Abstract
The cyclotides are a family of circular proteins with a range of biological activities and potential pharmaceutical and agricultural applications. The biosynthetic mechanism of cyclization is unknown and the discovery of novel sequences may assist in achieving this goal. In the present study, we have isolated a new cyclotide from Oldenlandia affinis, kalata B8, which appears to be a hybrid of the two major subfamilies (Möbius and bracelet) of currently known cyclotides. We have determined the three-dimensional structure of kalata B8 and observed broadening of resonances directly involved in the cystine knot motif, suggesting flexibility in this region despite it being the core structural element of the cyclotides. The cystine knot motif is widespread throughout Nature and inherently stable, making this apparent flexibility a surprising result. Furthermore, there appears to be isomerization of the peptide backbone at an Asp-Gly sequence in the region involved in the cyclization process. Interestingly, such isomerization has been previously characterized in related cyclic knottins from Momordica cochinchinensis that have no sequence similarity to kalata B8 apart from the six conserved cysteine residues and may result from a common mechanism of cyclization. Kalata B8 also provides insight into the structure–activity relationships of cyclotides as it displays anti-HIV activity but lacks haemolytic activity. The ‘uncoupling’ of these two activities has not previously been observed for the cyclotides and may be related to the unusual hydrophilic nature of the peptide.
Keywords: anti-HIV activity, cyclic backbone, cystine knot motif, haemolytic, kalata B8, NMR
Abbreviations: DQF-COSY, double-quantum-filtered COSY; EndoGluC, endoproteinase Glu C; ER, endoplasmic reticulum; LC-MS, liquid chromatography–MS; NOE, nuclear Overhauser effect; NTR, N-terminal repeat; RP, reverse phase
INTRODUCTION
The cyclotides are an expanding family of small proteins that are characterized by three disulphide bonds forming a cystine knot motif and a head-to-tail cyclized backbone [1]. They have been isolated from plants in the Rubiaceae, Violaceae and Cucurbitaceae families and display a range of biological activities, including uterotonic, antimicrobial and cytotoxic activities [2–7]. Insecticidal activity against Helicoverpa caterpillar species suggests that cyclotides are present in plants as a defence mechanism [8,9]. The cyclotides are highly stable proteins with compact three-dimensional structures that have potential applications as drug design templates [10,11]. Furthermore, their intrinsic insecticidal activity may also be useful in agricultural applications.
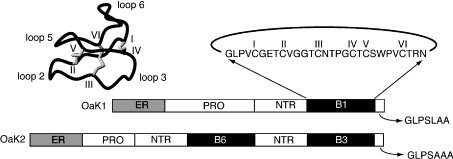
Understanding how the cyclotides are produced in vivo is an important question in their biology that is at present unanswered. However, it is known that they are gene-encoded [9,12] and the discovery of genes for a range of cyclotides has provided preliminary insights into the factors involved in processing of the mature circular peptides. The genes encoding the cyclotides were originally isolated from Oldenlandia affinis and contain an ER (endoplasmic reticulum) signal sequence, a pro-region and a mature cyclotide domain [9]. In some clones, multiple mature cyclotide domains are present. All cyclotide domains are preceded by a short conserved pro-sequence referred to as the NTR (N-terminal repeat) conserved domain. Figure 1 shows a summary of the gene organization for two of the clones isolated from a cDNA library prepared from O. affinis leaves [9] as well as the sequence of the prototypic cyclotide kalata B1 [13,14]. The mature peptide contains six conserved cysteine residues that form a cystine knot. The backbone sequences between the cysteine residues are referred to as loops, as shown in Figure 1. Based on the gene sequences, it appears that processing to form the mature circular proteins occurs in the region that becomes loop 6 in the mature peptide.
Figure 1. Gene organization of cyclotides and the three-dimensional structure of the prototypic cyclotide kalata B1.
A schematic representation of two of the genes from O. affinis is shown at the bottom of the Figure. The genes comprise a signal sequence labelled ER, a pro-region, a conserved repeated fragment labelled NTR and either one or multiple copies of the mature peptides, e.g. B1, B3 and B6. A short hydrophobic C-terminal tail is coded for at the end of the gene. The amino acid sequence of kalata B1 is shown with the cysteine residues labelled with roman numerals. The six inter-cysteine loops are labelled 1–6 on the structure in the top left.
The number of cyclotide sequences has been expanding recently, with more than 60 sequences now published [1]. Based on topological factors and sequence similarity, the cyclotides fall into two major subfamilies. This classification was originally proposed based on the presence or absence of a proline residue in loop 5, with those containing this proline referred to as Möbius cyclotides and those without referred to as bracelet cyclotides [15]. In two example cases, this proline has been shown to be in the cis conformation, introducing a conceptual twist in the otherwise all-trans backbone and leading to the name Möbius [8,16]. A third subfamily, which is referred to as the trypsin inhibitor cyclotides, currently contains only two members (MCoTI-I and MCoTI-II) that are quite different from the other cyclotides [17]. No sequence homology to either the Möbius or bracelet cyclotides is present in this subfamily, with the exception of the six conserved cysteine residues. The structure of MCoTI-II has been determined and reveals a similar cyclic cystine knot motif to other cyclotides. Interestingly, loop 6, putatively involved in the cyclization process, is disordered in the family of MCoTI-II structures [18,19], quite unlike the situation in all other cyclotides structurally characterized to date. The discovery of novel sequences in loop 6 may assist in finding clues to the mechanism of cyclization.
In the present study, we have characterized a novel cyclotide from O. affinis, kalata B8, which has sequence characteristics of both the Möbius and bracelet subfamilies, making it somewhat of a hybrid. Furthermore, kalata B8 displays disorder in loop 6 and isomerization of the cyclic peptide backbone similar to what is observed in MCoTI-II [17] but not in other cyclotides so far. The common theme of disorder, which presumably results from flexibility, and isomerization in the processing loops of both these peptides may be associated with the cyclization reaction and suggests a common processing mechanism.
EXPERIMENTAL
Isolation of kalata B8
Kalata B8 was extracted from the above-ground parts of O. affinis with dichloromethane/methanol (1:1; v/v) overnight at room temperature (23 °C). The extract was partitioned with water and the water/methanol layer was concentrated on a rotary evaporator prior to freeze-drying. The dried product was re-dissolved in water and purified on a preparative RP (reverse phase) C18 column (Vydac) at 8 ml/min.
Sequence determination of kalata B8
Kalata B8 (200 μl, 1 mg/ml) was reduced in 6 M guanidinium chloride in 0.1 M Tris buffer (pH 8.5) with 10 μl of 2-mercapto-ethanol by incubating at 50 °C for 4 h. The reduced peptide was alkylated with vinylpyridine in the dark at room temperature for 2 h prior to cleavage with EndoGluC (endoproteinase Glu C). Sequence determination was achieved using Edman degradation.
NMR spectroscopy
Samples for 1H NMR measurements contained approx. 1 mM peptide in 90% water/10% 2H2O (v/v) at approx. pH 3. 2H2O (99.9 and 99.99%) was obtained from Cambridge Isotope Laboratories (Woburn, MA, U.S.A.). Spectra were recorded between 280 and 320 K on a Bruker ARX-500 spectrometer equipped with a shielded gradient unit. Two-dimensional NMR spectra were recorded in phase-sensitive mode using time-proportional phase incrementation for quadrature detection in the second dimension [20]. The two-dimensional experiments consisted of a TOCSY [21] using an MLEV-17 spin lock sequence [22] with a mixing time of 80 ms, DQF-COSY (double-quantum-filtered COSY) [23], ECOSY (exclusive COSY) [24] and NOESY [25] with mixing times of 100–250 ms. Solvent suppression was achieved using a modified WATERGATE (water suppression by gradient-tailored excitation) sequence [26]. Spectra were acquired over 6024 Hz with 4096 complex data points in F2 and 512 increments in the F1 dimension. 3JHN-Hα coupling constants were measured from a one-dimensional spectrum or from the DQF-COSY spectrum.
Spectra were processed on a Silicon Graphics Indigo workstation using XWINNMR (Bruker) software. The second dimension was zero-filled to 1024 real data points, and 90° phase-shifted sine bell window functions were applied prior to Fourier transformation. Chemical shifts were referenced to internal 2,2-dimethyl-2-silapentane-5-sulphonate.
Structure calculations
Preliminary structures of kalata B8 were calculated using a torsion angle simulated annealing method within the program DYANA [27]. Final structures were calculated using simulated annealing and energy minimization methods within CNS version 1.1 [28]. The starting structures were generated using random (ϕ, ϕ) dihedral angles and energy-minimized to produce structures with the correct local geometry. A set of 50 structures was generated by a torsion angle simulated annealing method [29,30]. This method involves a high-temperature phase comprising 4000 steps of 0.015 ps of torsion angle dynamics, a cooling phase with 4000 steps of 0.015 ps of torsion angle dynamics during which the temperature is lowered to 0 K, and finally an energy minimization phase comprising 500 steps of Powell minimization. Structures consistent with restraints were subjected to further molecular dynamics and energy minimization in a water shell, as described by Linge and Nilges [31]. The refinement in explicit water involves the following steps. First, heating to 500 K via steps of 100 K, each comprising 50 steps of 0.005 ps of Cartesian dynamics. Secondly, 2500 steps of 0.005 ps of Cartesian dynamics at 500 K before a cooling phase where the temperature is lowered in steps of 100 K, each comprising 2500 steps of 0.005 ps of Cartesian dynamics. Finally, the structures were minimized with 2000 steps of Powell minimization. Structures were analysed using PROMOTIF [32] and PROCHECK-NMR [33].
Haemolytic assay
Human type A red blood cells were washed with PBS and centrifuged at 150 g for 30 s in a microcentrifuge several times until a clear supernatant was obtained. The assay was performed by adding 20 μl of peptide solution to 80 μl of a 1% suspension of red blood cells in PBS. Test concentrations ranged from 0.5 to 1400 μM. Synthetic melittin (Sigma) was used as a standard, as it is a well-established haemolytic agent. The mixtures were incubated at room temperature for 1 h and then centrifuged at 150 g for 1 min. The supernatant (35 μl) was diluted 1:30 in deionized (Milli-Q) water, and the absorbance was measured at 415 nm. The peptide concentration causing 50% haemolysis (HD50) was calculated.
Anti-HIV activity assays
An in vitro XTT [2,3-bis-(2-methoxy-4-nitro-5-sulphophenyl)-2H-tetrazolium-5-carboxanilide]-based anti-HIV assay was used as described previously [34] to examine the effect of kalata B8 and tricyclon A on virus-induced cell killing in HIV-infected cultures.
RESULTS
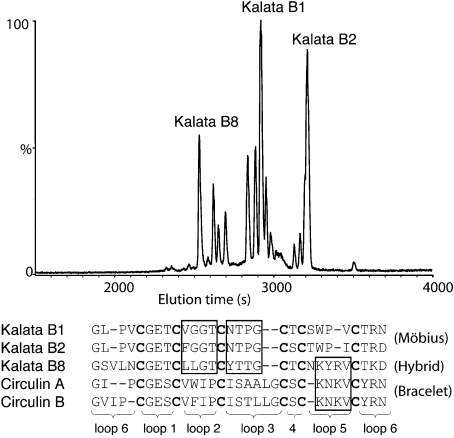
Kalata B8 was isolated from the above-ground parts of O. affinis and purified using RP-HPLC. An LC-MS (liquid chromatography–MS) trace is shown in Figure 2 and highlights the hydrophilic nature of kalata B8 compared with the prototypic cyclotides kalata B1 and B2, as reflected in its shorter retention time. Indeed, kalata B8 is the earliest eluting of the major cyclotide peaks present in O. affinis. Kalata B8 was reduced with dithiothreitol, alkylated with vinylpyridine and sequenced using Edman degradation. The sequence is shown in Figure 2 and compared with selected Möbius and bracelet cyclotides. Kalata B8 appears to be a hybrid between the two major subfamilies of cyclotides. Specifically, loops 2 and 3 of kalata B8 resemble those found in kalata B1 and B2, members of the Möbius subfamily, but loop 5 is similar to the bracelet cyclotides, as exemplified by circulins A and B and lacks the characteristic cis-proline present in the Möbius cyclotides.
Figure 2. LC-MS of a partially purified extract from O. affinis and sequence comparison of kalata B8 with representative known cyclotides.
Kalata B8 elutes significantly earlier than either kalata B1 or kalata B2, as shown at the top of the diagram. The sequence of kalata B8, elucidated by Edman sequencing, is shown at the bottom of the diagram and compared with previously characterized Möbius and bracelet cyclotides. Although the definition of Möbius cyclotides is based on the presence of a cis-proline in loop 5, there is also strong sequence similarity in certain loops within but not between the subfamilies for most known cyclotides. In contrast, kalata B8 represents a chimaera: loops 2 and 3 of kalata B8 resemble those in the Möbius cyclotides, whereas loop 5 resembles those in the bracelet cyclotides, as highlighted with boxes in the sequences.
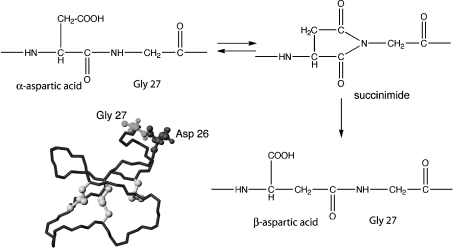
Close inspection of HPLC fractions and associated mass spectra of the partially purified extracts of O. affinis revealed two minor components eluting very close to kalata B8, one with the same mass (3282 Da) and one with a mass 18 Da lower. Incubation of the lower molecular mass form in 0.1 M ammonium bicarbonate buffer at pH 8 resulted in interconversion into kalata B8 and the isobaric form in an approx. 20:80 ratio, i.e. with the relative proportions of the two isobaric species reversed. Kalata B8 contains a single aspartate residue and the interconversion behaviour is consistent with the lower-molecular-mass form being a succinimide derivative and the two isobaric forms being α- and β-aspartic acid derivatives, as indicated in Figure 3. Conversion of succinimides into α- and β-aspartic acids generally favours the β-aspartic acid form [35,36], indicating that the minor form present in the O. affinis extract is the β-aspartic acid form. The major form in the extract was analysed using NMR spectroscopy and the assignments confirmed that it contains an α-aspartic acid.
Figure 3. Backbone isomerization in kalata B8.
It appears that the Asp-Gly sequence in loop 6 of kalata B8, highlighted on the structure, undergoes interconversion into succinimide and β-aspartic acid forms.
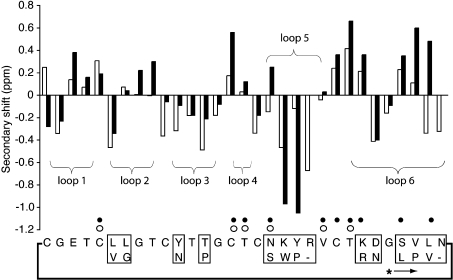
NMR spectral assignments for kalata B8 were made using established techniques [37] and the 1H chemical shifts are supplied as Supplementary data (http://www.BiochemJ.org/bj/393/bj3930619add.htm). A comparison of the αH secondary shifts of kalata B8 and kalata B1 is shown in Figure 4 and illustrates the broad structural similarities between the molecules. However, the shifts also indicate that there are substantial local structural differences, mainly associated with the loops where amino acid substitutions occur, in particular in loops 5 and 6. Figure 4 also shows that while kalata B8 has many of the same slowly exchanging amide protons as kalata B1, it is missing several slow- exchange amides in loop 6. This provides a first indication that loop 6 of kalata B8 is more solvent accessible and perhaps more flexible than that of kalata B1.
Figure 4. αH secondary-shift comparison of kalata B8 (white bars) and kalata B1 (black bars).
The secondary shifts were calculated by subtracting the random coil shifts [54] from the experimental αH chemical shift. The slowly exchanging amide protons for kalata B8 are highlighted with open circles on the sequence and those for kalata B1 with filled circles. The putative ligation site for cyclization of kalata B8 is highlighted with an asterisk and an arrow indicating the direction of the peptide chain from N- to C-terminus. The boxed regions show the sequence differences between kalata B1 (lower part of boxes) and B8.
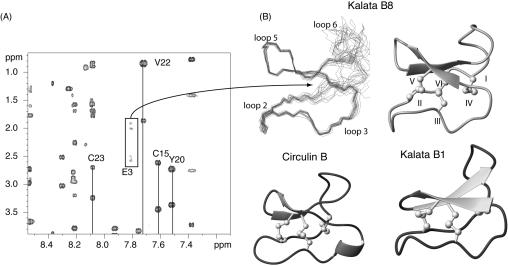
For kalata B8, the amide signals in the TOCSY and NOESY spectra for residues 1, 2 and 31 are weak and broadened in the temperature range analysed (280–320 K) and the amide signals of Glu3 are so broadened as to be visible only at temperatures >312 K, as shown in Figure 5. This provides further evidence of a degree of flexibility in kalata B8 that is not seen in kalata B1 and other cyclotides structurally characterized so far. Significantly, all of the broadened residues are in or near loop 1, which forms a segment of the embedded ring in the structure that is an integral part of the cystine knot. Despite the broadening of these residues, chemical shifts in the amide region are well dispersed and the large number of resolved cross peaks in the NOESY spectrum allowed determination of a well-defined structure for the majority of the molecule.
Figure 5. NMR spectra and three-dimensional structure of kalata B8.
(A) A region of a TOCSY spectrum of kalata B8 highlighting the weak and broadened nature of Glu3 signals at 312 K. The NH signal from Glu3 is not visible below this temperature. (B) A superposition of the 20 lowest energy structures of kalata B8 is shown at the top left of this panel and a ribbon representation is shown at the right. A comparison with kalata B1 and circulin B is shown in the lower half of the panel.
The three-dimensional structure of kalata B8 was calculated with 203 distance restraints and 23 angle restraints using a simulated annealing method in CNS. Ten restraints for five hydrogen bonds were included, based on the slowly exchanging amide protons and preliminary structures (5HN-21O, 16HN-24O, 18HN-22O, 22HN-18O and 24HN-16O). No hydrogen bond acceptor was apparent in the preliminary structures for the slowly exchanging Cys15 amide proton. The resulting family of structures had good structural and energetic statistics, as shown in Table 1. An ensemble and ribbon representation of the three-dimensional structure is shown in Figure 5 along with a comparison with kalata B1 and circulin B, two cyclotides belonging to the Möbius and bracelet cyclotides respectively. The major element of secondary structure in kalata B8 is a cystine knot motif. Interestingly, there is significant disorder in loops 1 and 6, consistent with the relative lack of medium or long-range NOEs (nuclear Overhauser effects) in loop 6, broadened resonances and lack of slow exchange amides in these regions. For example, in loop 6 there are only two medium-range NOEs between residues 28 and 30.
Table 1. NMR and refinement statistics for kalata B8.
| Parameter | Value |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 203 |
| Sequential (|i–j|=1) | 91 |
| Medium-range (|i–j|<4) | 67 |
| Long-range (|i–j>5) | 45 |
| Total dihedral angle restraints | |
| Φ | 13 |
| χ1 | 10 |
| Structure statistics | |
| Violations (mean±S.D.) | |
| Distance constraints (Å; 1 Å=0.1 nm) | 0.05±0.005 |
| Dihedral angle constraints (°) | 0.92±0.3 |
| Max. dihedral angle violation (°) | 4 |
| Max. distance constraint violation (Å) | 0.32 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.004±0.00012 |
| Bond angles (°) | 0.62±0.03 |
| Impropers (°) | 0.42±0.05 |
| Average pairwise R.M.S.D.* (Å) | |
| Heavy (residues 3–24) | 0.45±0.18 |
| Backbone (residues 3–24) | 1.32±0.36 |
| Ramachandran statistics (residues 3–24) | |
| Most favoured | 84% |
| Additionally allowed | 16% |
* Pairwise R.M.S.D. (root mean square deviation) was calculated among 20 refined structures.
Analysis of the structures with PROMOTIF [32] identified a type I β-turn between residues 5 and 8, a type II β-turn between residues 12 and 15 and a type I β-turn between residues 18 and 21. A β-hairpin is recognized between residues 16 and 24, as shown in Figure 5. This β-hairpin is invariably present in inhibitor cystine knot proteins [38,39]. The disulphide bond between Cys1 and Cys15 is characterized as a right-handed hook and the disulphide bonds between residues Cys5 to Cys 17 and Cys10 to Cys23 are lefthanded spirals [32]. The co-ordinates of the structures have been deposited in the Protein Data Bank with the PDB ID code 2B38.
A titration between pH 2.6 and 5.3 revealed that the chemical shifts of the amide protons of Tyr11 and Thr12 were significantly influenced by pH. As pH was increased, Thr12 was shifted downfield by >1.3 p.p.m. and Tyr11 was not detectable at pH 3.6 and above, presumably as a result of broadening. The amide protons of Tyr11 and Thr12 are in close proximity to the side chain of Glu3, which titrates in this pH range. The residues corresponding to Tyr11 and Thr12 in kalata B1 and cycloviolacin O1 have previously been shown to have amide protons whose shifts are highly sensitive to pH as a result of protonation/deprotonation of the Glu side chain [16].
Kalata B8 did not display haemolytic activity up to a tested concentration of 370 μM, despite the fact that such activity is routinely observed in other cyclotides at much lower concentrations (∼50 μM) [5,40]. Anti-HIV assays were performed on kalata B8 and tricyclon A, a cyclotide previously shown to lack haemolytic activity [41] with the sequence (cyclo-GGTIFDCGESCFLGTCYTKGCSCGEWKLCYGTN-). The highest concentration of sample tested for kalata B8 and tricyclon A was 4.5 and 5.8 μM respectively. Kalata B8 inhibited the cytopathic effects of HIV-1 infection in cultured human T-lymphoblast (CEM-SS) cells with an antiviral cytoprotective concentration (EC50) of approx. 2.5 μM, while the cytotoxic concentration (IC50) was >11 μM. Tricyclon A did not display anti-HIV or cytotoxic activity. In contrast, other cyclotides have much more potent anti-HIV activities. For example, kalata B1 and circulins A–F have EC50 values of approx. 140 nM [42] and 40–275 nM [4,43] respectively.
DISCUSSION
In the present study, we have determined the sequence of a novel circular protein (kalata B8) belonging to the family of plant cyclotides. The cyclic nature of the backbone was confirmed by MS analysis following EndoGluC cleavage and the sequence was determined using Edman sequencing. A significant difference between kalata B8 and most of the published cyclotide sequences is that it does not fall into either of the two subfamilies commonly found in cyclotides, namely Möbius or bracelet. Instead, it appears to be a hybrid between the two subfamilies. In addition to its hybrid characteristics, kalata B8 is the most hydrophilic of any of the cyclotides expressed by O. affinis and therefore is an interesting target for structural and bioactivity studies.
The discovery of kalata B8 suggests that the sequence homologies within the Möbius and bracelet subfamilies may not be as distinct as previously thought, and with the discovery of more cyclotides the boundaries between the two subfamilies may merge further. Although the cyclotide family is rapidly expanding, there still remains a significant degree of sequence similarity amongst known members. This is in contrast with other disulphide-rich peptide families such as the conotoxins, which display significant sequence variation and multiple disulphide bond frameworks [44,45]. In the case of the conotoxins, which are used for prey capture by marine snails, it is thought that the rich diversity in structures in the cocktail of venom peptides is important for the targeting of multiple receptor sites simultaneously, to result in the rapid immobilization of prey. The rationale for a suite of broadly similar host defence peptides such as the cyclotides in a single plant is less clear, but the existence of such suites is clear. In a recent report, at least 57 new cyclotide masses were seen in the single plant Viola hederaceae, for example [46].
The structure of kalata B8 contains the cyclic cystine knot motif characteristic of cyclotides but has some interesting differences, including disordered loops 1 and 6, most probably as a result of conformational flexibility that is manifest in the broadening of NMR signals from residues in these regions. It is apparent from the present study that the dynamics of cyclotides can vary significantly despite a high degree of sequence conservation. For example, kalata B1 and B8 contain 17 identical residues, including an identical loop 1, and their overall structures are very similar. However, without determining their structures experimentally, the differences in flexibility would not have been apparent. Clearly, only subtle differences in sequence are sufficient to result in different dynamic properties.
The apparent flexibility observed in kalata B8 is particularly interesting as it occurs in a loop directly involved in the cystine knot motif, previously shown to be critical in maintaining the structural stability of the cyclotides [47]. Such disorder in the cystine knot motif is unprecedented in the cyclotides that have been structurally characterized so far. The disorder in loop 1 of kalata B8 is associated with broadening of several amide protons, most notably from Glu3. In all cyclotides structurally characterized so far, the side-chain carboxyl of this absolutely conserved residue hydrogen-bonds to the backbone amides of the first two residues in loop 3, thus mutually stabilizing loops 1 and 3. The equivalent residues in loop 3 for kalata B8 are Tyr11 and Thr12. The chemical shifts of these residues are influenced by pH as occurs for other cyclotides but Tyr11 is broadened beyond detection at high pH, indicating conformational exchange. Thus it appears that the interaction between loops 1 and 3 is reduced in kalata B8 compared with other cyclotides and the Glu residue may be in conformational exchange between the hydrogen-bonded conformation and one or more other conformations.
More generally, the cystine knot motif is present in a wide range of peptides and proteins, including many toxins and in general is a very stable motif [38,39], making the apparent flexibility observed in kalata B8 a surprising result. Disorder in cystine knot-containing peptides has previously been observed in only a few cases, for example in the conotoxin MrVIB [48], but this disorder was restricted to one large loop and did not involve the loops directly involved in the cystine knot. Disorder in loop 6 has also been observed for the trypsin inhibitor cyclotide MCoTI-II [18,19]. In that case, several isoforms, involving isomerization of the backbone at an Asp-Gly sequence in loop 6, are present. Kalata B8 also contains Asp-Gly in loop 6 and this sequence is well known to be susceptible to backbone isomerization [35,36]. In the case of kalata B8, the native extract contained small amounts of a succinimide form and a β-aspartate form in addition to the predominant α-aspartate isomer. The presence of these forms raises the question of whether they are degradation products of the mature peptide, or possibly by-products from the cyclization process that may provide clues as to its mechanism.
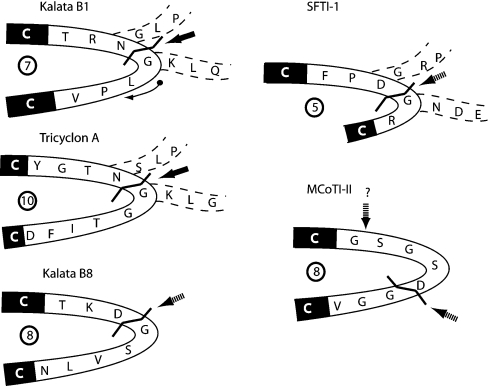
In support of the latter, the Asp-Gly sequence associated with the isomerization in kalata B8 is almost certainly located at the putative cyclization site, based on the gene sequences of a range of cyclotides [9,12]. In nearly all cyclotides sequenced so far, either an Asp-Gly sequence or, more commonly, an Asn-Gly sequence is present in loop 6 and it appears that this sequence is directly involved in the cyclization mechanism, with the Asn/Asp residue deriving from the C-terminus of the excised precursor fragment and the Gly from the N-terminus. Figure 6 shows the cyclization loops from a range of naturally occurring circular peptides and highlights the location of the processing site in kalata B8 by analogy with those in kalata B1 and tricyclon A, where the precursor sequences are known.
Figure 6. Processing loop sequences for plant-derived circular proteins.
The loop 6 sequences for kalata B1, kalata B8, tricyclon A and MCoTI-II are shown, along with the sequence of the single loop of SFTI-1. The arrow on the structure of kalata B1 indicates the direction of the peptide chain. The numbers of residues between the flanking cysteine residues in the various molecules are highlighted in boldface circles within the loops. All sequences contain an Asp/Asn-Gly sequence. Precursor sequences are available for kalata B1 [9], tricyclon A [41] and SFTI-1 [50], and the sequences that flank the mature peptide domain are indicated within dashed lines. Cyclization occurs at the Asp/Asn-Gly sequence, as highlighted with boldface arrows for these three sequences. By analogy, cyclization most likely occurs at the Asp-Gly site for kalata B8 (dashed arrow), although the precursor sequence is not yet known. The precursor sequence of MCoTI-II is not known, but by analogy with related acyclic trypsin inhibitors [17] it may occur between Gly and Ser, as shown with a question mark and dashed arrow. Alternatively, if a common mechanism of cyclization occurs in plant-derived circular proteins, the ligation site may be the Asp-Gly site.
Another example of a naturally occurring circular peptide containing an Asp-Gly sequence is the 14-amino acid sunflower trypsin inhibitor, SFTI-1 [49]. The recently discovered precursor protein for this peptide suggests that in vivo cyclization involves ligation to form an Asp-Gly sequence [50], as indicated in Figure 6. It has also been shown that, in vitro, an acyclic analogue of SFTI-1 is susceptible to cyclization and isomerization of the Asp-Gly sequence, confirming the high reactivity of this sequence [51]. Also shown in Figure 6 is loop 6 of MCoTI-II. Although the gene sequence of MCoTI-II has not been determined, loop 6 of the mature peptide (SGSDGGV) shares high homology with the corresponding part of the precursor (SGRHGGI) of a related acyclic trypsin inhibitor TGTI-II [52]. However, in this case, even though the loop contains an Asp-Gly sequence, alignment of the TGTI-II and MCoTI-II sequences suggests that cyclization may instead occur at a Gly-Ser sequence. In this case, the observation of both α- and β-aspartic acid linkages as well as a succinimide derivative in plant extracts is probably an indicator of post-cyclization degradation of the Asp-Gly bond rather than direct involvement in the cyclization mechanism. The flexible nature of loop 6 may facilitate this degradation.
The sizes of the processing loops for the various naturally occurring circular peptides shown in Figure 6 vary from 5 to 10 residues and the loops vary substantially in sequence. By definition, the loops are flanked by Cys residues at either end and it is interesting to note that the processing point in most cases appears to be three residues from the upstream Cys. If a common mechanism of cyclization is involved, then formation of a C-terminal succinimide intermediate via a side chain to backbone cyclization may provide the activated carbonyl groups for attack by the N-terminal residue once it is released from the precursor protein, presumably by proteolysis. Determination of the gene sequence for MCoTI-II is likely to be crucial for deciphering the mechanism.
The mode of biological action of the cyclotides is poorly understood but those with anti-HIV activity also display haemolytic activity, as reported for kalata B1 and circulins A and B [5,6,40,42]. The recently discovered cyclotide from Viola tricolor, tricyclon A, does not display haemolytic activity [41], and in the present study we found that it also lacks anti-HIV activity. In contrast, kalata B8 has anti-HIV activity but is not haemolytic up to concentrations of 370 μM. These results suggest that a different interaction occurs with the red blood cell membranes than occurs with HIV-infected mammalian cells.
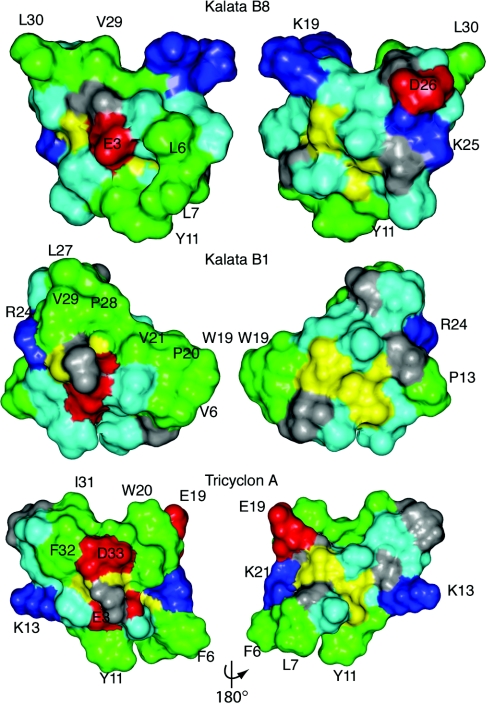
Typically, cyclotides display hydrophobic patches on their surface [16] and indeed it is this property that has facilitated their discovery based on late elution on RP-HPLC [15]. However, kalata B8 is more hydrophilic than most of the cyclotides, as evidenced by its early elution time on RP-HPLC. This hydrophilicity may account for the differences observed in activity because haemolytic activity has previously been shown to correlate with hydrophobicity [53]. A surface representation of the lowest-energy structure of kalata B8 is shown in Figure 7 and compared with kalata B1 and tricyclon A. The hydrophobic patch in kalata B1 is not as continuous in tricyclon A and this has been suggested to account for the lack of haemolytic activity in tricyclon A [41]. In kalata B8, the degree of surface-exposed hydrophobic residues is even less pronounced than tricyclon A, consistent with the lack of haemolytic activity for kalata B8. Further studies are required to determine the features important for anti-HIV activity.
Figure 7. Surface representations of kalata B8, kalata B1 and tricyclon A.
Lys and Arg residues are shown in blue, Glu and Asp in red, Gly in grey, Leu, Val, Ile, Pro, Trp, Tyr and Phe in green and the remaining residues in light blue.
In summary, we have characterized a novel cyclotide from O. affinis that has provided insights into structure–activity relationships and the mechanism of cyclization. Kalata B8 displays anti-HIV activity but not haemolytic activity, indicating that the mechanism by which cyclotides act is dependent on the composition of the membrane and is influenced by the hydrophilicity of the peptide. The structure of kalata B8 is disordered in loops 1 and 6, most likely as a result of flexibility, indicating that very similar sequences and structural motifs can display vastly different dynamic properties.
Online Data
Acknowledgments
N.L.D. is an NHMRC (National Health and Medical Research Council) Industry Fellow. D.J.C. is an ARC (Australian Research Council) Professorial Fellow. We thank Lillian Sando for performing the haemolytic assays, Kirk Gustafson for anti-HIV assays and Elizabeth Westbury for assistance with NMR analysis.
References
- 1.Craik D. J., Daly N. L., Mulvenna J., Plan M. R., Trabi M. Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 2.Gran L. On the effect of a polypeptide isolated from ‘Kalata-Kalata’ (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. 1973;33:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 3.Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 4.Gustafson K. R., Sowder R. C. I., Henderson L. E., Parsons I. C., Kashman Y., Cardellina J. H. I., McMahon J. B., Buckheit R. W. J., Pannell L. K., Boyd M. R. Circulins A and B: novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 1994;116:9337–9338. [Google Scholar]
- 5.Daly N. L., Love S., Alewood P. F., Craik D. J. Chemical synthesis and folding of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 6.Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svangard E., Göransson U., Hocaoglu Z., Gullbo J., Larsson R., Claeson P., Bohlin L. Cytotoxic cyclotides from Viola tricolor. J. Nat. Prod. 2004;67:144–147. doi: 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- 8.Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Mobius strips exist in nature? Biochemistry. 2005;44:851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 9.Jennings C., West J., Waine C., Craik D., Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craik D. J., Simonsen S., Daly N. L. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 2002;5:251–260. [PubMed] [Google Scholar]
- 11.Craik D. J. Plant cyclotides: circular, knotted peptide toxins. Toxicon. 2001;39:1809–1813. doi: 10.1016/s0041-0101(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 12.Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C. V., Anderson M. A., Craik D. J. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 13.Sletten K., Gran L. Some molecular properties of kalatapeptide B-1. A uterotonic polypeptide isolated from Oldenlandia affinis DC. Medd. Nor. Farm. Selsk. 1973;7–8:69–82. [Google Scholar]
- 14.Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 15.Craik D. J., Daly N. L., Bond T., Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 16.Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 2003;278:8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 18.Felizmenio-Quimio M. E., Daly N. L., Craik D. J. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 19.Heitz A., Hernandez J. F., Gagnon J., Hong T. T., Pham T. T., Nguyen T. M., Le-Nguyen D., Chiche L. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry. 2001;40:7973–7983. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 20.Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 21.Braunschweiler L., Ernst R. R. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 22.Bax A., Davis D. G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 23.Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 24.Griesinger C., Sørensen O. W., Ernst R. R. Practical aspects of the E.COSY technique, measurement of scalar spin-spin coupling constants in peptides. J. Magn. Reson. 1987;75:474–492. [Google Scholar]
- 25.Jeener J., Meier B. H., Bachmann P., Ernst R. R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 26.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 27.Guntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 28.Brünger A. T., Adams P. D., Rice L. M. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 29.Rice L. M., Brünger A. T. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- 30.Stein E. G., Rice L. M., Brünger A. T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 31.Linge J. P., Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson E. G., Thornton J. M. PROMOTIF – a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 34.Gulakowski R. J., McMahon J. B., Staley P. G., Moran R. A., Boyd M. R. A semiautomated multiparameter approach for anti-HIV drug screening. J. Virol. Methods. 1991;33:87–100. doi: 10.1016/0166-0934(91)90010-w. [DOI] [PubMed] [Google Scholar]
- 35.Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 36.Johnson B. A., Murray E. D., Jr, Clarke S., Glass D. B., Aswad D. W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J. Biol. Chem. 1987;262:5622–5629. [PubMed] [Google Scholar]
- 37.Wüthrich K. New York: Wiley-Interscience; 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 38.Pallaghy P. K., Nielsen K. J., Craik D. J., Norton R. S. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craik D. J., Daly N. L., Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 40.Barry D. G., Daly N. L., Clark R. J., Sando L., Craik D. J. Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry. 2003;42:6688–6695. doi: 10.1021/bi027323n. [DOI] [PubMed] [Google Scholar]
- 41.Mulvenna J. P., Sando L., Craik D. J. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure. 2005;13:691–701. doi: 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Daly N. L., Gustafson K. R., Craik D. J. The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 2004;574:69–72. doi: 10.1016/j.febslet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Gustafson K. R., Walton L. K., Sowder R. C. I., Johnson D. G., Pannell L. K., Cardellina J. H. I., Boyd M. R. New circulin macrocyclic polypeptides from Chassalia parvifolia. J. Nat. Prod. 2000;63:176–178. doi: 10.1021/np990432r. [DOI] [PubMed] [Google Scholar]
- 44.Dutton J. L., Craik D. J. α-Conotoxins: nicotinic acetylcholine receptor antagonists as pharmacological tools and potential drug leads. Curr. Med. Chem. 2001;8:327–344. doi: 10.2174/0929867013373453. [DOI] [PubMed] [Google Scholar]
- 45.Terlau H., Olivera B. M. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 46.Trabi M., Craik D. J. Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell. 2004;16:2204–2216. doi: 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colgrave M. L., Craik D. J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 48.Daly N. L., Ekberg J. A., Thomas L., Adams D. J., Lewis R. J., Craik D. J. Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons. J. Biol. Chem. 2004;279:25774–25782. doi: 10.1074/jbc.M313002200. [DOI] [PubMed] [Google Scholar]
- 49.Luckett S., Garcia R. S., Barker J. J., Konarev A. V., Shewry P. R., Clarke A. R., Brady R. L. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 1999;290:525–533. doi: 10.1006/jmbi.1999.2891. [DOI] [PubMed] [Google Scholar]
- 50.Mulvenna J. P., Foley F. M., Craik D. J. Discovery, structural determination and putative processing of the precursor protein that produces the cyclic trypsin inhibitor SFTI-1. J. Biol. Chem. 2005;280:32245–32253. doi: 10.1074/jbc.M506060200. [DOI] [PubMed] [Google Scholar]
- 51.Marx U. C., Korsinczky M. L., Schirra H. J., Jones A., Condie B., Otvos L., Jr, Craik D. J. Enzymatic cyclization of a potent Bowman-Birk protease inhibitor, sunflower trypsin inhibitor-1, and solution structure of an acyclic precursor peptide. J. Biol. Chem. 2003;278:21782–21789. doi: 10.1074/jbc.M212996200. [DOI] [PubMed] [Google Scholar]
- 52.Ling M. H., Qi H. Y., Chi C. W. Protein, cDNA, and genomic DNA sequences of the towel gourd trypsin inhibitor. A squash family inhibitor. J. Biol. Chem. 1993;268:810–814. [PubMed] [Google Scholar]
- 53.Chen Y., Mant C. T., Farmer S. W., Hancock R. E., Vasil M. L., Hodges R. S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.