Abstract
ARIP4 [AR (androgen receptor)-interacting protein 4] is a member of the SNF2-like family of proteins. Its sequence similarity to known proteins is restricted to the centrally located SNF2 ATPase domain. ARIP4 is an active ATPase, and dsDNA (double-stranded DNA) and ssDNA (single-stranded DNA) enhance its catalytic activity. We show in the present study that ARIP4 interacts with AR and binds to DNA and mononucleosomes. The N-terminal region of ARIP4 mediates interaction with AR. Kinetic parameters of the ARIP4 ATPase are similar to those of BRG-1 and SNF2h, two members of the SNF2-like protein family, but the specific activity of ARIP4 protein purified to >90% homogeneity is approximately ten times lower, being 120 molecules of ATP hydrolysed by an ARIP4 molecule per min in contrast with approx. 1000 ATP molecules hydrolysed per min by ATP-dependent chromatin remodellers. Unlike other members of the SNF2 family, ARIP4 does not appear to form large protein complexes in vivo or remodel mononucleosomes in vitro. ARIP4 is covalently modified by sumoylation, and mutation of six potential SUMO (small ubiquitin-related modifier) attachment sites abolished the ability of ARIP4 to bind DNA, hydrolyse ATP and activate AR function. We conclude that, similar to its closest homologues in the SNF2-like protein family, ATRX (α-thalassemia, mental retardation, X-linked) and Rad54, ARIP4 does not seem to be a classical chromatin remodelling protein.
Keywords: androgen receptor, androgen receptor-interacting protein 4 (ARIP4), ATPase, chromatin remodelling, co-regulator, sumoylation
Abbreviations: AR, androgen receptor; ARIP4, AR-interacting protein 4; ATRX, α-thalassemia, mental retardation, X-linked; dsDNA, double-stranded DNA; EMSA, electrophoretic mobility-shift assay; REA, restriction enzyme accessibility; RSC, remodelling the structure of chromatin; ssDNA, single-stranded DNA; SUMO-1, small ubiquitin-related modifier-1; TBE, 45 mM Tris/borate/1 mM EDTA
INTRODUCTION
ARIP4 [AR (androgen receptor)-interacting protein 4] was initially found through its interaction with the zinc finger region of the AR [1]. ARIP4 is an active DNA-dependent ATPase. Its ATPase domain is located in the central region of the protein, and it has high sequence similarity to ATPase domains of the SNF2-like proteins.
On the basis of their sequence similarity, the proteins of the SNF2 family can be divided into two major groups [2]. The first group includes active ATPases, such as BRG-1, hBRM and SNF2h, that utilize the energy from ATP hydrolysis to loosen contacts between DNA and histone octamers. As a result, DNA sequences become more accessible for transcriptional factors and/or histone-modifying enzymes [3]. In many instances, these proteins serve as the ATPase component of high-molecular-mass (∼2 MDa) protein complexes, such as yeast RSC (remodelling the structure of chromatin), and human and Drosophila SWI/SNF complexes. These complexes are known to act as co-activators or co-repressors for different transcription factors, including nuclear receptors [4,5]. Biochemical properties of the SNF2 family ATP-ases have been intensively studied, revealing functional differences between different ATP-dependent chromatin remodellers. Parameters of ATPase reaction, including values for Km and catalytic activity, have been estimated [6–8]. Chromatin remodellers differ also in their mononucleosome remodelling activity [9]. On the basis of in vitro assays, a general chromatin remodelling mechanism in which ATP-dependent chromatin remodellers act as anchored DNA translocases has been proposed [10].
The second group of proteins of the SNF2 family includes proteins, such as ATRX (α-thalassemia, mental retardation, X-linked), Rad26 and Rad54, that are less active in chromatin remodelling; rather, they are involved in DNA excision repair and homologous recombination. ARIP4 appears to belong to this latter group, with the ATRX protein exhibiting the highest sequence similarity to ARIP4 in the ATPase domain [2]. ATRX protein mutations cause X-linked mental retardation with α-thalassemia [11]. Through interaction with the apoptosis-related protein Daxx, the ATRX protein is involved in apoptosis regulation [12,13]. Recent studies have also suggested that ATRX is involved in mammalian sex differentiation [14].
We have previously shown that ARIP4 interacts with AR in yeast and mammalian two-hybrid assays and demonstrated that ARIP4 acts as an AR co-regulator in reporter gene assays. Wild-type ARIP4 stimulated AR activity on minimal promoters, whereas ATPase-deficient mutants behaved as trans-dominant-negative regulators of AR function [1]. Co-regulators can modulate AR activity by multiple mechanisms, including facilitated DNA binding, chromatin remodelling and recruitment of general transcription factors associated with RNA polymerase II complex to androgen-responsive promoters [15–19].
In the present study, we have characterized the biochemical properties of ARIP4 as a DNA-stimulated ATPase. We find that the kinetic parameters of the ARIP4 ATPase are similar to those of BRG-1 and SNF2h, whereas the catalytic activity of ARIP4 is ten times lower than that of the other two ATPases. In contrast with yeast RSC complex, ARIP4 is not active in REA (restriction enzyme accessibility) assay. We also show that the N-terminal region of ARIP4 mediates its interaction with AR and this interaction does not depend on DNA binding. Mutation of ARIP4 sumoylation sites renders the protein incapable of binding to DNA, catalysing ATP hydrolysis and activating AR function. Our results demonstrate that ARIP4 is not a classical chromatin remodelling protein, suggesting that it could be involved in homologous recombination and DNA repair.
MATERIALS AND METHODS
Materials
Mouse monoclonal anti-FLAG antibody M2 was purchased from Sigma and mouse monoclonal anti-GMP-1 was from Zymed Laboratories. Rabbit polyclonal anti-AR antibody K333 and anti-ARIP4 antibody K7991 have been described in [1,20]. Expression vectors pSG5hAR, pARE4-tk-LUC, pFLAG-ARIP4wt, pFLAG-ARIP4(1–1314) and pFLAG-ARIP4-K310A have been described previously [1]. pTPT plasmid was kindly provided by Dr Geeta Narlikar (University of California, San Francisco, CA, U.S.A.) and pSG5-His-SUMO-1 (where SUMO-1 is small ubiquitin-related modifier-1, also known as GMP-1) was a gift from Dr Anne Dejean (Institut Pasteur, Paris, France). Yeast chromatin remodelling complex RSC was a gift from Dr Tom Owen-Hughes (The Wellcome Trust Biocentre, University of Dundee, Scotland, U.K.).
Plasmid constructions
To generate pFLAG-ARIP4(1–276), pFLAG-ARIP4wt was digested with BamHI and re-ligated. To assemble pFLAG-ARIP4(1–620), pFLAG-ARIP4wt was digested with BglII/XbaI, treated with DNA polymerase I Klenow fragment (Promega) and religated. Expression vector pFLAG-ARIP4(1–889) was assembled in two steps. First, a fragment of ARIP4 cDNA was PCR-amplified with the primers 5′-GGAATTCTTATGATAACCTAGTGGAGTCCTTAGA-3′ and 5′-CCATCGATTCAATCCACTACCCGATCT-3′, digested with EcoRI/ClaI and inserted into the pFLAG-CMV2 (Sigma–Aldrich) vector digested with the same enzymes to assemble pFLAG-ARIP4(282–889). Secondly, the fragment of ARIP4 cDNA corresponding to residues 1–776 was cut out from pFLAG-ARIP4wt with HindIII/Bsp119I and cloned into the pFLAG-ARIP4(282–889) vector digested with the same enzymes. To assemble pFLAG-ARIP4(1–278/890–1309) and pFLAG-ARIP4(1–278/890–1206), a fragment of ARIP4 cDNA was amplified by PCR with the primers 5′-GAAGATCTTCGATCTAAATCCAATGCTGAACT-3′ and 5′-CGGATATCAAGTGAAGGGGGTGGTGAG-3′, digested with BglII/EcoRV or with BglII/SmaI and inserted into pFLAG-ARIP4wt digested with BamHI/SmaI. Consensus-like sumoylation sites were selected using SUMOplot analysis tool (http://www.abgent.com/doc/sumoplot). ARIP4 mutations for sumoylation analysis [K573R, K664R, K935R, K1013R, KK361/961RR, KK573/664RR, KKKK361/573/664/961RRRR (4K→4R), KKKKK361/573/664/961/1013RRRRR (5K→5R) and KKKKKK361/573/664/935/961/1013RRRRRR (6K6R)] were introduced into pFLAG-ARIP4wt using a site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Transfections and production of recombinant proteins
Transfections for transactivation assays were performed as described in [1]. For protein production, COS-1 cells (3.5×105) were transfected with 1.0 or 1.5 μg of an appropriate expression vector DNA using FuGene reagent (Roche). Cells were lysed 48 h after transfection in a buffer containing 20 mM Tris/HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P40, 0.5% Triton X-100, 1 mM dithiothreitol, 10% (v/v) glycerol and a protease inhibitor cocktail (Sigma–Aldrich). The lysates were clarified by centrifugation at 4 °C for 20 min at 16000 g and supernatants were adsorbed on 30 μl of anti-FLAG affinity matrix (Sigma–Aldrich) for 2 h at 4 °C. The matrix was subsequently washed with 3 ml of the above lysis buffer containing 500 mM NaCl. Bound proteins were eluted with 30 μl of lysis buffer containing 0.2 mg/ml FLAG peptide and used for EMSA (electrophoretic mobility-shift assay) and ATPase assays. Wild-type ARIP4 and the ATPase-deficient mutant ARIP4-K310A were produced in insect cells by the use of baculovirus vectors and purified, essentially as described previously [1].
ATPase activity assay
Fluorimetric ATPase assays were carried out as described in [21,22] with some modifications. Briefly, the reaction mixture (100 μl), containing 20 mM Tris/HCl (pH 7.5), 8 mM MgCl2, 40 mM KCl, 100 mM NaCl, 8% glycerol, 0.5 mM dithiothreitol, 100 μM NADH, 0.25 mM phosphoenolpyruvate (Roche), 15 units of pyruvate kinase/lactate dehydrogenase (Roche) and different concentrations of ATP, supercoiled plasmid DNA (pGL3Basic), ssDNA (single-stranded DNA) (ΦX174) or salmon sperm DNA, as indicated in the Figure legends, was preincubated at 37 °C for 15 min. NADH fluorescence was measured on a Varian Cary Eclipse fluorimeter (Varian) at 37 °C. The system was first calibrated by the addition of 200 pmol of ADP, then indicated amounts of ARIP4 protein were added and the course of the reaction was followed in real time.
ATPase activity of ARIP4 (200 ng), RSC (50 ng) and sumoylation mutants was assayed in a buffer containing 50 mM Tris/HCl (pH 7.5), 3 mM MgCl2, 50 mM NaCl, 3% glycerol (8% glycerol for sumoylation mutants), 1 mM ATP, 0.5 μCi of [γ-32P]ATP and 32 nM pFLAG-CMV2 plasmid. Samples (1 μl) of the reaction mixture were taken at different time points during the incubation at 30 °C. Reaction was stopped by the addition of 2 μl of 50 mM EDTA. Samples (1 μl) were spotted on to a poly(ethyleneimine) thin-layer plate that was developed in 1 M LiCl/1 M formic acid to resolve [32P]Pi from [γ-32P]ATP. The plates were subjected to autoradiography. The amount of released [32P]Pi was quantified by measuring the radioactivity of the spots using a Wallac 1409 liquid-scintillation counter (Wallac Oy, Finland) or scanning autoradiograms using Kodak Image station 440CF (Kodak).
Immunoprecipitation
Co-immunoprecipitation experiments were carried out as described previously [1], except that for the sumoylation assay, COS-1 cells were transfected with 0.7 μg of pFLAG-ARIP4wt or point mutants and 1.5 μg of pSG5-His-SUMO-1. Immunoblotting was performed as described in [23] by using mouse monoclonal anti-FLAG M2 (1:2000 dilution) or rabbit polyclonal anti-AR (K333; 1:4000) antibody.
Histone octamer purification, mononucleosome reconstitution and REA assay
Chicken blood was collected, and erythrocyte nuclei were isolated as described in [24]. Core histone octamers were purified as described in [25,26]. Briefly, nuclei (∼6 mg of DNA) were lysed for 30 min at 4 °C in 10 ml of lysis buffer containing 0.7 M NaCl, 50 mM sodium phosphate (pH 6.8), 0.5 mM PMSF and 1 mM 2-mercaptoethanol. Then, 7.5 g of hydroxyapatite DNA grade Bio-Gel powder (Bio-Rad) was added and the lysate was rotated for 15 min at 4 °C. The hydroxyapatite-immobilized chromatin was collected by centrifugation at 2000 g for 5 min at 4 °C and washed six times with 40 ml of lysis buffer. Core histone octamers were eluted in a buffer containing 2.5 M NaCl, 50 mM sodium phosphate (pH 6.8), 0.5 mM PMSF and 1 mM 2-mercaptoethanol.
The 202 bp DNA fragment was obtained by PCR amplification of pTPT [9] with TPT-for (5′-ACGCGTCGGTGTTAGAGCC-3′) and TPT-rev primers (5′-ACGCCAGGGTTTTCCCAGTCA-3′). Fragments were 32P-labelled as described previously [20]. Mononucleosomes were reconstituted by mixing 800 ng of core histone octamers with 500 ng of 32P-labelled 202 bp DNA in 25 μl of a buffer containing 50 mM Tris/HCl (pH 7.5), 1 mM EDTA and 2 M NaCl, after which the mixture was dialysed in 2 h steps at 4 °C against 400 ml of a buffer containing 50 mM Tris/HCl (pH 7.5), 1 mM EDTA and 1.25, 1.0, 0.8 and 0.6 M NaCl using Slide-A-Lyser Mini Dialysis Units (3500 Da molecular mass cut-off; Pierce). The final dialysis step was carried out overnight against 400 ml of 50 mM Tris/HCl (pH 7.5).
REA assay was carried out in 20 μl volume containing 50 ng of RSC or 200 ng of ARIP4, 0.75 unit/μl PstI, 1 mM Hepes (pH 7.4), 50 mM Tris/HCl (pH 7.5), 50 mM NaCl, 3% glycerol, 3 mM MgCl2, 0.25 mM dithiothreitol, 0.5 mM 2-mercaptoethanol, 10 μg/ml BSA (Sigma) and 1 mM ATP for 1 h at 30 °C. The reaction was stopped with 30 μl of a buffer containing 20 mM Tris/HCl (pH 7.8), 70 mM EDTA, 10% glycerol, 2% (w/v) SDS, 0.2 mg/ml Xylene Cyanol and 0.2 mg/ml Bromophenol Blue. Proteinase K was added to 1 mg/ml final concentration, and the samples were incubated for 1 h at 37 °C. DNA fragments were separated on a 6% (w/v) native polyacrylamide gel in 1×TBE (45 mM Tris/borate/1 mM EDTA).
Protein–DNA and protein–mononucleosome interactions in vitro
DNA fragments for EMSAs were 32P-labelled as described previously [20]. The binding reaction mixture (20 μl) contained either 30 ng of wild-type ARIP4 protein (or deletion mutant as indicated in the Figure legends) for DNA binding or 500 ng of wild-type ARIP4 for mononucleosome binding experiment, in a buffer containing 17 mM Tris/HCl (pH 7.8), 12 mM Hepes (pH 7.7), 6 mM MgCl2, 15 mM NaCl, 55 mM KCl, 0.3 mM EDTA, 5.5% glycerol, 1.25 mM dithiothreitol, 0.05% Nonidet P40, 0.05% Triton X-100, 0.25 mM PMSF, 0.3–2 mM ATP and 0–100 ng of unlabelled poly(dI-dC)·(dI-dC), as indicated in the Figure legends. After a preincubation on ice for 10 min, 0.5 ng of labelled oligonucleotides or mononucleosomes was added, and the incubation was continued at 37 °C for 20 min. Protein–DNA or protein–mononucleosome complexes were resolved on a 4% polyacrylamide gel containing 0.1% Nonidet P40 in 0.25×TBE. Gels were dried and exposed to Fuji X-ray films overnight at −70 °C.
Yeast two-hybrid screening
A Gal4 fusion to full-length wild-type ARIP4 was used to screen a human HeLa cDNA library in pGAD-GH (Clontech) according to the manufacturer's instructions. Positive clones were analysed by digestion and sequenced. Resulting sequences were used in BLASTN search (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) in the human sequences database. Most of the 36 positive clones contained fragments of Ubc9 or SUMO-1 cDNA.
RESULTS
Enzymatic characteristics of ARIP4 ATPase
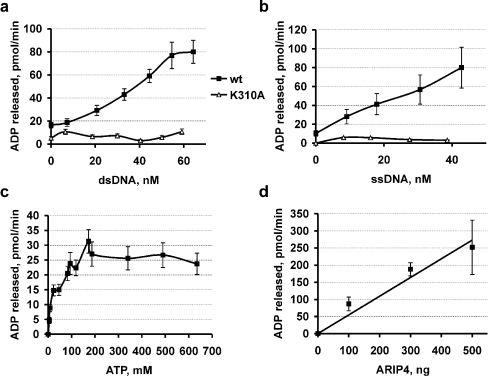
We have shown previously that ARIP4 binds ATP and has intrinsic ATPase activity [1]. We produced FLAG-tagged wild-type ARIP4 and its ATPase-mutated form ARIP4-K310A using a baculovirus expression system in insect cells. The proteins were purified by immunoaffinity chromatography with anti-FLAG affinity resin to >90% homogeneity. By titrating the concentration of one reaction component and monitoring ADP release rate in a fluorimetric ATPase assay, we estimated the kinetic parameters of ATP hydrolysis catalysed by ARIP4. First, we fixed ATP concentration at 250 μM and the amount of ARIP4 at 500 ng, and titrated the concentrations of dsDNA (double-stranded DNA) or ssDNA (Figures 1a and 1b). In both cases, ATPase activity was clearly enhanced. From the results presented in Figure 1(a), we have estimated the Km (app) for dsDNA (at 250 μM ATP) to be 40±10 nM. ARIP4-K310A (800 ng) exhibited only control level ATPase activity that could not be enhanced by the addition of either dsDNA or ssDNA (Figures 1a and 1b). Subsequently, we titrated the ATP concentrations while keeping ARIP4 (500 ng) and dsDNA (15 nM) concentrations constant (Figure 1c) and estimated the Km (app) for ATP (at 15 nM dsDNA) to be 25±10 μM. Finally, we fixed ATP at 250 μM and salmon sperm DNA at 600 μg/ml and titrated the ARIP4 amount (Figure 1d). The estimated activity of wild-type ARIP4 is 600±100 nmol of ADP released·min−1·(mg of ARIP4)−1. Assuming that ARIP4 has a molecular mass of approx. 200 kDa, this corresponds to approx. 120 molecules of ATP hydrolysed by a protein molecule per min. This value is approximately ten times lower than that of ATP-dependent chromatin remodelling enzymes, hydrolysing approx. 1000 ATP molecules per min [7].
Figure 1. ARIP4 is an active DNA-dependent ATPase.
ATPase activity was measured in a fluorimetric ATPase assay. (a, b) 500 ng of ARIP4wt (or 800 ng of ARIP4-K310A) and 250 μM ATP were incubated with different amounts of supercoiled plasmid DNA (pGL3Basic) or ssDNA (ΦX174). Estimated Km (app) for DNA (at 250 μM ATP) equals 40±10 nM. (c) 500 ng of ARIP4, 15 nM supercoiled plasmid DNA (pGL3Basic) were incubated with different amounts of ATP. Estimated Km for ATP (at 15 nM dsDNA) equals 25±10 μM. (d) For catalytic activity estimation, 250 μM ATP and 600 ng/ml salmon sperm DNA were incubated with different ARIP4 amounts. ARIP4 activity is estimated from the resulting line (R=0.97) and equals 600±100 nmol of ADP released·min−1·(mg of ARIP4)−1. Assuming that ARIP4 has a molecular mass of approx. 200 kDa, the catalytic activity is 120±20 ATP molecules hydrolysed per min.
N-terminal fragment of ARIP4 mediates interactions with AR
The ATPase domain of ARIP4 is located in the central region of the protein, whereas the N- and C-terminal regions of ARIP4 have no similarity to any known conserved protein domains. To define the region important for ARIP4 interaction with AR, we constructed several ARIP4 deletion mutants. The interaction of ARIP4 with AR was not dependent on the ATPase activity of the protein, as the ARIP4-K310A mutant recognized AR in co-immunoprecipitation experiments as well as wild-type ARIP4 (results not shown). When co-expressed with AR in COS-1 cells, an N-terminal fragment of ARIP4 (residues 1–620) co-immunoprecipitated with the receptor. Under the same conditions, a shorter N-terminal fragment (1–276) failed to co-immunoprecipitate with AR (Figure 2a), despite the fact that it is a part of this ARIP4 fragment (amino acid residues 91–230) that was initially recognized as a LexA fusion protein by the zinc finger region of AR in our yeast two-hybrid screen [1]. Since ARIP4(1–620) interacted with AR, but did not bind DNA (Figures 2a and 2b), we conclude that ARIP4 interaction with AR does not depend on ability of ARIP4 to bind DNA.
Figure 2. The N-terminal fragment of ARIP4 mediates interaction with AR and this interaction does not depend on DNA binding.
(a) The N-terminal (1–620) fragment of ARIP4 is sufficient to bind AR. COS-1 cells were transiently transfected with 100 ng of pSG5hAR and 300 ng of pFLAG-CMV2 (indicated as ‘mock’) or pFLAG-ARIP4wt, pFLAG-ARIP4(1–276), pFLAG-ARIP4(1–620), pFLAG-ARIP4(1–889) and pFLAG-ARIP4(1–1314), as indicated. Testosterone (T) was added to the culture medium as indicated for 2 h before harvesting the cells. Co-immunoprecipitation and immunoblotting were performed as described in the Materials and methods section. (b) Indicated ARIP4 deletion mutants were purified by affinity chromatography from transiently transfected COS-1 cells as described in the Materials and methods section. The proteins were incubated with a 32P-labelled 32 bp DNA fragment and protein–DNA complexes were resolved by EMSA. Asterisks indicate ARIP4-specific shifted bands. IP, immunoprecipitation.
Wild-type ARIP4 and ARIP4(1–1314) were able to bind 32 bp DNA fragment in EMSA experiments, whereas ARIP4(1–889) and shorter N-terminal ARIP4 fragments failed to bind DNA (Figure 2b). These results suggest that DNA binding domain is localized within ARIP4(889–1314). However, several N-terminal deletion mutants of ARIP4, including ARIP4(278–1314), ARIP4(428–1260), ARIP4(889–1260) and ARIP4(890–1316), failed to bind DNA in EMSA experiments (results not shown). Deletion of residues 1–277 resulted in substantially lower protein expression in transfected COS-1 cells, implying that these residues of ARIP4 are important for protein stability. Indeed, ARIP4(1–278/890–1206) and ARIP4(1–278/890–1309) were expressed in COS-1 cells to levels similar to that of wild-type ARIP4. However, neither ARIP4(1–278/890–1206) nor ARIP4(1–278/890–1309) could bind DNA in EMSA experiments (Figure 2b).
We have found no nucleotide sequence preference for DNA binding of ARIP4, as ARIP4 interacted with all DNA oligonucleotides tested by the EMSA technique. The binding of ARIP4 to DNA was efficiently competed for by unlabelled poly(dI-dC)· (dI-dC) (results not shown; see also Figure 3b).
Figure 3. ARIP4 binds, but does not remodel, reconstituted mononucleosomes.
(a) Mononucleosomes reconstituted with chicken erythrocyte core histone octamers. Mononucleosomes were reconstituted on a 202 bp 32P-labelled fragment containing nucleosome positioning sequence and PstI restriction site (55 bp from 5′-end) [9] by serial dialysis against buffers with progressively lower ionic strength and resolved on a 5% polyacrylamide gel in 0.5×TBE. (b) Wild-type ARIP4 was purified from Sf9 cells by affinity chromatography [1]. The protein was incubated with 32P-labelled mononucleosomes in the presence of indicated poly(dI-dC)·(dI-dC) amounts, and protein–mononucleosome complexes (marked as ‘complex’) were resolved by EMSA. (c) REA assay. Reconstituted mononucleosomes were incubated with 50 ng of RSC or 200 ng of ARIP4 in the presence of 0.75 unit/μl PstI and 1 mM ATP (where indicated) for 1 h at 30 °C. The reaction was stopped, proteins were removed by proteinase K treatment, and DNA fragments were resolved on a 6% polyacrylamide gel in 1×TBE. (d) ATPase activity of RSC and ARIP4 used in the REA assay. ATPase activity of 200 ng of ARIP4 and 50 ng of RSC was assayed using [γ-32P]ATP as described in the Materials and methods section. ATP hydrolysis is given as percentage of substrate hydrolysed.
ARIP4 binds, but does not remodel, reconstituted mononucleosomes
To test whether ARIP4 exhibits mononucleosome remodelling activity, we used histone octamers purified from chicken erythrocytes to reconstitute mononucleosomes on 32P-labelled 202 bp DNA fragments. These DNA fragments bear nucleosome positioning sequence and the PstI restriction site [9] (Figure 3a). As expected, wild-type ARIP4 bound to the reconstituted mononucleosomes, as judged by EMSA experiments. Once again, this binding failed to exhibit sequence specificity, as unlabelled poly(dI-dC)·(dI-dC) competed efficiently with mononucleosomes for ARIP4 complex formation (Figure 3b). To check whether mononucleosomes were remodelled during the interaction with ARIP4, we used the REA assay [9]. The restriction enzyme PstI cannot gain access to its recognition sequence in intact mononucleosomes. Should ARIP4 have remodelling activity and loosen histone–DNA contacts, PstI could then gain access to its cognate recognition sequence and digest DNA at this site. We incubated ARIP4 with mononucleosomes in the presence of ATP and PstI. Under our experimental conditions, PstI could not digest either intact mononucleosomes or those preincubated with ARIP4. The yeast chromatin remodelling complex RSC exhibited robust activity in the REA assay, when tested under identical conditions as a positive control (Figure 3c). Dissimilar catalytic ATPase activities of RSC and ARIP4 preparations cannot explain their differential behaviour in the REA assay, because ARIP4 had in fact higher ATPase activity than RSC (Figure 3d). It is of note that other previously identified ATPase-dependent chromatin remodellers (human BRG-1 and SNF2h) have been shown to possess clearly detectable activity in REA assay [9]. Therefore we conclude that ARIP4 can bind to mononucleosomes, but does not remodel them.
ARIP4 is sumoylated
In an attempt to find ARIP4 interaction partners, we performed yeast two-hybrid assay using full-length wild-type ARIP4 as the bait. This screen led to the identification of only two biologically meaningful interacting partners for ARIP4, namely SUMO-1 and Ubc9 (results not shown). SUMO-1 is covalently attached to lysine residue(s) of target protein in a process catalysed by the E1 activating enzyme Aos1/Uba2, E2 conjugating enzyme Ubc9 and different E3 ligating enzymes [27,28]. ARIP4 contains one consensus sequence for SUMO-1 attachment at Lys664 (VK664GE), and ARIP4 was indeed sumoylated in vivo (Figure 4a). Lysine-to-arginine mutation at Lys664 and mutations of five other consensus-like sites at Lys361 (SK361PE), Lys573 (AK573EE), Lys935 (LK935YP), Lys961 (TK961AE) and Lys1013 (LK1013GD) altered ARIP4 sumoylation pattern (Figure 4a). None of these mutations alone could prevent ARIP4 from being sumoylated. However, combining mutations of six lysine residues (6K→6R) resulted in a substantial decrease in ARIP4 sumoylation (Figure 4a). All sumoylation mutants of ARIP4 except ARIP4(6K→6R) had comparable ATPase activities that were not markedly different from wild-type ARIP4 activity (Figure 4b), and the activities of these mutants in AR-dependent reporter gene transactivation assays were similar (Figure 5a). The sumoylation-deficient ARIP4(6K→6R) mutant had, however, lost its ATPase activity (Figure 4b), AR co-activator activity (Figure 5a) and DNA binding (Figure 5b).
Figure 4. ARIP4 is sumoylated.
(a) Cells were transfected with 0.7 μg of empty pFLAG-CMV2 or pFLAG-ARIP4wt (or indicated mutants) and 1.5 μg of empty pSG5 vector or pSG5-His-SUMO-1. Cells were lysed 48 h post-transfection and immunoprecipitation was carried out as described in [36]. Asterisks indicate bands corresponding to sumoylated forms of ARIP4. (b) ATPase activity of sumoylation mutants. Wild-type (wt) ARIP4 or indicated mutants were produced in COS-1 cells. ATPase activity in the presence of dsDNA was assayed as described in the Materials and methods section. ATPase reaction was stopped after 15 and 30 min incubation at 30 °C. Autoradiograms were scanned and quantified using Kodak Image station 440CF (Kodak).
Figure 5. ARIP4 sumoylation mutants in AR-dependent transactivation and DNA binding.
(a) COS-1 cells were transiently transfected with 150 ng of pARE4-tk-LUC, 20 ng of pSGhAR and 30 ng of pFLAG-ARIP4wt or indicated sumoylation mutants of ARIP4. Testosterone (T; 100 nM) was added 24 h after transfection, as depicted by the+signs, and the cells were harvested 24 h later. The values are shown relative to that of AR in the presence of testosterone without ARIP4 expression plasmids (=100). Each bar represents the mean±S.E.M. values for three independent experiments. (b) Wild-type ARIP4 or indicated sumoylation mutants of ARIP4 were produced in COS-1 cells and purified by affinity chromatography. The proteins were incubated with a 32P-labelled 32 bp DNA fragment and protein–DNA complexes were resolved by EMSA. Asterisks indicate ARIP4-specific shifted bands.
DISCUSSION
Initial biochemical studies confirmed that ARIP4 is an active DNA-dependent ATPase. The rate of ATP hydrolysis was clearly enhanced in the presence of either dsDNA or ssDNA. Lysine-to-alanine mutation (K310A) in the potential ATP-binding site of the ARIP4 ATPase domain decreased the rate of ATP hydrolysis to almost baseline level and abolished DNA-dependent stimulation of the ATPase activity. Similar to several other members of the SNF2 protein superfamily, ARIP4 is able to generate superhelical torsion within linear DNA fragments in vitro [1]. We report here the kinetic parameters of ARIP4-catalysed ATP hydrolysis. Estimated Km (app) for ATP and Km for dsDNA were similar to those of BRG-1 and SNF2h proteins [8]; however, the catalytic activity of ARIP4 was approximately ten times lower than that of the other two proteins [7].
Regions other than the ATPase domain are important for the activity of proteins in the SNF2 family, in that the SANT (SWI3, ADA2, N-CoR and TFIIIB) domain is essential for substrate recognition by different chromatin remodelling enzymes [29,30] and bromodomains are important for gene activation by RSC [31]. Sequence similarity of ARIP4 to proteins of the SNF2 family is confined to the ATPase domain (residues 300–880) [1]. BLASTP search (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) did not detect any putative conserved domains in ARIP4 regions 1–300 and 880–1466 and found no significant similarity to any other protein sequence in the database.
To understand the function of these regions, we constructed several deletion mutants of ARIP4, and tested their ability to bind AR and DNA. In mammalian cells, the first 620 N-terminal residues of ARIP4 were sufficient to interact with AR, whereas shorter N-terminal fragment did not co-immunoprecipitate with AR. The interaction of ARIP4 with AR did not depend on DNA binding. The N-terminal fragment (1–620) co-immunoprecipitated with AR, but failed to bind DNA. The initial yeast two-hybrid screening placed the AR-interaction domain within residues 91–230 of ARIP4 [1]. The current results indicate, however, that the residues 231–620 of ARIP4 are important for proper folding of the AR-interaction domain in mammalian cells. Our results also suggest that ARIP4 DNA-binding domain localizes within residues 889–1314 of ARIP4. We could not, however, directly demonstrate DNA binding of ARIP4(890–1316) or any other ARIP4 N-terminal deletion mutant. Compared with wild-type ARIP4, N-terminal deletion mutants were expressed to substantially lower levels in transfected mammalian cells. Therefore we constructed mutants having N-terminal region of ARIP4 (residues 1–278) fused to putative DNA binding region (residues 890–1206 or 890–1309). These mutants were expressed to levels similar to that of wild-type ARIP4 in transfected mammalian cells, but were nevertheless unable to bind DNA in EMSA experiments. These results suggest that the presence of the ATPase domain (residues 300–880) within ARIP4 is important for correct folding of the DNA-binding domain.
The difference between ARIP4 and classical chromatin remodellers was revealed by the REA assay. This assay is a proven method to probe nucleosome stability and dynamics [32]. By using REA, it has been shown that chromatin remodelling proteins are capable of remodelling reconstituted nucleosomal arrays and mononucleosomes [6,8,9]. Similar to Fan et al. [9], we performed an REA assay with reconstituted mononucleosomes serving as a substrate. Our results revealed that ARIP4 was inactive in the REA assay; however, we cannot formally exclude the possibility that some specific histone modifications are required for the ability of ARIP4 to remodel mononucleosomes.
It is possible that ARIP4 needs the help of auxiliary proteins to remodel chromatin in vivo. ATPases of the SNF2-like family are frequently found to be members of large protein complexes [3,33–35]. However, under our experimental conditions, ARIP4 did not form stable high-molecular-mass protein assemblies. Gel filtration and immunoprecipitation studies failed to show the presence of high-molecular-mass complexes in F9 cellular extracts. Despite many different conditions tested, we have failed to find ARIP4 interaction partners in HeLa nuclear extracts. Yeast two-hybrid screening using HeLa cDNA library with full-length wild-type ARIP4 as bait revealed interactions with SUMO-1 and Ubc9 (results not shown). No other physiologically reasonable proteins were found in the two-hybrid screen. Thus ARIP4 seems to interact with its protein partners only weakly and/or transiently in such a manner that, at any given time, only a small fraction of ARIP4 pool is involved in the protein–protein interactions.
Ubc9 is the E2 conjugating enzyme in the protein sumoylation pathway. This protein modification is important for transcription regulation, intracellular protein localization, DNA repair and protein–protein interactions [23,27,28,36–38]. We confirmed in the present study that ARIP4 is indeed sumoylated. Mutation of consensus-like lysine residues within ARIP4 altered, but did not totally abolish, ARIP4 sumoylation pattern. It is known that mutation of consensus sumoylation site can promote SUMO attachment to other lysine residues in the protein [39]. Indeed, combined mutation of six lysine residues within ARIP4 greatly decreased the extent of ARIP4 sumoylation. Surprisingly, the sumoylation-deficient ARIP4 mutant had lost its ATPase activity and ability to co-activate AR-dependent transactivation. We assume that this effect is due to impaired DNA binding of ARIP4(6K→6R), which, in turn, may result from inappropriate folding of the protein as a consequence of lysine mutations.
Chromatin remodelling is not the only function of the proteins in the SNF2-like family. The two ARIP4 homologues of ARIP4, ATRX and Rad54, are not active chromatin remodellers. Even though Rad54 protein has a low chromatin remodelling activity, its main function is believed to be to catalyse D-loop formation during recombination [40,41] and promote homologous DNA pairing [42]. ATRX is active in a triplex-displacement assay, but not so efficient in remodelling nucleosomes [12]. Proteins of the SNF2 family have a role in regulation of cellular growth and proliferation [33], and DNA repair [43]. Recently, a proapoptotic protein Daxx has been shown to interact with ATRX [12,13] and AR [44].
In view of our current results, we have to conclude that ARIP4 is most likely not involved in chromatin remodelling. Considering the high sequence similarity of ARIP4 to ATRX and Rad54 proteins, unspecific DNA and mononucleosome binding, and low mononucleosome remodelling activity, we hypothesize that the physiological function of ARIP4 is involved in homologous recombination, DNA repair, and/or mediation of apoptosis. We cannot exclude the possibility that ARIP4 possesses a direct role in transcriptional regulation. However, ARIP4 appears to stimulate AR-dependent transactivation only from minimal promoters [1] and, unlike most other DNA-binding transcription factors, it has no sequence specificity in DNA binding. Recently, ARIP4 was reported to interact with Dyrk1A, a serine/threonine kinase involved in neuronal development and in adult brain physiology [45]. Given that ARIP4 orthologue is ubiquitously expressed in Xenopus laevis embryos [2], it is tempting to assign a role for ARIP4 in embryonal and neural development.
Acknowledgments
We thank Seija Mäki, Leena Pietilä, Saija Kotola and Katja Kiviniemi for skilful technical assistance, and Andrey Golubtsov and Pentti Somerharju for advice on the fluorimetric ATPase assay. This work was supported by grants from Academy of Finland, Finnish Foundation for Cancer Research, Helsinki University Central Hospital, Sigrid Jusélius Foundation and Helsinki Graduate School of Biotechnology and Molecular Biology.
References
- 1.Rouleau N., Domans'kyi A., Reeben M., Moilanen A. M., Havas K., Kang Z., Owen-Hughes T., Palvimo J. J., Jänne O. A. Novel ATPase of SNF2-like protein family interacts with androgen receptor and modulates androgen-dependent transcription. Mol. Biol. Cell. 2002;13:2106–2119. doi: 10.1091/mbc.01-10-0484.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder B., Cabot R. A., Schwickert T., Rupp R. A. The SNF2 domain protein family in higher vertebrates displays dynamic expression patterns in Xenopus laevis embryos. Gene. 2004;326:59–66. doi: 10.1016/j.gene.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 3.Lusser A., Kadonaga J. T. Chromatin remodeling by ATP-dependent molecular machines. BioEssays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 4.Lemon B. D., Freedman L. P. Nuclear receptor cofactors as chromatin remodelers. Curr. Opin. Genet. Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 5.Belandia B., Parker M. G. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell (Cambridge, Mass.) 2003;114:277–280. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- 6.Boyer L. A., Logie C., Bonte E., Becker P. B., Wade P. A., Wolffe A. P., Wu C., Imbalzano A. N., Peterson C. L. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 7.Peterson C. L. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 2000;476:68–72. doi: 10.1016/s0014-5793(00)01673-2. [DOI] [PubMed] [Google Scholar]
- 8.Aalfs J. D., Narlikar G. J., Kingston R. E. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 2001;276:34270–34278. doi: 10.1074/jbc.M104163200. [DOI] [PubMed] [Google Scholar]
- 9.Fan H. Y., He X., Kingston R. E., Narlikar G. J. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 10.Langst G., Becker P. B. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons R. J., Picketts D. J., Villard L., Higgs D. R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell (Cambridge, Mass.) 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang J., Wu S., Liu H., Stratt R., Barak O. G., Shiekhattar R., Picketts D. J., Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 14.Tang P., Park D. J., Marshall Graves J. A., Harley V. R. ATRX and sex differentiation. Trends Endocrinol. Metab. 2004;15:339–344. doi: 10.1016/j.tem.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Jänne O. A., Moilanen A., Poukka H., Rouleau N., Karvonen U., Kotaja N., Hakli M., Palvimo J. J. Androgen-receptor-interacting nuclear proteins. Biochem. Soc. Trans. 2000;28:401–405. [PubMed] [Google Scholar]
- 16.Lee J. W., Lee Y. C., Na S. Y., Jung D. J., Lee S. K. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell. Mol. Life Sci. 2001;58:289–297. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K. C., Lee Kraus W. Nuclear receptors, coactivators and chromatin: new approaches, new insights. Trends Endocrinol. Metab. 2001;12:191–197. doi: 10.1016/s1043-2760(01)00392-7. [DOI] [PubMed] [Google Scholar]
- 18.Heinlein C. A., Chang C. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 19.Kang Z., Jänne O. A., Palvimo J. J. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 2004;18:2633–2648. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y. B., Sui Y. P., Shan L. X., Palvimo J. J., Phillips D. M., Jänne O. A. Expression of androgen receptor in insect cells. Purification of the receptor and renaturation of its steroid- and DNA-binding functions. J. Biol. Chem. 1992;267:4939–4948. [PubMed] [Google Scholar]
- 21.Pullman M. E., Penefsky H. S., Datta A., Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 1960;235:3322–3329. [PubMed] [Google Scholar]
- 22.Gonzalo P., Sontag B., Guillot D., Reboud J. P. Fluorometric assay of GTPase activity: application to the couple elongation factor eEF-2-ribosome. Anal. Biochem. 1995;225:178–180. doi: 10.1006/abio.1995.1133. [DOI] [PubMed] [Google Scholar]
- 23.Poukka H., Aarnisalo P., Karvonen U., Palvimo J. J., Jänne O. A. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 24.von Holt C., Brandt W. F., Greyling H. J., Lindsey G. G., Retief J. D., Rodrigues J. D., Schwager S., Sewell B. T. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 25.Stein A. Reconstitution of chromatin from purified components. Methods Enzymol. 1989;170:585–603. doi: 10.1016/0076-6879(89)70066-5. [DOI] [PubMed] [Google Scholar]
- 26.Yager T. D., McMurray C. T., van Holde K. E. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 1989;28:2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]
- 27.Muller S., Ledl A., Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 28.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 29.Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., Peterson C. L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 30.Grune T., Brzeski J., Eberharter A., Clapier C. R., Corona D. F., Becker P. B., Muller C. W. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 31.Kasten M., Szerlong H., Erdjument-Bromage H., Tempst P., Werner M., Cairns B. R. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polach K. J., Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Methods Enzymol. 1999;304:278–298. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 33.Vignali M., Hassan A. H., Neely K. E., Workman J. L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith C. L., Horowitz-Scherer R., Flanagan J. F., Woodcock C. L., Peterson C. L. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 35.Asturias F. J., Chung W. H., Kornberg R. D., Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeler J. S., Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 38.Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature (London) 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 40.Alexeev A., Mazin A., Kowalczykowski S. C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 41.Jaskelioff M., Van Komen S., Krebs J. E., Sung P., Peterson C. L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 42.Petukhova G., Stratton S., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 43.Allard S., Masson J. Y., Cote J. Chromatin remodeling and the maintenance of genome integrity. Biochim. Biophys. Acta. 2004;1677:158–164. doi: 10.1016/j.bbaexp.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Lin D. Y., Fang H. I., Ma A. H., Huang Y. S., Pu Y. S., Jenster G., Kung H. J., Shih H. M. Negative modulation of androgen receptor transcriptional activity by Daxx. Mol. Cell. Biol. 2004;24:10529–10541. doi: 10.1128/MCB.24.24.10529-10541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sitz J. H., Tigges M., Baumgartel K., Khaspekov L. G., Lutz B. Dyrk1A potentiates steroid hormone-induced transcription via the chromatin remodeling factor Arip4. Mol. Cell. Biol. 2004;24:5821–5834. doi: 10.1128/MCB.24.13.5821-5834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]