells. Cancer Res. 1998;58:4468–4474. [PubMed] [Google Scholar]
- 26.Hortsch M. The L1 family of neural cell adhesion molecules: old proteins performing new tricks. Neuron. 1996;17:587–593. doi: 10.1016/s0896-6273(00)80192-0. [DOI] [PubMed] [Google Scholar]
- 27.Haspel J., Grumet M. The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front. Biosci. 2003;8:s1210–s1225. doi: 10.2741/1108. [DOI] [PubMed] [Google Scholar]
- 28.Schachner M. Neural recognition molecules and synaptic plasticity. Curr. Opin. Cell Biol. 1997;9:627–634. doi: 10.1016/s0955-0674(97)80115-9. [DOI] [PubMed] [Google Scholar]
- 29.Mechtersheimer S., Gutwein P., Agmon-Levin N., Stoeck A., Oleszewski M., Riedle S., Postina R., Fahrenholz F., Fogel M., Lemmon V., et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gast D., Riedle S., Schabath H., Schlich S., Schneider A., Issa Y., Stoeck A., Fogel M., Joumaa S., Wenger T., et al. L1 augments cell migration and tumor growth but not beta3 integrin expression in ovarian carcinomas. Int. J. Cancer. 2005;115:658–665. doi: 10.1002/ijc.20869. [DOI] [PubMed] [Google Scholar]
- 31.Gavert N., Conacci-Sorrell M., Gast D., Schneider A., Altevogt P., Brabletz T., Ben-Ze'ev A. L1, a novel target of {beta}-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J. Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogel M., Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Smirnov A., Edler L., Ben A. A., Huszar M., Altevogt P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003;362:869–875. doi: 10.1016/S0140-6736(03)14342-5. [DOI] [PubMed] [Google Scholar]
- 33.Gutwein P., Stoeck A., Riedle S., Gast D., Runz S., Condon T. P., Marme A., Phong M. C., Linderkamp O., Skorokhod A., et al. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clin. Cancer Res. 2005;11:2492–2501. doi: 10.1158/1078-0432.CCR-04-1688. [DOI] [PubMed] [Google Scholar]
- 34.Pupa S. M., Menard S., Morelli D., Pozzi B., De Palo G., Colnaghi M. I. The extracellular domain of the c-erbB-2 oncoprotein is released from tumor cells by proteolytic cleavage. Oncogene. 1993;8:2917–2923. [PubMed] [Google Scholar]
- 35.Codony-Servat J., Albanell J., Lopez-Talavera J. C., Arribas J., Baselga J. Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 1999;59:1196–1201. [PubMed] [Google Scholar]
- 36.Gadducci A., Ferdeghini M., Fanucchi A., Annicchiarico C., Cosio S., Prontera C., Bianchi R., Genazzani A. R. Serum assay of soluble CD44 standard (sCD44-st), CD44 splice variant v5 (sCD44-v5), and CD44 splice variant v6 (sCD44-v6) in patients with epithelial ovarian cancer. Anticancer Res. 1997;17:4463–4466. [PubMed] [Google Scholar]
- 37.Blanchard N., Lankar D., Faure F., Regnault A., Dumont C., Raposo G., Hivroz C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 2002;168:3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 38.Warner F. J., Lew R. A., Smith A. I., Lambert D. W., Hooper N. M., Turner A. J. Angiotensin-converting enzyme 2 (ACE2), but not ACE, is preferentially localised to the apical surface of polarised kidney cells. J. Biol. Chem. 2005 doi: 10.1074/jbc.M508914200. in the press. [DOI] [PubMed] [Google Scholar]
- 39.Schlondorff J., Becherer J. D., Blobel C. P. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE) Biochem. J. 2000;347:131–138. [PMC free article] [PubMed] [Google Scholar]
- 40.Borroto A., Ruiz-Paz S., de la Torre T. V., Borrell-Pages M., Merlos-Suarez A., Pandiella A., Blobel C. P., Baselga J., Arribas J. Impaired trafficking and activation of tumor necrosis factor-alpha-converting enzyme in cell mutants defective in protein ectodomain shedding. J. Biol. Chem. 2003;278:25933–25939. doi: 10.1074/jbc.M301673200. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura H., Suenaga N., Taniwaki K., Matsuki H., Yonezawa K., Fujii M., Okada Y., Seiki M. Constitutive and induced CD44 shedding by ADAM-like proteases and membrane-type 1 matrix metalloproteinase. Cancer Res. 2004;64:876–882. doi: 10.1158/0008-5472.can-03-3502. [DOI] [PubMed] [Google Scholar]
- 42.Savina A., Furlan M., Vidal M., Colombo M. I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 43.Lunn C. A., Fan X., Dalie B., Miller K., Zavodny P. J., Narula S. K., Lundell D. Purification of ADAM 10 from bovine spleen as a TNFalpha convertase. FEBS Lett. 1997;400:333–335. doi: 10.1016/s0014-5793(96)01410-x. [DOI] [PubMed] [Google Scholar]
- 44.Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 2003;102:1186–1195. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 45.Hawari F. I., Rouhani F. N., Cui X., Yu Z. X., Buckley C., Kaler M., Levine S. J. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1297–1302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacKenzie A., Wilson H. L., Kiss-Toth E., Dower S. K., North R. A., Surprenant A. Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 47.Taverna S., Ghersi G., Ginestra A., Rigogliuso S., Pecorella S., Alaimo G., Saladino F., Dolo V., Dell'Era P., Pavan A., et al. Shedding of membrane vesicles mediates fibroblast growth factor-2 release from cells. J. Biol. Chem. 2003;278:51911–51919. doi: 10.1074/jbc.M304192200. [DOI] [PubMed] [Google Scholar]
- 48.Chaput N., Taieb J., Schartz N. E., Andre F., Angevin E., Zitvogel L. Exosome-based immunotherapy. Cancer Immunol. Immunother. 2004;53:234–239. doi: 10.1007/s00262-003-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savina A., Fader C. M., Damiani M. T., Colombo M. I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6:131–143. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 50.Buxbaum J. D., Liu K. N., Luo Y., Slack J. L., Stocking K. L., Peschon J. J., Johnson R. S., Castner B. J., Cerretti D. P., Black R. A. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 51.Merlos-Suarez A., Fernandez-Larrea J., Reddy P., Baselga J., Arribas J. Pro-tumor necrosis factor-alpha processing activity is tightly controlled by a component that does not affect notch processing. J. Biol. Chem. 1998;273:24955–24962. doi: 10.1074/jbc.273.38.24955. [DOI] [PubMed] [Google Scholar]