Abstract
RRF (ribosome recycling factor) consists of two domains, and in concert with EF-G (elongation factor-G), triggers dissociation of the post-termination ribosomal complex. However, the function of the individual domains of RRF remains unclear. To clarify this, two RRF chimaeras, EcoDI/TteDII and TteDI/EcoDII, were created by domain swaps between the proteins from Escherichia coli and Thermoanaerobacter tengcongensis. The ribosome recycling activity of the RRF chimaeras was compared with their wild-type RRFs by using in vivo and in vitro activity assays. Like wild-type TteRRF (T. tengcongensis RRF), the EcoDI/TteDII chimaera is non-functional in E. coli, but both wild-type TteRRF, and EcoDI/TteDII can be activated by coexpression of T. tengcongensis EF-G in E. coli. By contrast, like wild-type E. coli RRF (EcoRRF), TteDI/EcoDII is fully functional in E. coli. These findings suggest that domain II of RRF plays a crucial role in the concerted action of RRF and EF-G for the post-termination complex disassembly, and the specific interaction between RRF and EF-G on ribosomes mainly depends on the interaction between domain II of RRF and EF-G. This study provides direct genetic and biochemical evidence for the function of the individual domains of RRF.
Keywords: domain function, domain swaps, Escherichia coli, elongation factor (EF-G), ribosome recycling factor (RRF), Thermoanaerobacter tengcongensis
Abbreviations: aaRRF, Aquifex aeolicus RRF; DTT, dithiothreitol; EcoDI/TteDII, chimaera of EcoRRF domain I and TteRRF domain II; EF-G, elongation factor-G; EcoEF-G, Escherichia coli EF-G; EcoRRF, E. coli RRF; GDPNP, guanidine 5′-(β,γ-imido)tri-phosphate; IF3, initiation factor 3; IPTG, isopropyl-β-D-thiogalactopyranoside; RF, release factor; RRF, ribosome recycling factor; TteDI/EcoDII, chimaera of TteRRF domain I and EcoRRF domain I; TteEF-G, Thermoanaerobacter tengcongensis EF-G; TteRRF, T. tengcongensis RRF; TthEF-G, Thermus thermophilus EF-G; TthRRF, T. thermophilus RRF
INTRODUCTION
Protein synthesis consists of initiation, elongation, termination and ribosome recycling. When translation of an mRNA has been completed on the ribosome, the peptidyl-tRNA with a nascent polypeptide is translocated into the ribosomal P site, and a stop codon is translocated into the ribosomal A site. Then either class I RF1 (release factor1) or RF2 binds to the ribosomal A site and induces the release of the nascent polypeptide. Afterwards class II RF3 binds to the ribosome, catalyses the release of RF1 or RF2, and dissociates in a GTP-dependent manner [1–3], leaving behind the post-termination ribosomal complex consisting of mRNA, deacylated tRNA in the P site, and an empty A site. Disassembly of the post-termination complex, and recycling of the ribosomal subunits back to a new round of initiation, is an essential step in protein synthesis, which is catalysed by RRF (ribosome recycling factor) and EF-G (elongation factor-G) [4–7]. Although the simplest explanation that RRF is a functional mimic of tRNA for disassembly of the post-termination complex has been ruled out [8–10], the mechanism of ribosomal recycling is still a debated issue. One point of dispute is whether disassembly of the post-termination complex depends on the translocation activity of EF-G. Kiel et al. demonstrated that, as part of the RRF-dependent post-termination complex disassembly reaction, EF-G released RRF from ribosomal complexes, and this release activity of EF-G was due to the translocation activity of EF-G [11]. By contrast, Fujiwara et al. pointed out that RRF disassembled the post-termination ribosomal complex independently of the ribosomal translocase activity of EF-G, based on the results showing that EF-G variants remain active in GTP hydrolysis but are defective in tRNA translocation fully activate RRF function in vivo and in vitro [10]. Recently, by using fluorescence and fluorescence resonance energy transfer assays to monitor sequence of steps in ribosome recycling Peske et al. also showed that RRF and EF-G together with GTP promoted the dissociation of 50S subunits from the post-termination complex without involving translocation or a translocation-like event [12]. Another point of dispute is whether IF3 (initiation factor 3) is required for splitting of the post-termination ribosome in to subunits. Experiments from Ehrenberg and co-workers demonstrated that RRF and EF-G rapidly split the post-termination ribosome into subunits in the absence of IF3 by a mechanism that requires GTP, as well as GTP hydrolysis, and that the primary role of IF3 is to keep the subunits apart after the splitting event [7,13]. Recently, Peske et al. also indicated that IF3 did not affect subunit dissociation but prevented re-association, thereby masking the dissociation effect of EF-G and RRF under certain experimental conditions [12]. By contrast, Kaji and co-workers showed that 70 S ribosomes, as well as the model post-termination complexes, were dissociated into stable subunits by the co-operative action of three translation factors: RRF, EF-G and IF3 [14]. However, they also indicated that RRF and EF-G alone could transiently dissociate 70 S ribosomes, and that IF3 stabilized the dissociation by binding to the transiently formed 30 S subunits, preventing re-association back to 70 S ribosomes [14]. Thus the two statements about mechanisms of post-termination complex disassembly seem to resemble each other except in small details.
As major protein factors for disassembly of the post-termination complex, all reported RRFs are highly similar in amino acid sequence [15,16]. Crystal and solution structures of RRF have been solved from six different bacterial species [15–21]. All of these structures display a similar architecture and consist of two highly conserved domains: domain I displays a three-stranded α helix bundle structure, and domain II exists as a three layer β/α/β sandwich structure. The two domains are arranged in an L-shaped conformation. Although the structure of RRF is well known, and disassembly of the post-termination complex by the concerted action of RRF and EF-G has been widely accepted, there still is not a full understanding of the RRF's structural features that are responsible for its mechanism of action. Recently, Rao and Varshney reported that although Mycobacterium tuberculosis RRF is non-functional in Escherichia coli, it regains activity upon the coexpression of M. tuberculosis EF-G [22]. Ito et al. also showed the same result when the function of Thermus thermophilus RRF was studied in E. coli [23]. These observations not only suggest the necessity for a specific interaction between the homologous RRF and EF-G in E. coli but also open a way to study interactions between the RRF and EF-G molecules. Three questions to be addressed are particularly interesting. Does each domain of RRF play a different role? If it does, which one is most responsible for the activity of disassembly of the post-termination complex? Is there any specific interaction between the domain II of RRF and EF-G? To answer these questions, two RRF chimaeras, EcoDI/TteDII and TteDI/EcoDII were created. EcoDI/TteDII consists of domain I from E. coli RRF (EcoRRF) and domain II from Thermoanaerobacter tengcongensis RRF (TteRRF) [24]. TteDI/EcoDII consists of domain I from T. tengcongensis and domain II from E. coli RRF. The ribosome recycling activity of the RRF chimaeras was compared with wild-type RRFs using in vivo and in vitro activity assays. The experiments showed that like wild-type TteRRF, the EcoDI/TteDII chimaera failed to complement the RRFts (temperature sensitive) phenotype of the E. coli LJ14 (frrts) strain. However, under the same conditions, the TteDI/EcoDII chimaera complemented the RRFts phenotype as efficiently as wild-type EcoRRF. It is noteworthy that although TteRRF and EcoDI/TteDII chimaeras are inactive in E. coli LJ14 (frrts), both of them regain activity upon coexpression of T. tengcongensis EF-G. These results indicate that domain II of RRF plays a crucial role in the function of RRF. The fact that TteDI/EcoDII expressed with EcoEF-G, and EcoDI/TteDII expressed with TteEF-G, regain their activity in E. coli suggests that domain II of RRF is involved in a specific interaction between RRF and EF-G.
MATERIALS AND METHODS
Strains, plasmids, growth conditions and chemicals
The various bacterial strains and plasmids used in this study are listed in Table 1. Luria–Bertani broth [25] was used for bacterial growth. The media was supplemented with various antibiotics at the following final concentrations: ampicillin 50 μg/ml, kanamycin 50 μg/ml and chloramphenicol 25 μg/ml, as required. IPTG (isopropyl-β-D-thiogalactopyranoside) was added to a final concentration of 0.5 mM for RRF protein expression. For complementation of the E. coli RRF defect, various plasmids were transformed in to the E. coli LJ14 strain (frrts). Cultures were started with a 0.06% (v/v) inoculum from freshly-grown (at 30 °C) overnight cultures, and growth at both permissive (30 °C) and non-permissive (42 °C) temperatures was monitored by recording culture turbidities at 600 nm at regular intervals. GDPNP [guanidine 5′-(β,γ-imido)tri-phosphate] (Sigma), a non-hydrolysable GTP analogue, was used for ribosome binding assays.
Table 1. Strains and plasmids.
Kmr, kanamycin resistant; Apr, ampicillin resistant; Cmr, chloramphenicol resistant.
| Strain | Characteristics | Source or reference |
|---|---|---|
| Bacteria | ||
| T. tengcongensis | MB4TA gram-negative anaerobic Eubacterium | [27] |
| E. coli | ||
| LJ14 | MC1061 frr14(ts) | [39] |
| DH5α | FΦ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK−mK+) | Nat. Lab. Biomacromolecules |
| phoA supE44λthi-1 gyra96relA1 | ||
| BL21(DE3)pLysS | F−ompT hadSB(rB−mB+) gal dcm (DE3)/pLysS; Cmr | Nat. Lab. Biomacromolecules |
| Plasmids | ||
| pET-DB | T7 promoter-driven high-efficiency protein expression vector, encodes Kmr | [26] |
| pET-DB-TteRRF | T. tengcongensis RRF gene cloned in to pET-DB, encodes Kmr | Present study |
| pET-28a(+) | T7 promoter-driven high-efficiency protein expression vector, encodes Kmr | Novagen |
| pET28-EcoRRF | E. coli RRF gene cloned in to pET-28a(+),encodes Kmr | Present study |
| pET28-EcoEF-G | E. coli EF-G gene cloned in to pET-28a(+),encodes Kmr | Present study |
| pET28-EcoDI/TteDII | EcoDI/TteDII cloned in to pET-28a(+),encodes Kmr | Present study |
| pET28-TteDI/EcoDII | TteDI/EcoDII cloned in to pET-28a(+),encodes Kmr | Present study |
| pET28-TteEF-G | T. tengcongensis EF-G gene cloned in to pET-28a(+),encodes Kmr | Present study |
| pQE-60 | T5 promoter/lac operator-driven protein expression vector, encodes Apr | Qiagen |
| pQE-EcoRRF | E. coli RRF gene cloned in to pQE-60, encodes Apr | Present study |
| pQE-TteRRF | T. tengcongensis RRF gene cloned in to pQE-60, encodes Apr | Present study |
| pQE-EcoDI/TteDII | EcoDI/TteDII cloned in to pQE-60, encodes Apr | Present study |
| pQE-TteDI/EcoDII | TteDI/EcoDII cloned in to pQE-60, encodes Apr | Present study |
| pSTV28 | Plasmid which is reconstructed from a replication start of pACYC184 and | TaKaRa |
| β-galactosidase gene including pUC118 multi-cloning site, encodes Cmr | ||
| pSTV28-TteEF-G | T. tengcongensis EF-G gene cloned in to pSTV28, encodes Cmr | Present study |
Cloning of the RRF gene from T. tengcongensis
The RRF gene from T. tengcongensis was cloned as described previously [24]. In brief, forward, 5′-GGTACCATGGGTAGCGATTATTTGAAAGACAGTG-3′ and reverse, 5′-GATTGGATCCTTAAATTTCCATTATCTCCTTTTC-3′ primers containing NcoI and BamHI restriction sites (bold) respectively, were designed to amplify the T. tengcongensis RRF gene by PCR using T. tengcongensis genomic DNA as the template DNA. The PCR product was inserted in to the NcoI and BamHI sites of pET-DB [26] after digestion with the same enzymes, resulting in pET-DB-TteRRF for expression of TteRRF. The RRF gene was also cloned into the pQE-60 vector between the NcoI and BamHI sites, resulting in pQE-TteRRF for the complementation assay.
Cloning of the EF-G gene from T. tengcongensis
Based on the putative EF-G gene sequence of T. tengcongensis [27] a forward, 5′-AGACAGCCATATGCCAAGGGATTTCAGCTTAGATAAAGTTAGG-3′ and a reverse, 5′-GCAAGGATCCTTATTTTTTTGCTGATAATATTTGCTCAGC-3′ primer containing NdeI and BamHI sites (bold) respectively, were designed to amplify the T. tengcongensis EF-G gene by PCR using T. tengcongensis genomic DNA as a template. The PCR product was inserted into the NdeI and BamHI sites of pET-28(+) after digestion with the same enzymes, resulting in pET-28-TteEF-G for expression of TteEF-G. The EF-G gene was also cloned into the pSTV-28 vector between the KpnI and BamHI sites, resulting in pSTV-TteEF-G for the complementation assay.
Cloning of the RRF gene from E. coli
Forward, 5′-GGCACCATGGGTATTAGCGATATCAGAAAAGATGC-3′ and reverse, 5′-GCTGCTCGAGGAACTGCATCAGTTCTGCTTCTTTG-3′ primers containing NcoI and XhoI restriction sites (bold), respectively, were used to amplify the E. coli RRF gene from E. coli genomic DNA [28]. The PCR product was digested with NcoI and XhoI and cloned into pET-28a(+) after digestion with the same enzymes, resulting in pET28-EcoRRF for expression of EcoRRF. The EcoRRF gene was also cloned into the pQE-60 vector between the NcoI and BamHI sites, resulting in pQE-EcoRRF for the complementation assay.
Cloning of the EF-G gene from E. coli
Based on the EF-G gene sequence of E. coli [29] forward, 5′-AGACAGCAGCCATATGGCTCGTACAACACCCATCGCACGCTACCG-3′ and reverse, 5′-GCAAGGATCCTTATTTACCACGGGCTTCAATTACGGCCTGAGCAACG-3′ primers containing NdeI and BamHI restriction sites (bold) respectively, were designed to amplify the E. coli EF-G gene by PCR using genomic DNA from E. coli DH5α as the template DNA. The PCR product was inserted into the NdeI and BamHI sites of pET-28(+) after digestion with the same enzymes, resulting in pET28-EcoEF-G for expression of EcoEF-G.
Cloning of chimaeric RRF variants
Two chimaeric RRF variants were constructed by using gene SOEing (gene splicing by overlap extension) [30]. For construction of the EcoDI/TteDII chimaera gene, six primers were designed. Primers 1 and 2 were the same as the forward and reverse primers used for cloning of the RRF gene from E. coli as described above. The other four primers were as follows: primer 3, 5′-AGGGCCGGATTCGCTCTACCCGTGCGTATTTTGCTG-3′; primer 4, 5′-CAGCAAAATACGCACGGGTAGAGCGAATCCGGCCCT-3′; primer 5, 5′-GTTCTTCCGTCAGCGGTGGAAGGACCAGTCTTAG-3′; primer 6, 5′-CTAAGACTGGTCCTTCCACCGCTGACGGAAGAAC-3′. Each primer contained a priming sequence for the amplification of the EcoRRF gene or the TteRRF gene and also has an overlapping sequence at its 5′ end (bold) that is complementary to a segment of the EcoRRF gene or the TteRRF gene. Five PCRs were performed using the pQE-EcoRRF or pQE-TteRRF plasmid as a template. The PCR product was digested with NcoI and XhoI and cloned into pET-28a(+) after digestion with the same enzymes, resulting in pET28-EcoDI/TteDII for expression of EcoDI/TteDII. The EcoDI/TteDII gene was also cloned into the pQE-60 vector between the NcoI and BamHI sites, resulting in pQE-EcoDI/TteDII, which was used for the complementation assay. The TteDI/EcoDII chimaera gene was created by using the same strategy, resulting in pET28-TteDI/EcoDII and pQE-TteDI/EcoDII for expression of TteDI/EcoDII and for the complementation assay respectively.
Purification of TteRRF, EcoRRF, EcoDI/TteDII, TteDI/EcoDII, EcoEF-G and TteEF-G
All proteins were expressed in a soluble form, details of the purification and characterization of the proteins were described previously [24]. In brief, each plasmid was transformed in to E. coli BL21 (DE3) pLysS cells respectively. Fresh cultures of transformants were induced with 0.5 mM IPTG, and the overexpressed proteins were purified by metal chelating affinity chromatography [24,31]. Each purified protein appeared as a single band on an SDS 15% polyacrylamide gel. The protein concentration was determined as described [32].
In vitro assays for RRF activity
For the in vitro polysome-breakdown assay, the tetracycline-treated polysome fraction was prepared from E. coli strain MRE600 [33,34] and used for the post-termination complex-disassembly reaction by RRF, as described previously [35,36]. A reaction mixture [250 μl; containing 10 mM Tris/HCl (pH 7.4), 8.2 mM MgSO4, 80 mM NH4Cl, 1 mM DTT (dithiothreitol), 0.16 mM GTP, 0.01 mM puromycin, 125 units of ribonuclease inhibitor, 1.0 unit of A260 polysomes, 60 μg of EF-G, and 30 μg of RRF] was incubated at 30 °C for 20 min, and fractionated on 15–30% sucrose density gradients by centrifugation at 190000 g for 1 h. The gradients were scanned from top to bottom at 254 nm using a Hitachi gradient fraction collector.
Binding of RRF to 70 S ribosome in the presence or absence of EF-G
E. coli 70 S ribosome was prepared as described previously [34]. Complexes of EF-G and 70 S ribosome were prepared by mixing EF-G and ribosomes (molar ratio of EF-G/ribosome=4) for 10 min at 30 °C before addition of RRF in an assay buffer with or without 3 μM GDPNP [10 mM Tris/HCl (pH 7.4), 8.2 mM MgSO4, 80 mM NH4Cl, 1 mM DTT and 50 μM puromycin]. Binding of RRFs to 70 S ribosome or the EF-G–ribosome complexes (with or without GDPNP) was measured using a filtering technique [16,37]. Briefly, various RRFs were incubated with 70 S ribosome or the complex (0.25 μM) in 40 μl of the assay buffer at 30 °C for 10 min. The mixture was applied on to Microcon YM-100 columns (Millipore) and centrifuged for 5 min at 3000 g to trap the ribosome-bound RRFs. Then 40 μl of the same buffer was loaded onto the column and centrifuged for 10 min at 3000 g to wash out the unbound RRFs. After two washes, the filter was incubated with 40 μl of the buffer for 1 min at room temperature. The ribosome-bound RRFs were collected from the inverted column by centrifugation. The recovered RRFs were detected by Western blotting with rabbit anti-(E. coli RRF) antibody (1:1000 dilution). Bound RRFs were quantified by the blotting of known amounts of RRF standards. To quantify non-specific binding between RRFs and the filter apparatus, control experiments without ribosomes were performed.
Release of RRFs from the RRF–ribosome complexes by EF-G
70 S ribosome (0.25 μM) and RRFs (4.0 μM) were incubated together for 10 min at 30 °C in 40 μl of assay buffer solution containing 0.5 mM GTP. Unbound RRFs were isolated by the Microcon YM-100 ultrafiltration method as described above. Various amounts of EF-Gs were added and incubated in the same buffer for 15 min at 30 °C. The mixture was subjected to Microcon YM-100 ultrafiltration to separate ribosomes from the released RRFs. The remaining ribosome-bound RRF was determined via quantitative Western blotting [16,37].
RESULTS
Construction of chimaeric RRF variants
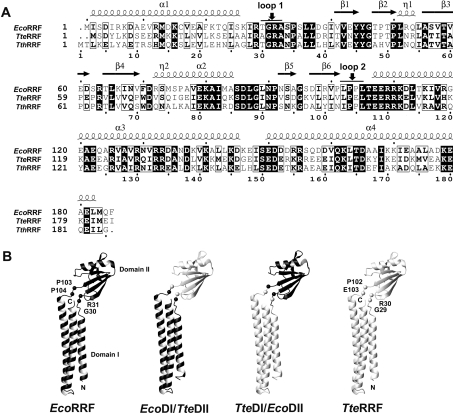
As described in the Materials and methods section, two RRF chimaeras were constructed. EcoDI/TteDII consists of domain I from E. coli and domain II from T. tengcongensis, whereas TteDI/EcoDII consists of domain I from T. tengcongensis and domain II from E. coli. As known, the RRFs from different bacteria are highly conserved in their primary structure and the three-dimensional structure of the RRFs from six bacterial organisms is also highly conserved [15–21], consisting of two domains. The two domains are connected to each other by two loops acting as a hinge region. Sequence alignment of TteRRF and EcoRRF (Figure 1A) shows that loop 1 of the hinge is composed of T29GRA32 for EcoRRF and A28GRA31 for TteRRF, loop 2 of the hinge is composed of L102PPL105 for EcoRRF and L101PEL104 for TteRRF. The points where the domain swaps were chosen between Gly–Arg of loop 1, and Pro–Pro or Pro–Glu of loop 2 (Figure 1A) ensure that the domains of the RRF chimaeras retain their intact amino acid sequence. In order to clearly show the domain swaps between EcoRRF and TteRRF, Figure 1(B) shows schematic drawings of the structures of EcoRRF, TteRRF, TteDI/EcoDII and EcoDI/TteDII. It is noteworthy that our previous study showed that the secondary CD profiles of TteRRF, EcoRRF, TteDI/EcoDII and EcoDI/TteDII are very similar [24]. This suggests that domain swaps between EcoRRF and TteRRF have no significant effect on their folded structures.
Figure 1. (A) Comparison of the primary structures of EcoRRF with TteRRF.
Amino acid sequences are shown as single letter codes. The numbering at the bottom is the amino acid sequence from T. thermophilus RRF. Identical residues are shadowed in black and similar residues are boxed. The corresponding secondary-structure elements of EcoRRF are indicated at the top. Vertical arrows indicate the points where the two domains were exchanged (G30 and R31, P103 and P104 for EcoRRF; G29 and R30, P102 and E103 for TteRRF respectively). (B) Schematic drawings of the structures of EcoRRF, TteRRF, TteDI/EcoDII, and EcoDI/TteDII.
TteRRF alone does not rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts)
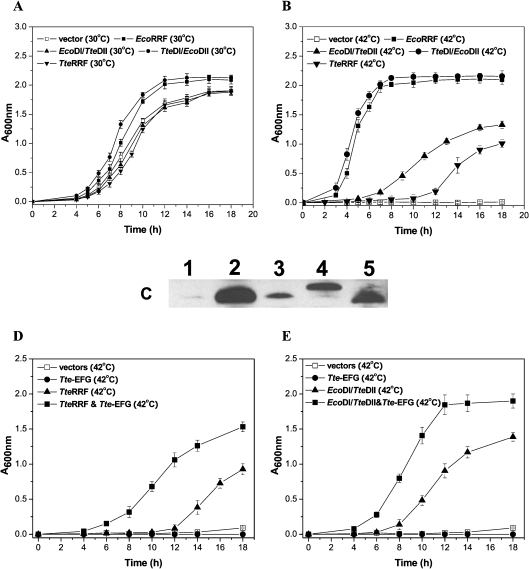
It is known that RRF is an essential protein for bacterial growth since deletion of the RRF gene is lethal in E. coli [38], as are temperature sensitive (frrts) mutations [39]. Complementation analysis allowed the in vivo activity of RRF to be tested. The TteRRF gene cloned in pQE-TteRRF was tested for intergeneric complementation of the E. coli LJ14 (frrts) strain. The growth rates of the various transformants in liquid cultures at the permissive (30 °C) and non-permissive (42 °C) temperatures were investigated. As shown in Figure 2(A), at the permissive temperature, transformants harbouring pQE-TteRRF, pQE-EcoRRF, pQE-EcoDI/TteDII or pQE-TteDI/EcoDII grew at the same rate as those harbouring the vector pQE-60 alone, suggesting that the expression of these RRFs was not toxic to E. coli. However, at the non-permissive temperature, the transformants harbouring pQE-EcoRRF grew well, whereas those harbouring either the vector pQE-60 or pQE-TteRRF did not. The growth of transformants harbouring pQE-TteRRF was significantly delayed (Figure 2B). This observation indicates that TteRRF alone does not rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts). It is noteworthy that immuno-blot analysis of the cell-free extracts of E. coli LJ14 harbouring pQE-TteRRF, pQE-EcoRRF, pQE-EcoDI/TteDII, pQE-TteDI/EcoDII or pQE-60 plasmids respectively, grown at both the permissive (30 °C) and the non-permissive temperature (42 °C) showed that all the RRFs were expressed in the LJ14 strain (Figure 2C). This result ruled out the lack of TteRRF expression as a possible reason for failure to rescue the RRFts phenotype of the LJ14 strain.
Figure 2. Complementation analysis of E. coli LJ14 with RRFs.
The E. coli LJ14 (frrts) strain was transformed with the respective plasmids and examined for growth-phenotype. (A) Growth curves of various transformants of E. coli LJ14 (frrts) at the permissive temperature (30 °C). Vector, transformants with pQE-60; EcoRRF, transformants with pQE-EcoRRF; EcoDI/TteDII, transformants with pQE-EcoDI/TteDII; TteDI/DII, transformants with pQE-TteDI/EcoDII; TteRRF, transformants with pQE-TteRRF. (B) Growth curves of various transformants of E. coli LJ14 (frrts) at the non-permissive temperature (42 °C). The transformants are the same as those indicated in (A). (C) Detection of RRF expression in E. coli LJ14 by immunoblotting using anti-EcoRRF antibodies. Protein extracts were prepared from transformants harbouring the respective plasmids. Bands: 1, pQE-60 vector; 2, PQE-EcoRRF; 3, pQE-TteRRF; 4, pQE-EcoDI/TteDII; 5, pQE-TteDI/EcoDII. (D) Growth curves of various transformants of E. coli LJ14 (frrts) at the non-permissive temperature (42 °C). Vectors, transformants with pQE-60 and pSTV-28; TteEF-G, transformants with pQE-60 and pSTV-TteEF-G; TteRRF, transformants with pQE-TteRRF and pSTV-28; TteRRF and Tte-EFG, transformants with pQE-TteRRF and pSTV-TteEF-G. (E) Growth curves of various transformants of E. coli LJ14 (frrts) at the non-permissive temperature (42 °C). Vectors, transformants with pQE-60 and pSTV-28; Tte-EFG, transformants with pQE-60 and pSTV-TteEF-G; EcoDI/TteDII, transformants with pQE-EcoDI/TteDII and pSTV-28; EcoDI/TteDII&Tte-EFG, transformants with pQE-EcoDI/TteDII and pSTV-TteEF-G. The experimental conditions are described in the text.
TteDI/EcoDII RRF rescues the temperature-sensitive phenotype of E. coli LJ14 (frrts)
TteDI/EcoDII is a chimaeric RRF in which the domain I and II are from T. tengcongensis and E. coli respectively. Complementation analysis of E. coli LJ14 (frrts) strain with the TteDI/EcoDII RRF gene, which was cloned into pQE-TteDI/EcoDII, was performed. As shown in Figure 2(B), transformants harbouring pQE-EcoRRF or pQE-TteDI/EcoDII grew at the same rate at the non-permissive temperature. This observation indicates that TteDI/EcoDII RRF does rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts) as efficiently as EcoRRF.
EcoDI/TteDII RRF does not efficiently rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts)
EcoDI/TteDII is another chimaeric RRF in which the domains I and II are from E. coli and T. tengcongensis respectively. The EcoDI/TteDII gene cloned into pQE-EcoDI/TteDII was also tested for complementation of the E. coli LJ14 (frrts) strain. Figure 2(B) also showed the growth rate of transformants harbouring pQE-EcoDI/TteDII at the non-permissive temperature. Compared with the growth rate of E. coli LJ14 harbouring pQE-EcoRRF and pQE-TteDI/EcoDII at the non-permissive temperature, the growth rate of E. coli LJ14 harbouring pQE-EcoDII/TteDII is much slower and is close to that of TteRRF, indicating that like TteRRF, EcoDI/TteDII does not rescue the temperature-sensitive phenotype of E. coli LJ14.
Like T. thermophilus RRF [40], in spite of the apparent defect in complementation, slow growth of the transformants harbouring pQE-TteRRF was observed after prolonged growth (Figure 2B), suggesting that weak residual activity may remain in TteRRF in E. coli. It is noteworthy that compared with TteRRF, the transformant harbouring pQE-EcoDI/TteDII shows more activity for complementation of the E. coli LJ14 (frrts) strain (Figure 2B). This suggests that the interaction between domain I of E. coli RRF and domain II of T. tengcongensis RRF may allow EcoDI/TteDII to gain some activity.
Both TteRRF and EcoDI/TteDII require TteEF-G to rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts)
To examine whether TteRRF or EcoDI/TteDII regains activity upon coexpression of TteEF-G in E. coli, the E. coli strain LJ14 (frrts) harbouring pQE-TteRRF or pQE-EcoDI/TteDII was transformed with pSTV-TteEF-G expressing TteEF-G, and the growth of transformants was monitored at the non-permissive temperature (42 °C). As shown in Figures 2(D) and 2(E), the growth rate of transformants harbouring pQE-TteRRF and pSTV-TteEF-G, or pQE-EcoDI/TteDII and pSTV-TteEF-G was significantly improved, compared with that of transformants harbouring pQE-TteRRF or pQE-EcoDI/TteDII alone. These observations indicate that both TteRRF and EcoDI/TteDII can rescue the temperature-sensitive phenotype of E. coli LJ14 (frrts) upon coexpression of TteEF-G.
It can be seen that compared with the TteRRF (Figure 2D), the transformant harbouring pQE-EcoDI/TteDII also shows more activity for complementation of the E. coli LJ14 (frrts) strain (Figure 2E), even upon coexpression of TteEF-G. Thermostability of TteRRF, EcoRRF and EcoDI/TteDII was studied by using heat induced denaturation. The thermostability of EcoDI/TteDII is close to EcoRRF, whereas TteRRF is still stable at above 85 °C [24]. This observation suggests that EcoDI/TteDII should have more flexibility than TteRRF for its functional expression in E. coli at 42 °C. This may be another reason why the transformant harbouring pQE-EcoDI/TteDII always shows more activity on complementation of the E. coli LJ14 (frrts) strain than that harbouring TteRRF (Figures 2B, 2D and 2E).
Polysome-breakdown activity of wild-type RRFs and their chimaeric RRFs
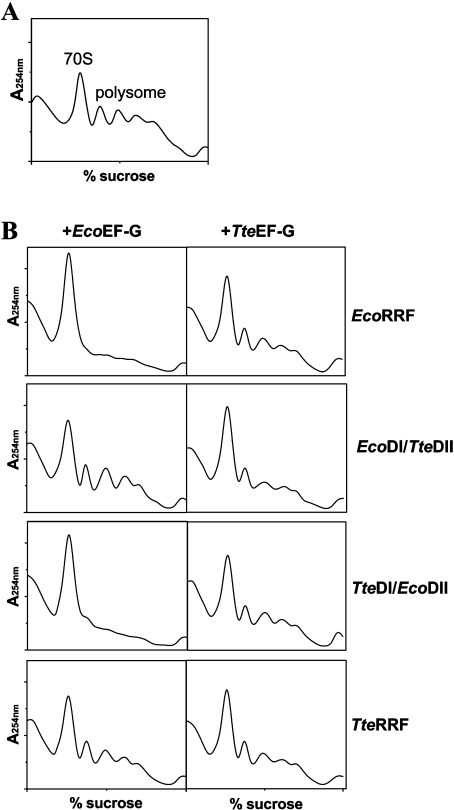
To further confirm ribosome recycling activities of wild-type RRFs and their chimaeric RRFs, in vitro polysome breakdown assays were performed using factor-free polysomes prepared from E. coli MRE600 according to the method described by Kaji and co-workers [35,36]. As shown in Figure 3(B), EcoRRF and TteDI/EcoDII RRF show polysome-breakdown activity in the present assay system containing E. coli polysomes and EcoEF-G. By contrast, TteRRF and EcoDI/TteDII RRF do not show any polysome-breakdown activity under the same conditions. However, when EcoEF-G was replaced with TteEF-G in the assay system, all four RRFs showed some polysome-breakdown activity [Figure 3B, right column compare with Figure 3A (polysomes alone)]. However, TteRRF, and especially EcoDI/TteDII showed more polysome-breakdown activity.
Figure 3. Polysome-breakdown assays of RRFs.
Reactions were set up as described in the Materials and methods section. (A) Polysomes alone; (B) left column, RRFs with EcoEF-G; right column, RRFs with TteEF-G. Each reaction mixture contains 30 μg of RRF and 60 μg of EcoEF-G or TteEF-G.
Release of RRF by EF-G from E. coli 70 S ribosomes was also performed. Complexes of 70 S ribosomes and RRFs were prepared as described in the Materials and methods section. As shown in Figure 4(A), EcoEF-G releases EcoRRF and TteDI/EcoDII from ribosomes in a dose-dependent manner, but does not release TteRRF and EcoDI/TteDII from ribosomes. By contrast, TteEF-G has more activity for the release of TteRRF, especially for the release of EcoDI/TteDII from ribosomes (Figure 4B). These results correspond well with the above in vitro polysome-breakdown activity of wild-type RRFs and their chimaeric RRFs, suggesting a specific interaction between RRF and EF-G.
Figure 4. EF-Gs release RRFs from 70 S ribosomes in a dose-dependent manner.
Complexes of EcoRRF, EcoDI/TteDII, TteDI/EcoDII or TteRRF (4 μM) with 70 S ribosomes (0.25 μM) were formed in 40 μl of assay buffer solution as described in the Material and methods section. (A) Various amounts of EcoEF-G (0.05, 0.125, 0.25, 0.5, 0.75 and 1.0 μM) and GTP (0.5 mM) were then added and incubated at room temperature for 15 min. The mixture was subjected to Microcon-100 ultrafiltration to separate the ribosomes from the released RRFs. The remaining ribosome-bound RRFs were determined by quantitative Western blotting. The RRFs bound to ribosomes in the absence of EF-Gs was taken as 100%. (B) Various amounts of TteEF-G (0.05, 0.125, 0.25, 0.5, 0.75 and 1.0 μM) and GTP (0.5 mM) were applied to the assay system. Others are the same as in (A).
Binding of wild-type RRFs and their chimaeric RRFs to ribosomes
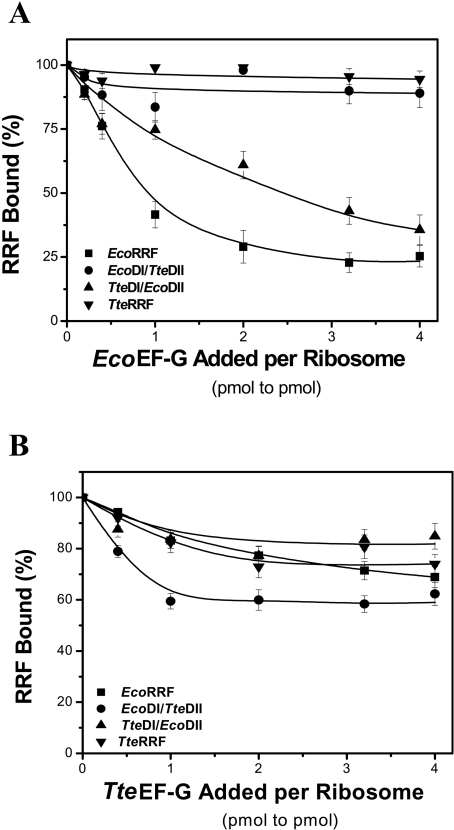
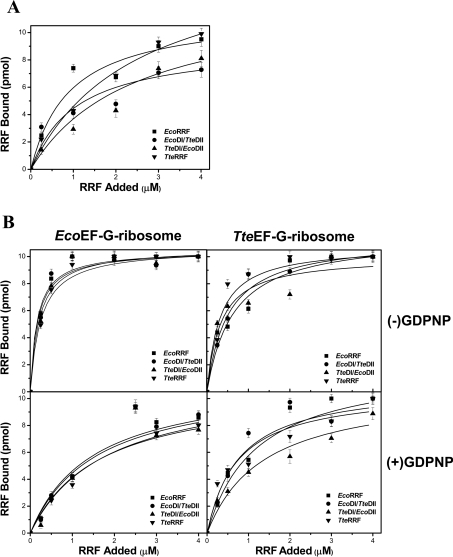
To test whether different RRFs have different binding capacities for the E. coli ribosome and whether the lack of activity of TteRRF and EcoDI/TteDII in E. coli polysome breakdown is due to the loss of binding capacity to the E. coli ribosome, the ability of EcoRRF, TteRRF, EcoDI/TteDII and TteDI/EcoDII to bind to the E. coli 70 S ribosome was tested under different conditions as described in Materials and methods. Approximate dissociation constant (Kd) values were revealed from the dose-response curves according to the amounts of bound RRF determined by quantitative Western blotting (Figure 5). Although these measured Kd values may be overestimated because the filter technique is a non-equilibrium technique, these values should be relatively meaningful, as discussed by Hirokawa et al. [37]. Figure 5(A) shows the dose-response curves for binding of various amounts of RRFs to ribosomes in the absence of EF-G and GDPNP. The apparent Kd values of EcoRRF, EcoDI/TteDII, TteRRF, and TteDI/EcoDII release from the 70 S ribosome were estimated to be in the range (1.0–2.4)×10−6 M (Table 2). The apparent Kd values for all four RRFs in the presence of EcoEF-G are in the range of (0.20–0.25)×10−6 M (Table 2), which are slightly lower than in the presence of TteEF-G, (0.31–0.64)×10−6 M (Table 2). Compared with the apparent Kd values in the absence of EF-G, this observation suggests that binding of EF-G to 70 S ribosome could facilitate RRF binding. However, in the presence of EcoEF-G and GDPNP, the apparent Kd values for all four RRFs are in the range (1.49–1.79)×10−6 M (Table 2), which are slightly higher than that in the presence of TteEF-G and GDPNP [(0.73–1.36)×10−6 M, Table 2]. Compared with the apparent Kd values in the absence of GDPNP, the data presented above suggest the presence of EF-G–GDPNP slows down the binding of RRF to the 70 S ribosome. This result is consistent with the previous finding that EF-G in the GTP form, and RRF bind with negative co-operativity to the 70 S ribosome [11,13].
Figure 5. Binding of RRFs to 70 S E. coli ribosomes under different conditions.
(A) The dose-response curve for binding of various amounts of EcoRRF, EcoDI/TteDII, TteDI/EcoDII or TteRRF to ribosomes in the absence of EF-Gs and GDPNP. The concentration of 70 S ribosomes is 0.25 μM. The RRF concentrations were from 0–4 μM. (B) The left column shows the binding curve of EcoRRF, EcoDI/TteDII, TteDI/EcoDII or TteRRF to EcoEF-G–ribosome complexes without (top) or with (bottom) 3.0 μM GDPNP; the right column shows the binding curve of EcoRRF, EcoDI/TteDII, TteDI/EcoDII or TteRRF to TteEF-G–ribosome complexes without (top) or with (bottom) 3.0 μM GDPNP. The concentrations of EcoEF-G and TteEF-G were 1 μM respectively. Other conditions were similar to those in (A).
Table 2. The apparent Kd values of EcoRRF, EcoDI/TteDII, TteRRF and TteDI/EcoDII from the 70 S ribosome under different conditions.
| Kd (×10−6 M) | |||||
|---|---|---|---|---|---|
| EF-G (−) | EcoEF-G (+) | TteEF-G (+) | |||
| RRF | GDPNP (−) | GDPNP (−) | GDPNP (+) | GDPNP (−) | GDPNP (+) |
| EcoRRF | 0.92±0.26 | 0.18±0.04 | 1.55±0.40 | 0.64±0.20 | 1.19±0.29 |
| EcoDI/TteDII | 1.06±0.31 | 0.16±0.06 | 1.49±0.39 | 0.50±0.10 | 0.78±0.32 |
| TteDI/EcoDII | 2.45±0.73 | 0.21±0.06 | 1.65±0.68 | 0.32±0.12 | 1.36±0.50 |
| TteRRF | 2.48±0.42 | 0.25±0.04 | 1.79±0.72 | 0.31±0.10 | 0.73±0.18 |
It is noteworthy that when the apparent dissociation constants obtained under the same conditions for all four RRFs were compared, the Kd values are of the same order of magnitude. This similarity of Kd values for all four RRFs suggests that: (i) all four RRFs have the same binding capacity to the 70 S ribosome; (ii) TteRRF and EcoDI/TteDII are non-functional in E. coli which is not due to the loss of their binding capacity to the E. coli ribosomes.
DISCUSSION
Role of RRF domain II and its implications for disassembly of the post-termination ribosome complex
Although disassembly of the post-termination complex by the concerted action of RRF and EF-G has been widely accepted, how RRF interacts with EF-G and disassembles the post-termination complex is not fully known. Many details of the sequential events responsible for the disassembly of the post-termination complex remain to be established. Functional studies utilizing heterologous RRF proteins in E. coli have made significant contributions towards understanding the structure-function relationship of this protein [15,22,23,40]. With genetic approaches, Nakamura and colleagues studied the viability of E. coli temperature-sensitive-lethal strains expressing heterologous RRF (T. thermophilus RRF) and wild-type or chimaeric EF-G variants. They identified a large number of mutations in both EF-G and RRF that could restore the recycling function of heterologous RRF in E. coli [10,23]. These findings not only confirm the necessity for a specific interaction between homologous EF-G and RRF as pointed out by Rao and Varshney [22], but also indicate that direct interaction between RRF and domain IV of EF-G is a prerequisite for ribosome disassembly. It is understandable from the view point of bio-evolution that a specific interaction between homologous EF-G and RRF is more favourable for their functional expression. However, Nakamura and colleagues demonstrated that Aquifex aeolicus RRF (aaRRF) shows a unique feature in heterologous expression and complementation in E. coli compared with other heterologous RRFs from M. tuberculosis [22] and T. thermophilus [23]. Like the latter two RRFs, aaRRF is non-functional in E. coli, but it remains inactive even in the presence of homologous A. aeolicus EF-G [41]. A chimaeric EF-G; however, composed of E. coli domains I–III and A. aeolicus domains IV–V in EF-G, was able to activate aaRRF in E. coli. Considering that domain IV of EF-G serves as a site for contact with RRF [23], and domain V is known to interact with the ribosome and to be essential for the positioning of domain IV [42], and domains I–III serve as contact sites with the ribosome [43], this finding might suggest aaRRF with EF-G through domain IV and the functional co-ordination with the ribosome through EF-G domains I–III. Hence the chimaeric EF-G, EcoI–III/aaIV–V, resolves both problems. Of course, this result suggests that in addition to EF-G domain IV, which serves as the site of contact with RRF [23], the interaction of EF-G domains I–III with the ribosome also plays a pivotal role in post-termination ribosome disassembly.
The above results demonstrate that clarification of the function of the individual domains of EF-G is very important for fully understanding the mechanism of the post-termination ribosome complex disassembly. In this report, to clarify the function of the individual domains of RRF, two RRF chimaeras, EcoDI/TteDII and TteDI/EcoDII, were created by domain swaps between the E. coli and T. tengcongensis RRF proteins. The ribosome recycling activity of the RRF chimaeras was compared with wild-type RRFs using in vivo and in vitro activity assays. TteRRF and EcoDI/TteDII are non-functional in E. coli as compared with EcoRRF (Figures 2B and 3B). To test whether the lack of activity of TteRRF and EcoDI/TteDII in E. coli is due to the loss of binding capacity to the E. coli ribosome, we tested the binding ability of EcoRRF, TteRRF, EcoDI/TteDII and TteDI/EcoDII to the E. coli ribosome. The latter three RRFs bind in vitro to the E. coli 70 S ribosome as efficiently as EcoRRF (Figure 5). This observation suggests that TteRRF and EcoDI/TteDII do not interact properly with the E. coli EF-G. Interestingly, like EcoRRF, TteDI/EcoDII is fully functional in E. coli (Figures 2B and 3B), suggesting that TteDI/EcoDII interacts with the E. coli EF-G as efficiently as EcoRRF, and the domain II of E. coli RRF directly interacts with the E. coli EF-G. To validate this scenario, we further tested whether TteRRF and EcoDI/TteDII could be activated by coexpression of their homologous EF-G (TteEF-G) in E. coli. As expected, both TteRRF and EcoDI/TteDII can be activated by coexpression of TteEF-G in E. coli. The ability of TteEF-G to activate both TteRRF and EcoDI/TteDII in E. coli further confirms the necessity for a specific interaction between the domain II of TteRRF and the TteEF-G. This is consistent with the structural model for RRF action on the 70 S ribosome, provided by Wilson et al. [44], in which domain II of RRF makes contact with domain IV of EF-G, whereas the hinge region of RRF nestles against domain III of EF-G. While preparing this manuscript, we became aware of a recent study on the mechanisms for the disassembly of the post-termination complex inferred from cryo-electron microscopy studies [45]. Using this technique, Gao et al. obtained two density maps: one of the RRF bound post-termination complex and one of the 50 S subunit bound with both EF-G and RRF. Comparing the two maps, Gao et al. found domain I of RRF to be in the same orientation, while domain II in the EF-G-containing 50 S subunit is extensively rotated (approx. 60°) compared with its orientation in the 70 S complex. The rotation is induced by the presence of EF-G, since domain I of RRF is firmly anchored in a pocket of rRNA helices. Mapping the 50 S conformation of RRF on to the 70 S post-termination complex suggests that the domain II movement can disrupt the intersubunit bridges B2a and B3, and could therefore be the first step in ribosome splitting into subunits [44]. Taking these results into consideration, it could be concluded that domain II of RRF plays a crucial role in the concerted action of RRF and EF-G for the posttermination complex disassembly.
Domain I of the RRF molecule mainly contributes to ribosome binding
To elucidate how the RRF molecule interacts with the ribosome and which part of the RRF molecule is mainly responsible for binding to the ribosome is important for fully understanding the mechanism of post-termination ribosome disassembly. Interaction of RRF with E. coli ribosomes, first studied by the surface plasmon resonance technique, showed that RRF interacts with 70 S ribosomes as well as 50 S and 30 S subunits, but RRF interacts preferentially with 50 S subunits [46]. Then binding of Vibrio parahaemolyticus RRF and its domain I (RRF-DI) to E. coli 70 S ribosome or its subunits was measured by a filtering technique, in which non-specific binding of wild-type RRF and RRF-DI to filter apparatus was found to be negligible [16]. This experiment showed that both wild-type RRF and RRF-DI of V. parahaemolyticus are bound to the 70 S ribosome and the 50 S subunit but not to the 30 S subunit. Polysome-breakdown assay of RRF-DI showed that RRF-DI does not show any ribosome recycling activity, but it inhibits wild-type RRF activity. These results suggest that both RRF-DI and wild-type RRF share the same binding site [16]. It can be inferred that the reason for RRF-DI inhibition of its wild-type RRF activity is due to competition at the binding-site. Since the affinity of RRF-DI to 50 S subunits is equivalent to that of wild-type RRF, binding between RRF and the ribosome depends on the interaction between domain I of RRF and the 50 S subunit [16]. A recent study further showed that amino acid residues Glu122–Arg133 and Gln161–Asp165 within E. coli RRF domain I appear to form the most stable connection with 23 S rRNA [9], which corresponds well with the previous observation that R132H and R132G mutants were structurally similar to wild-type RRF but are non-functional, they failed to bind to 50 S subunits or 70 S ribosome [46]. Taking all the observations into consideration, it can be concluded that domain I of the RRF molecule mainly contributes to ribosome binding.
However, all the above observations were made in vitro. Further clarification was sought through the in vivo experiments carried out in this study. Since binding of RRF to ribosomes is a prerequisite for its function, comparison of the ribosome recycling activity of EcoRRF, TteDI/EcoDII, TteRRF and EcoDI/TteDII in vivo provides useful evidence for the ribosome binding mechanism of the RRF molecule. As shown above, TteDI/EcoDII and EcoRRF have the same ribosome recycling activity in vivo, as do EcoDI/TteDII and TteRRF (Figure 2), suggesting that both TteRRF domain I (TteDI) and EcoRRF domain I (EcoDI) should have the same binding affinity for the E. coli ribosome. The in vitro ribosome binding assays also show that all four RRFs can bind E. coli 70 S ribosome with almost the same apparent Kd values under the same conditions. Taken together, we could suggest that there is apparently not any species-specific binding to E. coli ribosomes for EcoRRF or TteRRF. This notion is further supported by the latest works from Yamami et al. [41] and Gao et al. [45]. Yamami et al. showed that although A. aeolicus RRF is non-functional in E. coli and it remains inactive even upon coexpression of A. aeolicus EF-G, A. aeolicus RRF binds in vitro to E. coli ribosomes as efficiently as EcoRRF. As described above, Gao et al. showed domain I of RRF to be in the same orientation when bound to either the post-termination complex or the 50 S subunit, it is very tightly anchored in a pocket formed by several 23 S rRNA helices (His69, His71, His80, His89, His92 and His93) of the peptidyl transfer centre. This observation is consistent with the fact that most of the conserved residues among the helix bundle are basic amino acids [15]. The positively charged armpit region may facilitate ‘functional’ loading of RRF on to the ribosome, in relation to EF-G, by electrostatic interaction with the negatively charged phosphate group of rRNA.
It is noteworthy that to highlight the major function of the individual domains of RRF does not mean that the other function of each domain is ruled out. The RRF molecule as a whole is involved in proper interactions with EF-G and ribosomes for the post-termination ribosome disassembly. Two key processes are required for post-termination ribosome recycling to become functional: interaction of RRF with EF-G through domain IV and the functional co-ordination with the ribosome through EF-G domains I-III. TteRRF and EcoDI/TteDII are non-functional in E. coli, but they can be activated by coexpression of TteEF-G in E. coli. This observation suggests that both TteRRF and EcoDI/TteDII interact incorrectly with E. coli EF-G. To find the structural basis for the interaction between TteRRF and EcoEF-G or TteEF-G, the structural features of TteRRF were compared with its highly similar proteins, EcoRRF and TthRRF from E. coli and T. thermophilus respectively. Figure 1(A) shows the primary and secondary structures of RRFs. The deduced amino acid sequence of TteRRF shows a 51.4% identity and 68.1% similarity with that of EcoRRF, and a 50.5% identity and 66.7% similarity with that of TthRRF. More importantly, the conserved or conservatively substituted residues on the molecular surface and in hydrophobic core residues are very similar to each other [15]. The structural similarity suggests that TteRRF folds with a similar topology to other RRFs. However, amino acid sequence diversity between TteRRF and EcoRRF is also observed. According to the docking simulation results of the 50 S–EF-G–GDPNP–RRF complex [45], EcoRRF residues A94GSD97 in β-sheet 5 of RRF domain II may interact with a fragment, E559QLKAGPLAGY569, in domain IV of EcoEF-G. However, the equivalent residues for TteRRF, D93GKV96, are significantly different from those of EcoRRF. Moreover, the equivalent fragment, E546AMQNGVLGGY556 in domain IV of TteEF-G is also different from that in EcoEF-G. Interestingly, the equivalent amino acid sequences for both TthRRF and TthEF-G are similar to those in TteRRF and TteEF-G respectively. The difference in amino acid residues suggests that proper interactions between RRF and EF-G at their interfaces are essential for ribosome recycling. This may be one of the reasons why TteRRF and EcoDI/TteDII alone are non-functional in E. coli.
Acknowledgments
We thank Professor A. Kaji, Department of Microbiology, School of Medicine, University of Pennsylvania, PA, U.S.A. for providing E. coli LJ14 and MRE600 strains. We thank Dr S. Perrett (Institute of Biophysics, China) for critical reading of the manuscript and for valuable comments. We are also grateful to Z.N. Zhou and M.R. Zhang (Institute of Biophysics, China) for technical assistance. This work was supported by grants (G1999075608 and 3017021) from the China Committee for Science and Technology.
References
- 1.Freistroffer D. V., Pavlov M. Y., MacDougall J., Buckingham R. H., Ehrenberg M. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zavialov A. V., Buckingham R. H., Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 3.Zavialov A. V., Mora L., Buckingham R. H., Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 4.Janosi L., Hara H., Zhang S., Kaji A. Ribosome recycling by ribosome recycling factor (RRF)- an important but overlooked step of protein synthesis. Adv. Biophys. 1996;32:121–201. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- 5.Janosi L., Ricker R., Kaji A. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 6.Pavlov M. Y., Freistroffer D., MacDougall J., Buckingham R. H., Ehrenberg M. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karimi R., Pavlov M. Y., Buckingham R. H., Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 8.Lancaster L., Kiel M. C., Kaji A., Noller H. F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal R. K., Sharma M. R., Kiel M. C., Hirokawa G., Booth T. M., Spahn C. M., et al. Visualization of ribosome-recycling factor on the Escherichia coli 70 S ribosome: function implications. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara T., Ito K., Yamami T., Nakamura Y. Ribosome recycling factor disassembles the posttermination ribosomal complex independent of the ribosomal translocase activity of elongation factor G. Mol. Microbiol. 2004;53:517–528. doi: 10.1111/j.1365-2958.2004.04156.x. [DOI] [PubMed] [Google Scholar]
- 11.Kiel M. C., Raj V. S., Kaji H., Kaji A. Release of ribosome-bound ribosome recycling factor by elongation factor G. J. Biol. Chem. 2003;278:48041–48050. doi: 10.1074/jbc.M304834200. [DOI] [PubMed] [Google Scholar]
- 12.Peske F., Rodnina M. V., Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Zavialov A. V., Hauryliuk V. V., Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa G., Nijman R. M., Raj V. S., Kaji H., Igarashi K., Kaji A. The role of ribosome recycling factor in dissociation of 70 S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyoda T., Tin O. F., Ito K., Fujiwara T., Kumasaka T., Yamamoto M., Garber M. B., Nakamura Y. Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA. 2000;6:1432–1444. doi: 10.1017/s1355838200001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano H., Yoshida T., Uchiyama S., Kawachi M., Matsuo H., Kato T., Ohshima A., Yamaichi Y., Honda T., Kato H., et al. Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium. J. Biol. Chem. 2003;278:3427–3436. doi: 10.1074/jbc.M208098200. [DOI] [PubMed] [Google Scholar]
- 17.Selmer M., Al-Karadaghi S., Hirokawa G., Kaji A., Liljas A. Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science (Washington D.C.) 1999;286:2349–2352. doi: 10.1126/science.286.5448.2349. [DOI] [PubMed] [Google Scholar]
- 18.Kim K. K., Min K., Suh S. W. Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J. 2000;19:2362–2370. doi: 10.1093/emboj/19.10.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano H., Uchiyama S., Yoshida T., Ohkubo T., Kato H., Yamagata Y., Kobayashi Y. Crystallization and preliminary X-ray crystallographic studies of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly. Acta Crystallogr. D Biol. 2002;58:124–126. doi: 10.1107/s0907444901019941. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida T., Uchiyama S., Nakano H., Kashimori H., Kijima H., Ohshima T., Saihara Y., Ishino T., Shimahara H., Yoshida T., et al. Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry. 2001;40:2387–2396. doi: 10.1021/bi002474g. [DOI] [PubMed] [Google Scholar]
- 21.Saikrishnan K., Kalapala S. K., Varshney U., Vijayan M. X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications. J. Mol. Biol. 2005;345:29–38. doi: 10.1016/j.jmb.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Rao A. R., Varshney U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 2001;20:2977–2986. doi: 10.1093/emboj/20.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito K., Fujiwara T., Toyoda T., Nakamura Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell. 2002;9:1263–1272. doi: 10.1016/s1097-2765(02)00547-6. [DOI] [PubMed] [Google Scholar]
- 24.Guo P., Zhang L. Q., Qi Z., Chen R. S., Jing G. Z. Expression in Escherichia coli, purification and characterization of Thermoanaerobacter tengcongensis ribosome recycling factor. J. Biochem. (Tokyo) 2005;138:89–94. doi: 10.1093/jb/mvi102. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 26.Zhang X. L., Guo P., Jing G. Z. A vector with the downstream box of the initiation codon can highly enhance protein expression in Escherichia coli. Biotech. Lett. 2003;25:755–760. doi: 10.1023/a:1023563600459. [DOI] [PubMed] [Google Scholar]
- 27.Bao Q., Tian Y., Li W., Xu Z., Xuan Z., Hu S., Dong W., Yang J., Chen Y., Xue Y., et al. A complete sequence of the. T tengcongensis genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichikawa S., Kaji A. Molecular cloning and expression of ribosome releasing factor. J. Biol. Chem. 1989;264:20054–20059. [PubMed] [Google Scholar]
- 29.Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene coding for elongation factor G. Nucleic Acids Res. 1984;12:2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton R. M., Cai Z., Ho S. N., Pease L. R. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 31.Feng Y. M., Zhang Y. M., Jing G. Z. Soluble expression in Escherichia coli, purification and characterization of a human TF-1 cell apoptosis-related protein TFAR19. Protein Exp. Purif. 2002;25:323–329. doi: 10.1016/s1046-5928(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhou B., Jing G. Z. Determination of protein extinction coefficients by alkaline hydrolysis. Progr. Biochem. Biophys. 1998;26:384–387. [Google Scholar]
- 33.Tai P., Davis B. Isolation of polysomes free of initiation factors. Methods Enzymol. 1979;59:362–371. doi: 10.1016/0076-6879(79)59097-1. [DOI] [PubMed] [Google Scholar]
- 34.Jelenc P. C. Rapid purification of highly active ribosomes from Escherichia coli. Anal. Biochem. 1980;105:369–374. doi: 10.1016/0003-2697(80)90472-8. [DOI] [PubMed] [Google Scholar]
- 35.Hirashima A., Kaji A. Factor-dependent release of ribosomes from messenger RNA: requirement for two heat-stable factors. J. Mol. Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- 36.Ohnishi M., Janosi L., Shuda M., Matsumoto H., Terawaki T., Kaji A. Molecular cloning, sequencing purification and characterization of Pseudomonas aeruginosa ribosome recycling factor. J. Bacteriol. 1999;181:1281–1291. doi: 10.1128/jb.181.4.1281-1291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirokawa G., Kiel M. C., Muto A., Selmer M., Raj V. S., Liljas A., Igarashi K., Kaji H., Kaji A. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janosi L., Shimizu I., Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janosi L., Mottagui-Tabar S., Isaksson L. A., Sekine Y., Ohtsubo E., Zhang S., Goon S., Nelken S., Shuda M., Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujiwara T., Ito K., Nakayashiki T., Nakamura Y. Amber mutations in ribosome recycling factors of Escherichia coli and Thermus thermophilus: evidence for C-terminal modulator element. FEBS Lett. 1999;447:297–302. doi: 10.1016/s0014-5793(99)00302-6. [DOI] [PubMed] [Google Scholar]
- 41.Yamami T., Ito K., Fujiwara T., Nakamura Y. Heterologous expression of Aquifex aeolicus ribosome recycling factor in Escherichia coli is dominant lethal by forming a complex that lacks functional co-ordination for ribosome disassembly. Mol. Microbiol. 2005;55:150–161. doi: 10.1111/j.1365-2958.2004.04387.x. [DOI] [PubMed] [Google Scholar]
- 42.Rodnina M. V., Savelsbergh A., Katunin V. I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature (London) 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 43.Agrawal R. K., Penczek P., Grassucci R. A., Frank J. Visualization of elongation factor G on the Escherichia coli 70 S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson D. N., Schluenzen F., Harms J. M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao N., Zavialov A. V., Li W., Sengupta J., Valle M., Gursky R. P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Ishino T, Atarashi K., Uchiyama S., Yamami T., Saihara Y., Yoshida T., Hara H., Yokose K., Kobayashi Y., Nakamura Y. Interaction of ribosome recycling factor and elongation factor EF-G with E. coli ribosomes studied by the surface plasmon resonance technique. Genes to Cells. 2000;5:953–963. doi: 10.1046/j.1365-2443.2000.00382.x. [DOI] [PubMed] [Google Scholar]